Abstract
About 10% to 30% of patients with ataxia-telangiectasia (A-T) develop leukemias or lymphomas. There is considerable interpatient variation in the age of onset and leukemia/lymphoma type. The incomplete penetrance and variable age of onset may be attributable to several factors. These include competing mortality from other A-T-associated pathologies, particularly neurodegeneration and interstitial lung disease, and allele-specific effects of ataxia-telangiectasia mutated (ATM) gene mutations. There is also limited evidence from clinical observations and studies using Atm knockout mice that modifier genes may account for some variation in leukemia/lymphoma susceptibility. We have introgressed the Atmtm1Awb knockout allele (Atm-) onto several inbred murine strains and observed differences in thymic lymphoma incidence and latency between Atm-/- mice on the different strain backgrounds and between their F1 hybrids. The lymphomas that arose in these mice had a pattern of sequence gains and losses that were similar to those previously described by others. These results provide further evidence for the existence of modifier genes controlling lymphomagenesis in individuals carrying defective copies of Atm, at least in mice, and the characterized Atm- congenic strain set provides a resource with which to identify these genes. In addition, we found that fewer than expected Atm-/- pups were weaned on two strain backgrounds and that there was no correlation between body weight of young Atm-/- mice and lymphoma incidence or latency.
Introduction
Cancer follows respiratory injury as the major cause of mortality in ataxia-telangiectasia (A-T), and 10% to 30% of patients with A-T develop leukemias or lymphomas, generally of T cell origin. Non-Hodgkin lymphoma is the most common malignancy, followed by T cell acute lymphocytic leukemia [1]. T-prolymphocytic leukemia has been reported in older patients. However, most patients with A-T do not develop leukemia or lymphoma, and the age of onset varies for those who do. The reason for this incomplete penetrance is unknown, but there is some evidence that modifier genes may play a role. Concordance between members of the same family for leukemia or lymphoma type and for the age of onset has been reported [2,3]. This concordance could also be explained by differing phenotypic effects of specific ataxia-telangiectasia mutated (ATM) mutations, and indeed, missense mutations and small, in-frame deletions are found in some concordant families. However, in other families with concordant members, both ATM alleles are inactivated by truncating mutations, and ATM protein is not detectable. In addition, a particular ATM mutation may cause leukemia or lymphoma in some families yet lead to different leukemia/lymphoma types with different ages of onset in other families.
Since the cloning of the gene defective in A-T, ATM, in 1995 [4], at least five laboratories have created mice with Atm knockout alleles [5–9]. Mice with a conditional Atm knockout allele [10] and at least two Atm knock-in alleles have also been engineered [11,12]. Mice homozygous for the Atm knockout alleles (referred to here as Atm-/- mice) develop malignant thymic lymphomas resulting in diminished life spans. On the basis of a literature survey, Reliene and Schiestl [13] found that different laboratories report differing life spans for Atm-/- mice. To explain the discrepancies, they focused on differences in husbandry conditions (specific pathogen-free as opposed to conventional facilities) and differences in commercial rodent chows. The possibility that the strain background for the Atm knockout allele may be a factor in varying life spans was also considered but somewhat discounted because Atm-/- mice on the 129S6/SvEvTac background survive for about the same time as Atm-/- mice on mixed backgrounds.
To determine if genetic background influences thymic lymphoma susceptibility or latency in Atm-/- mice, we introgressed the Atmtm1Awb knockout allele onto three additional inbred strain backgrounds. We then monitored lymphoma development in the four different Atm-/- inbred strains and in three Atm-/- F1 hybrid strains. Here, we report strain-specific differences in lymphoma susceptibility and latency in Atm-/- mice that suggest a role for modifier genes in these phenotypes. In addition, we surveyed the lymphomas for recurrent sequence gains and losses and for granzyme (Gzm) gene rearrangements. We found sequence gains and losses that were largely in agreement with those previously reported [14,15] and no evidence for previously reported rearrangements of the GzmB and GzmC genes [16].
Materials and Methods
Generation of Atmtm1Awb Congenic Strains
129S6/SvEvTac Atmtm1Awb mice originally created by Barlow et al. [5] served as the donor strain for the Atm knockout allele. The A/J Atmtm1Awb and C57BL/6J Atmtm1Awb congenic strains were generated by five marker-directed backcrosses using a chromosome elimination “speed congenic” strategy we have previously described [17]. The marker-directed backcrosses were followed by two conventional backcrosses and five intercross generations. BALB/cByJ Atmtm1Awb congenic mice were generated by 13 generations of conventional backcrosses, followed by 5 intercross generations. These congenic strains are available from the Jackson Laboratory (Bar Harbor, ME). Atm-/- mice are infertile, so the congenic strains are maintained with Atm+/- breeders. For the remainder of this report, we will refer to the background strains as 129S6, C57BL/6, BALB/c, and A/J. F1 hybrids were generated by matings of 129S6 and C57BL/6 mice, 129S6 and A/J mice, and C57BL/6 and A/J mice. We will refer to these hybrids as 129SB6F1, 129SAF1, and B6AF1 regardless of the maternal and paternal strains used in the crosses.
Atm-/- and Atm+/+ mice used in this study were littermates generated from crosses of Atm+/- mice. The mice on all strain backgrounds were bred contemporaneously and were weaned into common cages. Consequently, mice of any given strain were housed in the same cages as similarly aged mice of other strains.
Genotyping
Genotyping was performed on DNA isolated from tail snips and amplified using Atm-F (5′-GACTTCTGTCAGATGTTGCTGCC-3′), Atm-R (5′-CGAATTTGCAGGAGTTGCTGAG-3′), and Atm-Neo (5′-GGGTGGGATTAGATAAATGCCTG-3′). This three-primer set yields a 161-bp amplimer from the wild-type Atm allele and a 441-bp amplimer from the knockout allele.
