Abstract
Breast cancer preferentially metastasizes to lung, lymph node, liver, bone, and brain. However, it is unclear whether properties of cancer cells, properties of organ microenvironments, or a combination of both is responsible for this observed organ tropism. We hypothesized that breast cancer cells exhibit distinctive migration/growth patterns in organ microenvironments that mirror common clinical sites of breast cancer metastasis and that receptor-ligand interactions between breast cancer cells and soluble organ-derived factors mediate this behavior. Using an ex vivo model system composed of organ-conditioned media (CM), human breast cancer cells (MDA-MB-231, MDA-MB-468, SUM149, and SUM159) displayed cell line-specific and organ-specific patterns of migration/proliferation that corresponded to their in vivo metastatic behavior. Notably, exposure to lung-CM increased migration of all cell lines and increased proliferation in two of four lines (P < .05). Several cluster of differentiation (CD) 44 ligands including osteopontin (OPN) and L-selectin (SELL) were identified in lung-CM by protein arrays. Immunodepletion of SELL decreased migration of MDA-MB-231 cells, whereas depletion of OPN decreased both migration and proliferation. Pretreatment of cells with a CD44-blocking antibody abrogated migration effects (P < .05). “Stemlike” breast cancer cells with high aldehyde dehydrogenase and CD44 (ALDHhiCD44+) responded in a distinct chemotactic manner toward organ-CM, preferentially migrating toward lung-CM through CD44 receptor-ligand interactions (P < .05). In contrast, organ-specific changes in migration were not observed for ALDHlowCD44- cells. Our data suggest that interactions between CD44+ breast cancer cells and soluble factors present in the lung microenvironment may play an important role in determining organotropic metastatic behavior.
Introduction
Breast cancer remains a leading cause of morbidity and mortality in women [1], mainly due to the propensity of primary breast tumors to metastasize to distant sites and the failure of most therapies in the metastatic setting. Further insight into the biology of metastasis is therefore essential to gain a greater understanding of this process and to develop better cancer therapies.
Metastasis is a complex process, and tumor cells must successfully negotiate a series of sequential steps to establish clinically relevant macrometastases. These steps include dissemination from the primary tumor through blood or lymphatic systems, survival within the circulation, extravasation into secondary sites, initiation of growth into micrometastases, and maintenance of growth as vascularized macro-metastases [2]. Clinical observations indicate that many cancers show an organ-specific pattern of metastasis, termed organ tropism, and it is well established that breast cancer favors metastasis to the lung, liver, bone, lymph node (LN), and brain [2–4]. In the 1920s, James Ewing first proposed that blood flow patterns alone were sufficient to account for both physical delivery of tumor cells to secondary organs and for patterns of organ-specific metastasis [5]. However, several theories have challenged this idea by proposing that there are additional, molecular-level mechanisms that explain why and how cancer cells can travel to and grow in “favorite” metastatic sites. Among these is Paget's seminal “seed and soil” hypothesis, first proposed in 1889 [6]. This predicts that a cancer cell (“seed”) can survive and proliferate only in secondary sites (“soil”) that produce appropriate molecular factors. A meta-analysis of published autopsy data [7] demonstrated that, in some cases, metastases detected at autopsy were in proportion to blood flow from the primary tumor to the secondary organ. However, in many cases, more or fewer metastases than would be expected by blood flow alone were detected, indicating that the microenvironment is likely very important for metastatic dissemination and growth.
For the past several years, elegant work by Joan Massagué and colleagues has focused on defining specific genes that mediate organ-specific metastasis in breast cancer [4,8–10]. Using in vivo selection and genetic analysis of the MDA-MB-231 human breast cancer cell line, this group demonstrated that particular genes can mediate experimental breast cancer metastasis in an organ-specific manner to lung [10], bone [9], and brain [8] and validated that these genes reflect organ-specific metastatic disease in patients with breast cancer. Although these studies contribute valuable knowledge regarding the contribution of the cancer cell (“seed”) to organ tropism of breast cancer, the factors contributed by the metastatic microenvironment (“soil”) still remain poorly understood. In addition, these studies do not take into account the concepts of tumor cell heterogeneity and the cancer stem cell hypothesis.