Histopathology
Tissues were fixed in 10% buffered formaldehyde for 48 to 72 hours, followed by transfer into and storage in 70% ethanol until processed and embedded in paraffin. Sections (6 µm) were stained with hematoxylin and eosin and examined on a Nikon Eclipse 51E microscope (Nikon Instruments Inc, Melville, NY) equipped with a Nikon DS-Fi1 camera with a DS-U2 unit and NIS-Elements F software.
Gzm Gene Rearrangement
Reverse transcription-polymerase chain reaction (PCR) was used to detect fusion transcripts resulting from rearrangements of the GzmB and GzmC genes. Approximately 2 µg of total RNA prepared using an RNeasy Kit (Qiagen Sciences, Germantown, MD) was reverse transcribed (SuperScript II; Invitrogen, Grand Island, NY), and the first strand cDNA was amplified using different primer pair combinations of GzmB forward primers (GzmB-F1 to GzmB-F7) and GzmC reverse primers (GzmC-R1 to GzmC-R7, listed in Table W1). To confirm the expression of GzmB and GzmC genes and the suitability of the primers, primers of GzmB-F1 through GzmB-F7 were used in PCR control reactions with GzmB-R1, and GzmC-R1 through GzmC-R7 were assessed with GzmC-F1.
Array Comparative Genomic Hybridization
Genomic DNA was extracted from five tumors isolated from 129SAF1 mice and six tumors isolated from 129SB6F1 mice using a DNeasy Blood and Tissue Kit (Qiagen Sciences) according to the manufacturer's instructions. DNA isolated from the tail of each respective animal was used as reference DNA. DNA concentration and sample buffer quality was determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). Samples containing contaminants as indicated by low 260:230 ratios were repurified using a DNA clean-up and concentrator kit (Zymo Research, Irvine, CA). Labeling reactions were prepared using the Roche NimbleGen Labeling Protocol (Roche NimbleGen, Madison, WI) for comparative genomic hybridization (aCGH; version 7.0) with 1 µg of total DNA input. One microgram of test DNA and reference DNA was labeled with Cyanine 5-Random Nonamers and Cyanine 3-Random Nonamers, respectively, by Exo-Klenow fragment. Labeled DNA was then purified by isopropanol precipitation, and the labeling efficiency was determined using a NanoDrop spectrophotometer. Thirty-one micrograms of the labeled DNA was prepared for hybridization, placed on the mouse 720K whole-genome array, and hybridized at 42°C for ∼72 hours using a Maui Hybridization System (BioMicro Systems, Salt Lake City, UT). The arrays were then scanned at 2 µM resolution on a NimbleGen MS 200 high-resolution scanner.
Data were processed through the NimbleScan 2.6 software package (Roche NimbleGen) using the mouse 100718_MM9_WG_CGH design files. Data were analyzed as cyanine 3 (Cy3) versus cyanine 5 (Cy5), and all samples passed quality control (QC) metrics cutoffs as determined by Roche NimbleGen. Processed data were subsequently loaded into Nexus Copy Number analysis software version 6 (Bio-Discovery, El Segundo, CA) for Fast Adaptive States Segmentation Technique segmentation analysis and copy number variation.
Analyses
GraphPad Prism software (GraphPad Software, Inc, La Jolla, CA) was used for t tests, χ2 analyses to test for the expected ratio of Atm-/- to Atm+/+ and Atm+/- pups, and Pearson correlation testing of weight and lymphoma latency. Kaplan-Meier survival analyses of overall survival and tumor-free survival were done using SigmaPlot 11.2 software (Systat Software Inc, San Jose, CA).
Results
In Utero and Neonatal Survival of Atm-/- Mice
Atm+/+ and Atm-/- mice were generated on all seven genetic backgrounds by mating Atm+/- mice of the appropriate strains. We tested whether the expected ratios of Atm wild-type (Atm+/+), heterozygous (Atm+/-), and null (Atm-/-) pups were weaned and found fewer Atm-/- pups than were expected on the C57BL/6 and A/J backgrounds (χ2 = 8.567, P = .0138, df = 2, and χ2 = 31.152, P < .0001, df = 2, respectively). We do not know whether the loss of these pups occurred in utero or shortly after birth. There were also fewer BALB/c Atm-/- weanlings than might be expected, but the difference was not quite statistically significant (χ2 = 5.886, P = .0527, df = 2). The expected numbers of Atm-/- pups were weaned on the 129S6, 129SB6F1, 129SAF1, and B6AF1 backgrounds. In addition, the expected ratio of male to female Atm-/- mice was found for all the strain backgrounds indicating that Atm loss does not lead to preferential in utero or neonatal loss of one sex.
Interestingly, fewer A/J Atm+/- pups were weaned than expected (χ2 = 6.682, P = .0097, df = 1) on the basis of the expected ratio of 2:1 for Atm+/- to Atm+/+ pups. This raises the possibility that some A/J Atm+/- mice were lost in utero or shortly after birth.
Atm-/- Mice Weigh Less than Atm+/+ Littermates
The mice were weighed weekly from 5 to 12 weeks of age (Figure W1). On average, Atm-/- pups were smaller than Atm+/+ pups of the same age and sex throughout this period. At 5 weeks, Atm-/- pups weighed, on average, 72% to 87% as much as Atm+/+ mice of the same sex and strain. As might be expected from the dearth of A/J Atm-/- weanlings, these pups were smallest relative to their Atm wild-type littermates.