Despite the deadly nature of metastasis, it is an inherently inefficient process [2,11]. This suggests that only a small subset of cells can successfully navigate the metastatic cascade. We believe that these metastasis-initiating cells may in fact be cells with “stemlike” properties [12]. In breast cancer, tumor-initiating cells have been isolated from primary tumors and pleural effusions on the basis of a cluster of differentiation (CD) 44-positive-CD24-negative (CD44+CD24-) phenotype [13] and/or high aldehyde dehydrogenase (ALDH) activity [14]. Our group and others have demonstrated that breast cancer cells with an ALDHhiCD44+ phenotype show enhanced metastatic behavior in vitro and in vivo compared to their ALDHlowCD44- counterparts [15–17]. However, the role of such cells in mediating organ-specific metastasis has not been investigated.
In the current study, we hypothesized that breast cancer cells exhibit distinctive growth and migration patterns in organ microenvironments that mirror common clinical sites of breast cancer metastasis and that receptor-ligand interactions between breast cancer cells and specific soluble organ-derived factors can mediate this behavior. We first developed and validated a comprehensive ex vivo model system for investigating the influence of organ-specific soluble factors on metastatic behavior of human breast cancer cells. Our results indicate that human breast cancer cells with varying genetic backgrounds exhibit differential migration and growth patterns toward specific organ conditions. Notably, these patterns reflect the known metastatic dissemination patterns of these cell lines in vivo and highlight the lung as an important source of soluble factors that mediate metastatic behavior. Furthermore, our results suggest for the first time that interactions between subpopulations of CD44-expressing breast cancer cells (including ALDHhiCD44+ cells) and soluble ligands present in the lung microenvironment may play an important role in determining organotropic metastatic behavior.
Materials and Methods
Cell Culture and Reagents
MDA-MB-231 cells [18] were obtained from American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM)/F12 + 10% FBS. SUM159 and SUM149 cells [19] were obtained from Asterand Inc (Detroit, MI) and maintained in HAMS:F12+ 5% FBS+5 µg/ml insulin + 1 µg/ml hydrocortisone + 10 mM Hepes. MDA-MB-468 cells were obtained from Dr Janet Price (MD Anderson Cancer Center, Houston, TX [20]) and maintained in α minimum essential medium + 10% FBS. Cell lines were authenticated through third-party testing (CellCheck; IDEXX RADIL, Columbia, MO) in January 2012. All Media/supplements were from Invitrogen (Carlsbad, CA); FBS was from Sigma-Aldrich (St Louis, MO).
Organ-Conditioned Media
Healthy female nude mice (Hsd:Athymic Nude-Foxn1nu; Harlan Sprague-Dawley, Indianapolis, IN) were maintained as per the Canadian Council of Animal Care under a protocol approved by the Western University Animal Use Subcommittee (No. 2009-064). Mice (6-12 weeks old) were euthanized, and individual organs (lung, liver, and brain), femurs, and axillary/brachial/inguinal LNs were aseptically removed, washed, and cut into ∼1-mm3 fragments. Liver-conditioned media (CM) were isolated in the presence of 1X Halt protease inhibitor (aprotinin, bestatin, E-64, leupeptin, NaF, Na3VO4, Na4P2O7, and β-glycerophosphate) (Pierce, Nepean, Ontario). Lung, liver, and brain tissues were weight normalized by resuspending in 4:1 media to tissue (vol/wt) ratio in DMEM/F12 + 1X MITO+ (BD Biosciences, Mississauga, Ontario) + penicillin (50 U/ml)/streptomycin (50 µg/ml) (pen/strep; Invitrogen). Organs were cultured for 24 hours before collecting CM for storage at -20°C.
Due to the smaller cellular content of LN and bone marrow (BM) relative to other organs, a different approach was taken to generate CM from these tissues. LNs were mechanically dissociated, washed, and plated at a density of 5 x 106 cells per well in six-well dishes in Roswell Park Memorial Institute Medium 1640 + 10% FBS + pen/strep + 5 x 10-5 M β-mercaptoethanol (BioShop, Burlington, Ontario) as previously described [21]. BM was collected by flushing femur cavities as previously described [22]. Aspirates were dissociated by pipetting and washed, and ∼1 x 107 cells were plated in T75 flasks in DMEM + 10% FBS + pen/strep. Resulting adherent LNSCs or BM stromal cells (BMSCs) were passaged two to three times, washed, and exposed to DMEM/F12 + MITO+ + pen/strep. CM were collected after 72 hours and stored at -20°C. To account for mouse-to-mouse variability, organ-CM from multiple mice were pooled before use in functional experiments.
Migration Assays
Transwells (6.5 mm, 8-µm pore size; Becton Dickinson, Franklin Lakes, NJ) were coated with 6 µg of gelatin per well as previously described [16]. Organ-CM [weight normalized to 0.0156 g of tissue per milliliter of media; determined by dose-response experiments (Figure W1)], basal media (DMEM/F12 + MITO+), or positive control media (basal media + 10% FBS) were placed in the bottom portion of 24-well dishes (n = 3 wells per condition). Human breast cancer cells (5 x 104 cells per well) were plated on top of transwells. In experiments involving functional blocking of CD44 or ALDH, cells were pre-incubated at room temperature (RT) for 30 minutes in the presence or absence of a broad-spectrum rat anti-human CD44 antibody (10 µg/ml; clone A020; Calbiochem, Mississauga, Ontario) or 100 µM diethylaminobenzaldehyde (DEAB) as previously described [17,23]. After 24 hours, transwells were removed, fixed, and stained with hematoxylin. Nonmigrated cells on the inner surface of transwells were removed. Five high-powered fields (HPFs) were counted for each transwell, and mean numbers of migrated cells were calculated using ImageJ software [National Institutes of Health (NIH), Bethesda, MD]. Results are expressed as fold increase from negative control (basal media) (N = 3).
Bromodeoxyuridine Assays
Human breast cancer cells (1.5 x 104 per well) were plated onto eight-well chamber slides (Lab-Tek; Thermo Fisher Scientific, Waltham, MA) and allowed to adhere for 24 hours (n = 3 wells per condition) before synchronizing cells into G0 in serum-free DMEM/F12 for 72 hours. In experiments involving CD44 blocking, cells were pretreated as described above. Media were changed to organ-CM [weight normalized to 0.0624 g of tissue per milliliter of media; determined by dose-response experiments (Figure W2)], basal media, or positive control media for 24 hours. Cells were pulsed with bromodeoxyuridine (BrdU) (5 µl/ml; Amersham Cell Proliferation Labeling Reagent; GE Healthcare, Piscataway, NJ) for 30 minutes, washed, fixed, permeabilized, and denatured. Slides were stained with Anti-BrdU (1:75; BD Biosciences) for 2 to 3 days in a humid slide chamber. Secondary antibody [1:100; fluorescein isothiocyanate (FITC) anti-mouse IgG; Vector Laboratories, Burlington, Ontario] was added for 1 hour at RT. Slides were mounted with ProLong Gold with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) and allowed to cure for 24 hours in the dark at RT. Images (five HPFs per well) were taken, and nuclei were enumerated using ImageJ. Results are normalized to negative control (basal media) and expressed as a percentage of total nuclei that were BrdU+ (N = 3).