Survival for Atm-/- Mice to 18 Months
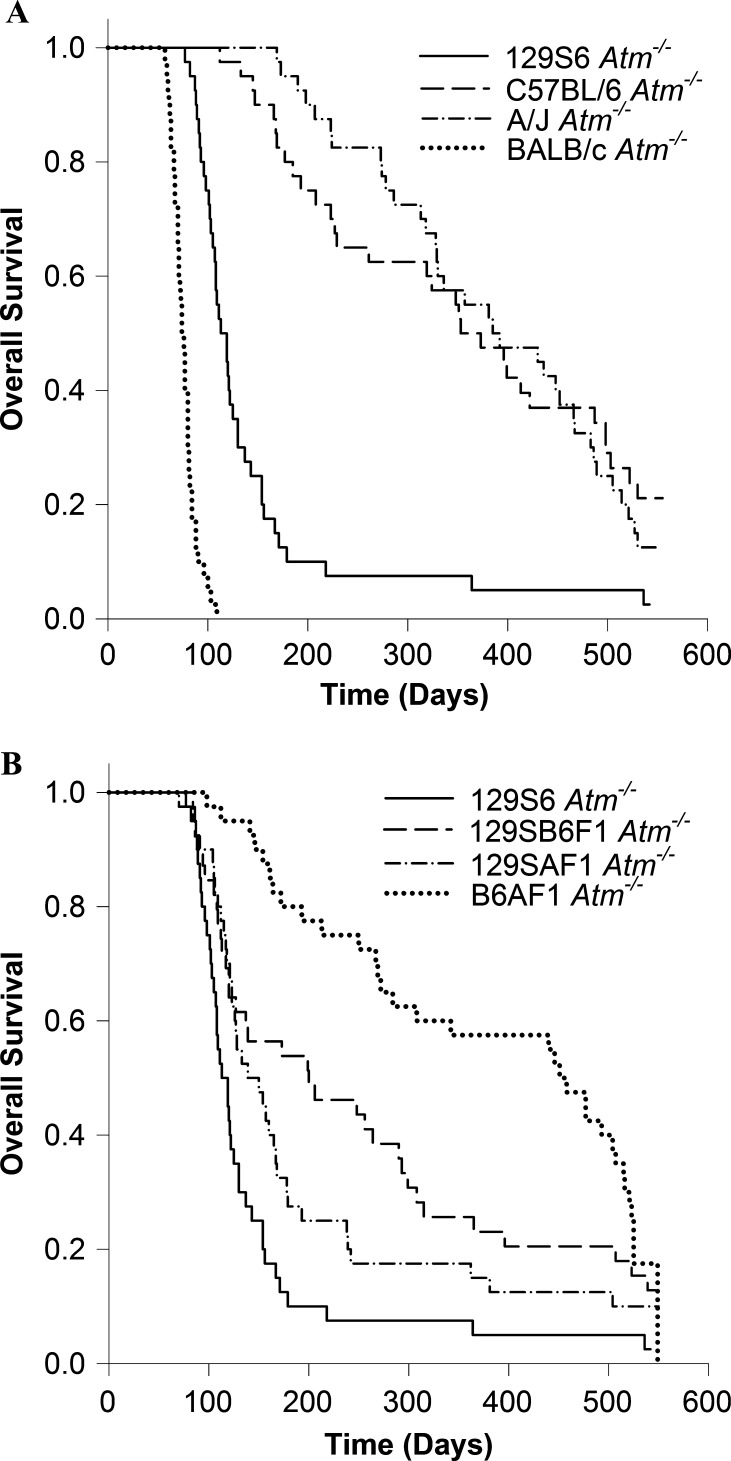
We monitored 20 male and 20 female Atm-/- mice from each strain and F1 hybrid for 18 months. The survival of Atm-null mice depended on their strain background (Figure 1, A and B). Atm-/- mice on the founder 129S6 background had a median survival time of 113 days with only 1 of the 40 mice surviving to 18 months of age. Mice on the BALB/c background had even shorter survival times with a median survival of 74 days, and none of the mice on this background lived longer than 109 days. Median survival was longer for mice on the A/J and C57BL/6 backgrounds (385 and 353 days, respectively). Four of 40 A/J mice survived to 18 months. Eight of 40 C57BL/6 mice survived to 18 months; 1 was killed at 383 days for brain tissue samples.
Figure 1.
Overall survival of Atm-/- mice on four inbred strain backgrounds (A) and three F1 hybrid backgrounds with the survival of 129S6 Atm-/- mice for comparison (B).
Atm-/- mice on 129SB6F1 and 129SAF1 hybrid backgrounds survived longer (median survival of 200 days and 139 days, respectively) than Atm-null mice on the 129S6 background but not as long as on the A/J or C57BL/6 backgrounds. Atm-/- mice on the B6AF1 background had a median survival of 451 days, longer than that of any of the other strain backgrounds tested. Five of 39 129SB6F1 mice survived to 18 months. Four 129SAF1 mice survived to 18 months. Six B6AF1 mice survived to 18 months.
Five Atm+/+ mice from each strain and sex were also followed for survival to 18 months of age. As expected, most of these mice survived the entire time, though in some strains, individual mice were lost. However, there were two exceptions: all of the BALB/c male mice died between 197 and 385 days of age, one from a thymic lymphoma; and three A/J female mice became moribund between 405 and 497 days of age. These losses cannot be attributed to cage effects because cage assignments at weaning were not based on strain or genotype.