Protein Arrays
To identify soluble factors present within lung-CM, RayBio AAM-BLM-1 label-based mouse antibody arrays were used to simultaneously assess expression of 308 soluble murine target proteins (RayBiotech Inc, Norcross, GA). Postdialysis protein concentration of lung-CM or basal media (N = 3 per condition) was determined using the DC protein assay (Bio-Rad Laboratories, Mississauga, Ontario), and 40 µl of internal array control was spiked into 100 µg of total protein, labeled, and incubated with protein arrays as per manufacturer's instructions. Results were visualized using chemiluminescence and film exposure (CL-XPosure Film; Pierce).
Densitometric analysis was conducted using ImageJ with the MicroArray Profile Macro (OptiNav Inc, Bellevue, WA), and results (N = 3 per media condition) were analyzed using the RayBiotech analysis tool for AAM-BLM-1. Seventy confirmed protein hits were identified as having values >1 after background subtraction and validation across three replicates. Due to differences in antibody affinities for target antigens, quantitative comparison between different proteins was not feasible using this platform. The resulting protein list was cross-referenced with the Ensembl gene database (European Bioinformatics Institute and Wellcome Trust Sanger Institute, Hinxton, United Kingdom; www.ensembl.org), and gene symbols were queried using Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA).
Immunodepletion
Target proteins were immunodepleted from lung-CM using antibodies against mouse L-selectin (SELL) (2 µg/ml) or osteopontin (OPN) (10 µg/ml) (R&D Systems, Burlington, Ontario). Antibodies were incubated for 20 minutes at RT with Dynabeads Protein G (Invitrogen) and washed. Bead-antibody complexes were incubated with lung-CM or basal media (N = 3 per condition) for 30 minutes at RT. Resulting bead-antibody-antigen complexes were removed using a DynaMag-2 magnet (Invitrogen), and depleted media were assessed by Quantikine ELISA kits specific for mouse SELL or OPN (R&D Systems). Negative controls included media exposed to beads only (no antibody) or beads + nonspecific IgG antibody.
In Vivo Metastasis Assays
Procedures were conducted in accordance with the recommendations of Canadian Council of Animal Care, under an approved protocol (No. 2009-064). Using established models of experimental metastasis, 1 x 106 MDA-MB-231 cells/100 µl sterile Hank's buffered salt solution were injected into the lateral tail vein of 7- to 8-week-old female nude mice (n = 5) as described previously [16]. Lung metastases were allowed to develop for 8 weeks. Lung tissues collected at necropsy were formalin fixed, paraffin embedded, serially sectioned (4 µm), stained with hematoxylin and eosin, and evaluated by microscopy to identify regions of metastatic involvement. Alternatively, embedded mouse lung tissue was deparaffinized, rehydrated, and immersed in sodium citrate buffer at 100°C for 20 minutes. Slides were blocked by BLOXALL Endogenous Peroxidase and Alkaline Phosphatase (AP) Blocking Solution (Vector Laboratories) for 10 minutes at RT and stained by Polink DS-MR-Hu A2 kit (GBI Labs, Burlington, Ontario) according to the manufacturer's instructions. Primary antibodies included rabbit anti-mouse OPN (1:500; American Research Products, Inc, Waltham, MA), rabbit anti-mouse SELL (1:500; Bioss, Burlington, Ontario), and/or mouse anti-human CD44 (1:1000; Abcam, Cambridge, MA) for 1 hour, followed by incubation with 1:1 HRP-polymer anti-rabbit IgG and AP-polymer anti-mouse IgG for 30 minutes. Color was developed using DAB (brown) and AP-Red (Thermo Fisher Scientific). Slides were counterstained with hematoxylin, mounted, and imaged using an Image ScanScope (Aperio, Vista, CA).
Fluorescence-Activated Cell Sorting
Cell subpopulations (ALDHhiCD44+CD24- and ALDHlow-CD44low/-CD24+) were isolated from the MDA-MB-231 cell line as described previously [16,17] and in the Supplemental Materials and Methods section. FACS-isolated cells were used immediately for in vitro migration assays.