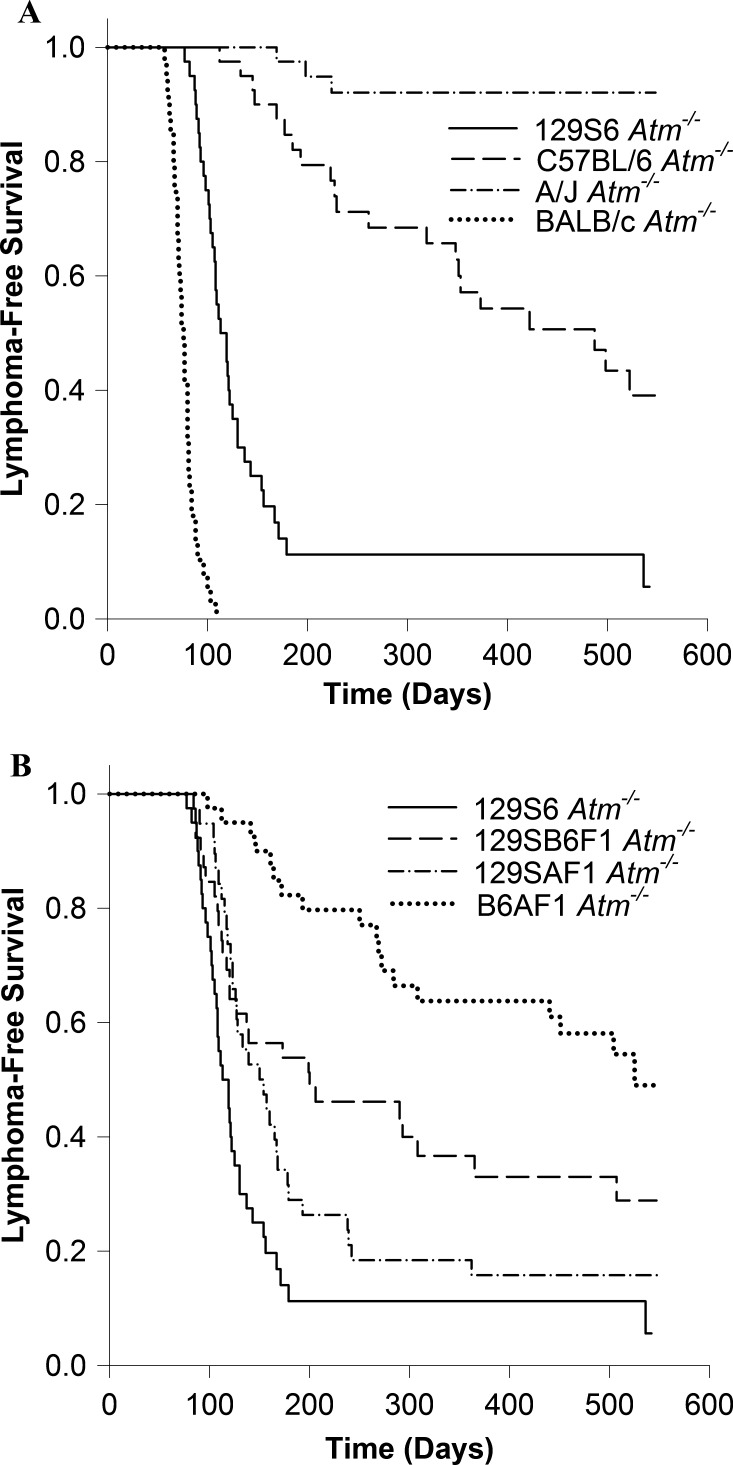
Almost all of the Atm-/- mice on the BALB/c, 129S6, 129SB6F1, and 129SAF1 backgrounds died of thymic lymphomas (Figure 2, A and B). Thirty-nine of 40 BALB/c, 37 of 40 129S6, 26 of 39 129SB6F1, and 32 of 40 129SAF1 Atm-/- mice died of thymic lymphomas. C57BL/6 Atm-/- mice and B6AF1 Atm-/- mice were less susceptible. Of the 40 C57BL/6 Atm nullizygous mice that became moribund or reached 18 months of age, 21 had developed thymic lymphomas. In the B6AF1 hybrids, 5 of 20 males and 13 of 20 females died of thymic lymphomas. However, Atm-/- mice on the A/J background appear to be resistant to thymic lymphomas. Only 3 of the 40 A/J Atm-/- mice that were followed until they became moribund or reached 18 months of age developed thymic lymphomas. With the exception of the B6AF1 hybrids, there were no significant differences within the strains in lymphoma-free survival between males and females, so the results for both sexes are combined in Figure 2.
Figure 2.
Lymphoma-free survival of Atm-/- mice on four inbred strain backgrounds (A) and three F1 hybrid backgrounds with the survival of 129S6 Atm-/- mice for comparison (B).
Because few of the A/J Atm-/- mice died of thymic lymphomas but nevertheless had a median survival shorter than C57BL/6 Atm-/- mice, we considered the possibility that the deaths might be due to other causes related to Atm loss. The necropsies we performed were limited to detecting tumors, and no tumors were detected in 22 of the 40 A/J Atm-/- mice. The most frequently observed tumors in the remaining mice were hepatocellular carcinoma (four tumors and one tentative) and bronchioalveolar carcinoma (four tumors). Spontaneous bronchioalveolar carcinomas have been described in A/J mice, and one was also observed in one of the A/J Atm+/+ controls. However, we are unaware of any reports that hepatocellular carcinomas are common in A/J mice, and they were only observed in Atm-/- mice, not the control mice. All the affected mice were male. In addition to the three thymic lymphomas found in A/J Atm-/- mice, we observed three lymphomas that were not classified as thymic lymphomas because they contained no recognizable thymic structure.
Intriguingly, two of the A/J Atm-/- mice developed rhabdomyosarcomas, a tumor type that has been associated with loss or mutation of ATM in humans [18]. However, rhabdomyosarcoma was also found in three of the control mice.
Decreased Body Weight Is Not Correlated with Lymphoma Latency or Incidence
Atm-/- pups varied considerably in body size. For example, at 6 weeks of age, female 129S6 Atm-/- mice weighed from 13 to 19.1 g. We looked for a correlation between body weight at weekly intervals between 5 and 12 weeks of age and the latency to lymphoma in 18 male and 18 female 129S6 Atm-/- mice that developed lymphoma. There was no correlation for the female mice, but there was for male mice. However, examination of a scatterplot of the data indicated that the correlation was due to one mouse with exceptionally long survival (Figure W2). When this outlier was excluded, there was no correlation between weight and lymphoma latency. We also tested for an association between weight and lymphoma incidence and weight and latency in 20 male and 20 female C57BL/6 Atm-/- mice. On this strain background, 50% of the male mice and 55% of the female mice developed thymic lymphomas by 18 months of age; the remainder died of other causes or were killed at 18 months. Once again, there was no correlation between body weight at 5 to 12 weeks of age and lymphoma latency. At 5 weeks of age, female mice that went on to develop thymic lymphomas had a mean body weight (± SEM) of 14.58 ± 0.20 g; those that did not weighed 14.32 ± 0.32 g. For male mice that developed or did not develop lymphoma, the mean body weights were 16.64 ± 0.36 g and 17.35 ± 0.35 g, respectively, at 5 weeks of age. These differences were not significant, nor were there any significant differences in weight between affected and unaffected mice at weeks 6 through 12.
Gzm Rearrangements and Genomic Copy Number Changes
Winrow previously reported GzmB-GzmC fusion transcripts in lymphoma cell lines originating in Atm-/- mice [16]. We tested RNA from 18 thymic lymphomas that arose in 129S6 Atm-/- mice for the presence of these fusion transcripts by reverse transcription-PCR using multiple primer sets. Though the primers we used readily detected GzmB and GzmC transcripts, fusion transcripts were not detected.
Genomic Gains and Losses
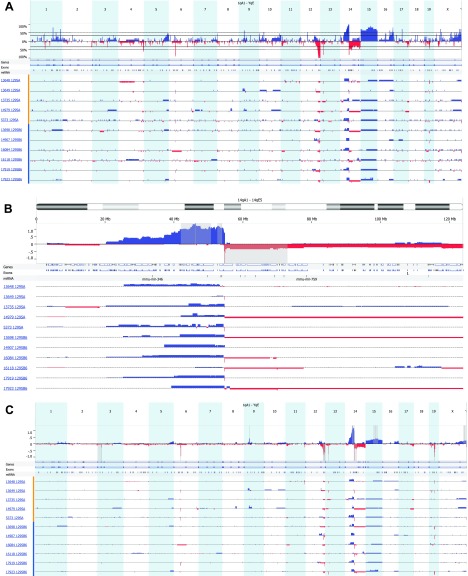
Figure 3A summarizes the aCGH profiles of sequence gains and losses in the thymic lymphomas from each of five 129SAF1 Atm-/- and six 129SB6F1 Atm-/- mice (summarized in Table W1). Most of these copy number alterations are similar to those reported by Zha et al. [15]. Ten of 11 lymphomas assayed had deletions of Tcell receptor gene β (Tcrβ; chromosome 6) with single-copy loss in seven cases and loss of both copies in three others. All of the cases had hemizygous deletions of distal chromosome 12 from about 109 Mb to the termini; in individual tumors, these deletions extended proximally as far as 88 Mb. As noted by Zha et al., the deleted region contains immunoglobulin heavy locus gene (Igh; 10 of 11 lymphomas), B cell leukemia/lymphoma 11B (Bcl11b; 8 of 11 lymphomas), and T cell lymphoma breakpoint 1 (Tcl1; 5 of 11 lymphomas). Six of 11 lymphomas had hemizygous deletions of Tcrγ on chromosome 13. Most of the lymphomas had amplification of proximal chromosome 14 sequences extending from 20 to 43 Mb to the Tcrα/δ locus. At, and distal to, Tcrα/δ, the sequence copy number abruptly changed from gain to loss. This region of loss also includes GzmB to GzmG. In three of the lymphomas, the deleted region extended to the end of the chromosome (Figure 3B). Trisomy of chromosome 15 was noted by Liyanage et al. [14] and Zha et al. [15]. The entirety of chromosome 15 was amplified in 6 of 11 cases, whereas large regions of amplification were seen in 3 of the remaining 5 cases. Four of 11 thymic lymphomas had single-copy deletions within chromosome 19 containing 38 genes and microRNAs (miRNAs), including phosphatase and tensin homolog (Pten; identified as homozygously deleted by Zha), TNF receptor superfamily member 6 gene, and microRNA-107, which regulates cell cycle likely through p53 activation.
Figure 3.
aCGH profiles of sequence gains and losses in thymic lymphomas from five 129SAF1 Atm-/- mice (designated by mouse number and 129SA) and six 129SB6F1 Atm-/- mice (designated by mouse number and 129SB6) (A). The top panel represents the frequency of gain or loss of the 21 chromosomal regions of mouse. The lower panel describes gain or loss for each sample by mouse ID and strain. Most regions of gain or loss are single copy, whereas regions of gain or loss greater than 1 show a block of double the height of the single-loss regions. (B) A representation of regions of sequence gain and loss specifically on chromosome 14 as per the description in A. In addition, the gray regions were identified by GISTIC analysis as a highly overrepresented alteration associated with this sample set. (C) Identification of highly overrepresented regions of copy number gain and loss by GISTIC analysis. Within the upper panel are 14 regions where there is a statistically significant high frequency of aberrations over the background level in this sample set. A q-bound of 0.05 and G-score of 1.0 was used. The G-score is a measure of frequency of occurrence and the magnitude of that occurrence. The dark gray bar represents the peak region, maximal G-score, and nominal q-bound. The surrounding lighter gray shaded areas represent boundaries generated by a leave-one-out recalculation of the peak region.