Data Analysis
In vitro experiments were performed a minimum of three times with at least three technical replicates included within each experiment. In vivo studies were carried out using multiple mice (N = 5–20 per experiment). In all cases, quantitative data were compiled from all experiments. Unless otherwise noted, data are presented as the means ± SEM. Statistical analysis was performed using GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA) using analysis of variance (ANOVA) with Dunnett posttests (for comparison to basal media control) or Tukey posttests (for comparison between all media conditions). Values of P ≤ .05 were regarded as being statistically significant.
Results
Development of an Ex Vivo Model System for Investigating the Influence of Organ-Specific Soluble Factors on Metastatic Behavior of Human Breast Cancer Cells
Previous studies have uncovered several tumor cell-specific characteristics that can influence organ tropism of breast cancer metastasis [4,8–10]. However, much less is known about the organ-specific factors contributed by the secondary microenvironment itself, and this has remained challenging to investigate. We began to address this by establishing a comprehensive ex vivo model system for investigating the influence of organ-specific soluble factors on the metastatic behavior of human breast cancer cells. Using in vivo xenograft models of metastasis as a foundation, organs representing common sites of clinical breast cancer metastasis were isolated from female nude mice and used to generate CM. Organ-CM was successfully generated from lung, liver, brain, and LNSCs and BMSCs. Stromal cells (LNSCs and BMSCs) were observed to be adhesive in culture, with LNSCs demonstrating an elongated and fibroblastic phenotype (Figure W3A) and BMSCs demonstrating a smaller and more mesenchymal appearance (Figure W3C). Flow cytometric analysis demonstrated that LNSCs were largely CD45- and glycoprotein 38-positive (gp38+); with 69.2 ± 8.5% of cells possessing a CD45- gp38+ phenotype indicative of LNSCs [24] (Figure W3B). The BMSCs were observed to be positive for CD44 and CD29, weakly positive for CD105 and stem cell antigen-1 (Sca-1), and negative for CD79 (Figure W3, D–H), in close agreement with previous studies [25,26].
Human Breast Cancer Cells Demonstrate Cell Line-Specific Chemotactic and Proliferative Behavior in Response to Organ-CM
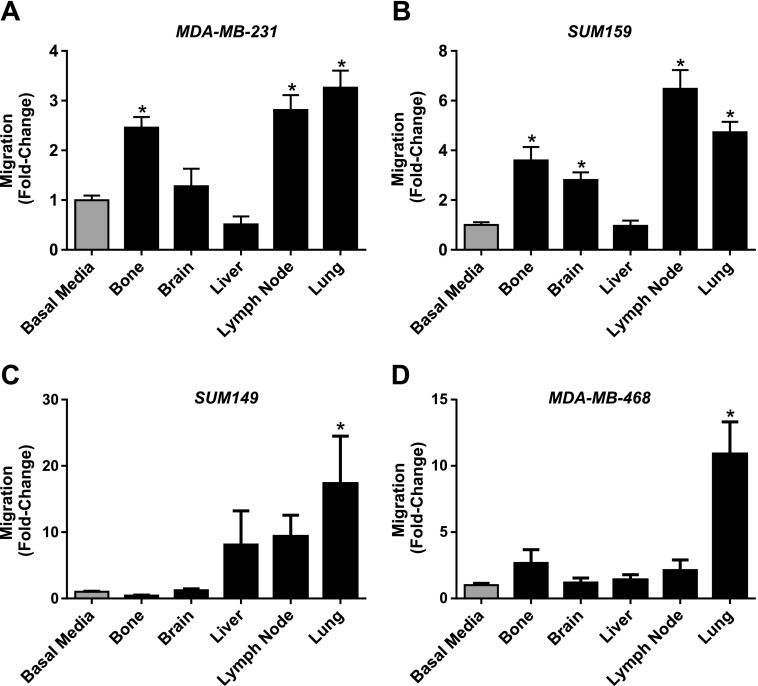
We used our newly developed ex vivo model system to investigate organ-specific metastatic cell behaviors of human breast cancer cells in response to soluble factors present in organ-CM. The breast cancer cell lines used in this study have been previously classified as being either an estrogen receptor (ER-)/progesterone receptor (PR-)/human epidermal growth factor receptor-positive (HER2+) subtype (SUM159 and SUM149) [19] or a triple-negative ER-/PR-/HER2- subtype (MDA-MB-231 and MDA-MB468) [18,20]. We employed transwell migration