There are some differences between our results and those of Zha et al. [15]. Zha noted amplification of Notch1 (chromosome 2) in 5 of 18 cases; however, we only found one single-copy amplification of the genomic region containing Notch1 in the 11 lymphomas we assayed. We did not, however, screen for the activating Notch1 mutations found by Zha in a number of lymphomas that lacked Notch1 amplification. Hemizygous loss of approximately 37 kb of chromosome 3 sequence encompassing the neuroligin 1 gene (Nlgn1) was detected in three lymphomas. Nlgn1 is expressed in a variety of tissues, but the characterized role of its encoded protein is in the formation of synaptic junctions, and its relevance to thymic lymphoma is unknown.
Fourteen regions of recurrent sequence gain or loss were identified by Genomic Identification of Significant Targets in Cancer (GISTIC) analysis: loss of sequence on chromosomes 3, 6, 12, 13, 14, and 19 and gains of sequence on chromosomes 9, 14, and 15 (q-bound < 0.05, G-score > 1.0). See Figure 3C and Table W2 for specific regions and associated genes and microRNAs. An arbitrary cutoff of q-bound < 0.05 and G-score of 4.0 was used to highlight only those regions more closely identified with the lymphoma phenotype. After eliminating those regions that did not contain genes or miRNA, there were 257 genes and miRNA associated with regions of copy number gain, whereas there were 459 genes and miRNA associated with regions of chromosomal loss.
Comparative analysis between lymphomas from the two hybrid strains found only three regions overrepresented in a given strain. Of those three regions, only one contained either a gene or miRNA, that being the friend leukemia integration 1 (Fli1) region of chromosome 9 (P < .015). Fli1, a proto-oncogene originally identified as the integration site of the Friend murine leukemia virus [19], is amplified in a 70-kb region of chromosome 9 in the 129SA strain but not in the 129B6 strain. There are no nonsynonymous coding region differences in Fli1 between A/J and C57BL/6J reported in dbSNP (build 128), but the Fli1 transcript is known to be differentially expressed in hematopoietic stem cells between strains of the BXD recombinant inbred strain set (Supplementary Table 2 of reference [20]).
Discussion
The identification of A-T modifier genes may lead to a better understanding of the molecular events that lead to leukemias and lymphomas in patients with A-T and may point to strategies to prevent their occurrence. To detect the presence of modifier genes affecting the incidence or latency of lymphoma in a murine model of A-T, we followed mice homozygous for an Atm knockout allele on several inbred strain and F1 hybrid backgrounds for the lymphoma development. We observed strain differences in lymphoma latency and incidence that we propose are due to modifier genes. There is some evidence that changes in husbandry might affect lymphoma latency in Atm-/- mice [13]. However, in the study reported here, all the mice were housed in the same room and fed the same chow. In addition, the mice were not assigned to separate cages by strain background; mice of different strains were cohoused in the same cages. Thus, the differences between strains in lymphoma latency and incidence we observed are likely due to genetic background rather than husbandry effects.
We found decreased lymphoma latency when the Atmtm1Awb knockout allele was moved from the 129S6 background on which it was originally generated to a BALB/c background. Protein kinase, DNA-activated, catalytic polypeptide (Prkdc) is a candidate modifier gene of the Atm-null allele in this strain. The BALB/c variant of Prkdc, PrkdcBALB, is a hypomorphic allele that has been linked to susceptibility to radiation-induced mammary tumors and thymic lymphomas [21,22]. It is found in all BALB/c substrains but not the other strains used in this study (data not shown). Prkdc encodes DNA-dependent protein kinase catalytic subunit (DNA-PKcs), a protein involved in repair of DNA double-strand breaks and V(D)J recombination. Scid mice have a truncating mutation in Prkdc that leads to severe defects in both DNA double-strand break repair and V(D)J recombination. Crosses between scid mice and Atm knockout mice designed to generate offspring that are deficient in both Atm and DNA-PKcs do not yield viable Atm-/- Prkdcscid/scid pups; the pups die in utero [23,24]. In our study, it is possible that some Atm-/- PrkdcBALB/BALB pups also died in utero, though the deficit of Atm-/- pups was not statistically significant on this background.
A role for Prkdc as a potential modifier gene of lymphomagenesis in Atm-/- mice is particularly interesting because some epidemiological studies are beginning to suggest that PRKDC polymorphisms may be involved in human cancer. Auckley et al. [25] found lower DNA-PKcs activity in the peripheral blood lymphocytes (PBLs) of newly diagnosed patients with lung cancer than in cancer-free controls. They also demonstrated a correlation between DNA-PKcs activity in PBL and bronchial epithelial cells in the same individuals, thus validating the use of DNA-PKcs activity in PBL as a surrogate for activity in other tissue types. Someya et al. [26] obtained similar results, finding low DNA-PKcs activity in PBL collected from patients with breast and uterine cervix cancers before treatment. Both the Auckley and Someya studies are retrospective, raising the possibility that decreased DNA-PKcs activity is a consequence of cancer rather than a contributor to carcinogenesis. However, a study by Bhatti et al. [27] found an association between sequence polymorphisms in PRKDC and breast cancer in radiologic technicians, and Danoy et al. [28] found a potential association between a PRKDC sequence variant and head and neck cancers in smokers. Of course, constitutional sequence polymorphisms are not affected by cancer, but it is not known if the variant sequences in either study have any effect on DNA-PKcs activity.
Unlike BALB/c Atm-/- mice, C57BL/6 Atm-/- and A/J Atm-/- mice had decreased lymphoma incidence and increased latency compared to 129S6 Atm-/- mice. F1 hybrids between these various strains had lymphoma latencies intermediate between those of the parental strains. Reliene and Schiestl, referencing unpublished data, noted a 50% survival at 12.5 months for Atm-/- mice on a C57BL/6 background [13]. This is consistent with our results. A/J Atm-/- mice are resistant to thymic lymphoma development, but their overall survival is not significantly different from C57BL/6 Atm-/- mice. Thus, some of the resistance in this strain may actually be attributable to competing causes of mortality.
Regardless of strain background, Atm-/- mice were smaller and shorter lived than their Atm+/+ littermates. Growth retardation is a clinical feature of A-T [29] and has also been reported in Atm-targeted knockout mice [5–7,9]. In this study, we found that lymphoma incidence or latency and decreased weight are dissociable phenotypes. That is, within a strain, low weight is not predictive of leukemia latency or incidence. We also noted fewer than expected Atm-/- weanlings on the C57BL/6 and A/J backgrounds. Whereas we are unaware of any evidence that A-T can cause fetal loss in humans, it is difficult to determine the proportions of affected and unaffected siblings in A-T families (discussed in reference [30]). The mice in our study were genotyped at or shortly after weaning, so we cannot distinguish if the losses occurred in utero or during the neonatal period. Because the loss only occurred in two strains, it may result from background genetic effects intrinsic to the Atm-/- fetuses, or it could indicate that C57BL/6 and A/J dams provide poor maternal care to smaller or less healthy pups. Recently, Daniel et al. [31] noted embryonic lethality in Atm-/- mice carrying transgenes encoding kinase-dead Atm, and a similar observation was made by Yamamoto et al. [12] in embryos homozygous for a knock-in allele for kinase-dead Atm. The embryonic lethality did not occur with Atm-null alleles.
Unlike Winrow [16], we did not detect GzmB-GzmC fusion transcripts in any of the 18 lymphoma samples tested. There are several possible reasons for the disparate findings. Our assay may fail to detect fusion transcripts that are actually present in the lymphomas or the fusion transcripts detected by Winrow might be an artifact of their assay. It should be pointed out that Winrow's results demonstrate up-regulation of a sequence that hybridizes with cloned GzmC cDNA, though the size of the transcript detected may be larger or smaller than expect for a GzmB-GzmC fusion gene message. Our samples consisted of primary lymphomas, whereas Winrow assayed lymphoma cell lines, so it is also possible that the Gzm gene rearrangements occur during acclamation to tissue culture.
In conclusion, our studies provide further evidence for the existence of modifier genes controlling lymphomagenesis in individuals carrying defective copies of Atm. Furthermore, we have characterized an Atm- congenic strain set that provides a resource with which to identify these genes.
Supplementary Materials and Methods
Abbreviations
- A-T
ataxia-telangiectasia
- ATM
ataxia-telangiectasia mutated
- CGH
comparative genomic hybridization
- GISTIC
Genomic Identification of Significant Targets in Cancer
- Gzm
granzyme
- PBL
peripheral blood lymphocyte
- Prkdc
protein kinase, DNA-activated, catalytic polypeptide
- Tcr
T cell receptor gene
Footnotes
This work was supported by National Institutes of Health grant R03 CA135528 (to M.M.W.) and the Ataxia-Telangiectasia Children's Project (to M.M.W.). The authors disclose no potential conflicts of interest.
This article refers to supplementary materials, which are designated by Tables W1 to W3 and Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Hecht F, Hecht BK. Cancer in ataxia-telangiectasia patients. Cancer Genet Cytogenet. 1990;46:9–19. doi: 10.1016/0165-4608(90)90003-s. [DOI] [PubMed] [Google Scholar]
- 2.Stankovic T, Kidd AM, Sutcliffe A, McGuire GM, Robinson P, Weber P, Bedenham T, Bradwell AR, Easton DF, Lennox GG, et al. ATM mutations and phenotypes in ataxia-telangiectasia families in the British Isles: expression of mutant ATM and the risk of leukemia, lymphoma, and breast cancer. Am J Hum Genet. 1998;62:334–345. doi: 10.1086/301706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor AM, Metcalfe JA, Thick J, Mak YF. Leukemia and lymphoma in ataxia telangiectasia. Blood. 1996;87:423–438. [PubMed] [Google Scholar]
- 4.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 5.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 6.Borghesani PR, Alt FW, Bottaro A, Davidson L, Aksoyn S, Rathbun GA, Roberts TM, Swat W, Segal RA, Gu Y. Abnormal development of Purkinje cells and lymphocytes in Atm mutant mice. Proc Natl Acad Sci USA. 2000;97:3336–3341. doi: 10.1073/pnas.050584897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elson A, Wang Y, Daugherty CJ, Morton CC, Zhou F, Campos-Torres J, Leder P. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc Natl Acad Sci USA. 1996;93:13084–13089. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herzog KH, Chong MJ, Kapsetaki M, Morgan JI, McKinnon PJ. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 1998;280:1089–1109. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 10.Zha S, Jiang W, Fujiwara Y, Patel H, Goff PH, Brush JW, Dubois RL, Alt FW. Ataxia telangiectasia-mutated protein and DNA-dependent protein kinase have complementary V(D)J recombination functions. Proc Natl Acad Sci USA. 2011;108:2028–2033. doi: 10.1073/pnas.1019293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spring K, Cross S, Li C, Watters D, Ben-Senior L, Waring P, Ahangari F, Lu SL, Chen P, Misko I, et al. Atm knock-in mice harboring an in-frame deletion corresponding to the human ATM 7636del9 common mutation exhibit a variant phenotype. Cancer Res. 2001;61:4561–4568. [PubMed] [Google Scholar]
- 12.Yamamoto K, Wang Y, Jiang W, Liu X, Dubois RL, Lin CS, Ludwig T, Bakkenist CJ, Zha S. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J Cell Biol. 2012;198:305–313. doi: 10.1083/jcb.201204098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reliene R, Schiestl RH. Differences in animal housing facilities and diet may affect study outcomes—a plea for inclusion of such information in publications. DNA Repair (Amst) 2006;5:651–653. doi: 10.1016/j.dnarep.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Liyanage M, Weaver Z, Barlo C, Coleman A, Pankratz DG, Anderson S, Wynshaw-Boris A, Ried T. Abnormal rearrangement within the α/δ T-cell receptor locus in lymphomas from Atm-deficient mice. Blood. 2000;96:1940–1946. [PubMed] [Google Scholar]
- 15.Zha S, Bassing CH, Sanda T, Brush JW, Patel H, Goff PH, Murphy MM, Tepsuporn S, Gatti RA, Look AT, et al. ATM-deficient thymic lymphoma is associated with aberrant tcrd rearrangement and gene amplification. J Exp Med. 2010;207:1369–1380. doi: 10.1084/jem.20100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winrow CJ, Pankratz DG, Vibat CR, Bowen TJ, Callahan MA, Warren AJ, Hilbush BS, Wynshaw-Boris A, Hasel KW, Weaver Z, et al. Aberrant recombination involving the granzyme locus occurs in Atm-/- T-cell lymphomas. Hum Mol Genet. 2005;14:2671–2684. doi: 10.1093/hmg/ddi301. [DOI] [PubMed] [Google Scholar]
- 17.Weil MM, Brown BW, Serachitopol DM. Genotype selection to rapidly breed congenic strains. Genetics. 1997;146:1061–1069. doi: 10.1093/genetics/146.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang P, Bhakta KS, Puri PL, Newbury RO, Feramisco JR, Wang JY. Association of ataxia telangiectasia mutated (ATM) gene mutation/deletion with rhabdomyosarcoma. Cancer Biol Ther. 2003;2:87–91. doi: 10.4161/cbt.231. [DOI] [PubMed] [Google Scholar]
- 19.Ben-David Y, Giddens EB, Bernstein A. Identification and mapping of a common proviral integration site Fli-1 in erythroleukemia cells induced by Friend murine leukemia virus. Proc Natl Acad Sci USA. 1990;87:1332–1336. doi: 10.1073/pnas.87.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T, Su AI, Vellenga E, Wang J, Manly KF, et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using “genetical genomics”. Nat Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- 21.Mori N, Matsumoto Y, Okumoto M, Suzuki N, Yamate J. Variations in Prkdc encoding the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) and susceptibility to radiation-induced apoptosis and lymphoma-genesis. Oncogene. 2001;20:3609–3619. doi: 10.1038/sj.onc.1204497. [DOI] [PubMed] [Google Scholar]
- 22.Yu Y, Okayasu R, Weil MM, Silver A, McCarthy M, Zabriskie R, Long S, Cox R, Ullrich RL. Elevated breast cancer risk in irradiated BALB/c mice associates with unique functional polymorphism of the Prkdc (DNA-dependent protein kinase catalytic subunit) gene. Cancer Res. 2001;61:1820–1824. [PubMed] [Google Scholar]
- 23.Gurley KE, Kemp CJ. Synthetic lethality between mutation in Atm and DNA-PKcs during murine embryogenesis. Curr Biol. 2001;11:191–194. doi: 10.1016/s0960-9822(01)00048-3. [DOI] [PubMed] [Google Scholar]
- 24.Sekiguchi J, Ferguson DO, Chen HT, Yang EM, Earle J, Frank K, Whitlow S, Gu Y, Xu Y, Nussenzweig A, et al. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proc Natl Acad Sci USA. 2001;98:3243–3248. doi: 10.1073/pnas.051632098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auckley DH, Crowell RE, Heaphy ER, Stidley CA, Lechner JF, Gilliland FD, Belinsky SA. Reduced DNA-dependent protein kinase activity is associated with lung cancer. Carcinogenesis. 2001;22:723–727. doi: 10.1093/carcin/22.5.723. [DOI] [PubMed] [Google Scholar]
- 26.Someya M, Sakata K, Matsumoto Y, Yamamoto H, Monobe M, Ikeda H, Ando K, Hosoi Y, Suzuki N, Hareyama M. The association of DNA-dependent protein kinase activity with chromosomal instability and risk of cancer. Carcinogenesis. 2006;27:117–122. doi: 10.1093/carcin/bgi175. [DOI] [PubMed] [Google Scholar]
- 27.Bhatti P, Struewing JP, Alexander BH, Hauptmann M, Bowen L, Mateus-Pereira LH, Pineda MA, Simon SL, Weinstock RM, Rosenstein M, et al. Polymorphisms in DNA repair genes, ionizing radiation exposure and risk of breast cancer in US radiologic technologists. Int J Cancer. 2007;122:177–182. doi: 10.1002/ijc.23066. [DOI] [PubMed] [Google Scholar]
- 28.Danoy P, Michiels S, Dessen P, Pignat C, Boulet T, Monet M, Bouchardy C, Lathrop M, Sarasin A, Benhamou S. Variants in DNA double-strand break repair and DNA damage-response genes and susceptibility to lung and head and neck cancers. Int J Cancer. 2008;123:457–463. doi: 10.1002/ijc.23524. [DOI] [PubMed] [Google Scholar]
- 29.Sedgwick RP, Boder E. Vinken PJ, Bruyn GW. Handbook of Clinical Neurology. Amsterdam, The Netherlands: North-Holland Publishing Company; 1972. Ataxia-telangiectasia; pp. 267–339. [Google Scholar]
- 30.Tadjoedin MK, Fraser FC. Heredity of ataxia-telangiectasia (Louis-Bar syndrome) Am J Dis Child. 1965;110:64–68. doi: 10.1001/archpedi.1965.02090030070009. [DOI] [PubMed] [Google Scholar]
- 31.Daniel JA, Pellegrini M, Lee BS, Guo Z, Filsuf D, Belkina NV, You Z, Paull TT, Sleckman BP, Feigenbaum L, et al. Loss of ATM kinase activity leads to embryonic lethality in mice. J Cell Biol. 2012;198:295–304. doi: 10.1083/jcb.201204035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.