assays as a surrogate for metastatic dissemination (“getting there”) and BrdU proliferation assays as a surrogate for metastatic growth (“growing there”). Analysis of MDA-MB-231, SUM159, SUM149, and MDA-MB-468 cell lines indicated that human breast cancer cells demonstrate cell line-specific chemotactic behavior in response to different organ-CM conditions (Figure 1). The MDA-MB-231 cell line, arguably the most metastatic cell line of our panel in vivo (Table 1), displayed increased migration toward bone-, LN-, and lung-CM relative to basal media (P < .05) (Figure 1A). The second most metastatic cell line in vivo, SUM159 (Table 1), demonstrated enhanced migration to bone-, brain-, LN-, and lung-CM relative to basal media (P < .05)(Figure 1B). SUM149 cells demonstrate less metastasis in vivo than SUM159 cells (Table 1) and were only observed to have an increase in migration toward lung-CM when compared to basal media (P < .05) (Figure 1C). MDA-MB-468 cells, the least metastatic in vivo of the four lines tested (Table 1), similarly only demonstrated increased migration to lung-CM compared to basal media (P < .05) (Figure 1D).
Figure 1.
Human breast cancer cells demonstrate cell line-specific chemotactic behavior in response to organ-CM. (A) MDA-MB-231, (B) SUM 159, (C) SUM 149, or (D) MDA-MB-468 cells (5 x 104 per well) were plated in triplicate (n = 3) on top of gelatin-coated trans-wells (8-µm pore size) before placement into either basal media (DMEM/F12 + MITO+) or organ-CM (bone, brain, liver, LN, or lung). Migration was allowed to occur for more than 18 hours at 37°C (5% CO2). Transwells were then fixed with glutaraldehyde, stained with Harris' hematoxylin, and developed with ammonium hydroxide. Five HPFs of view were captured per transwell, and migrated cells were enumerated using ImageJ software (NIH). Data are presented as means ± SEM (N = 3; fold change from negative control of basal media). *, significantly different from basal media (ANOVA with Dunnett posttest; P < .05).
Table 1.
In Vivo Organ-Specific Metastatic Dissemination Patterns of Different Human Breast Cancer Cell Lines in Xenograft Nude Mouse Models of Metastasis.
| LN* | Lung† | Liver‡ | Bone§ | Brain§ | |
| MDA-MB-231 | ✓✓✓[47]¶ | ✓✓✓✓[10,48]¶,# | ✓✓ [47]# | ✓✓✓[9,49]¶ | ✓✓ [8,50] |
| SUM-159 | ✓✓ [51]¶ | ✓✓ [51]¶ | ✓ [51] | ✓✓¶,** | ✓✓¶,** |
| SUM-149 | ✓ [52] | ✓✓ [52]¶ | |||
| MDA-MB-468 | ✓✓ [53] | ✓✓ [48,53]¶,# |
✓✓✓ to ✓✓✓✓, macroscopic metastases (grossly observable on necropsy)
✓ to ✓✓, microscopic metastases (observable by microscopy).
Following mammary fat pad injection.
Following mammary fat pad or tail vein injection.
Following mammary fat pad or mesenteric vein injection.
Following mammary fat pad or intracardiac injection.
Significantly increased migration to corresponding organ-CM in this study (P < .05).
Significantly increased BrdU incorporation in presence of corresponding organ-CM in this study (P < .05).
Author's unpublished data.
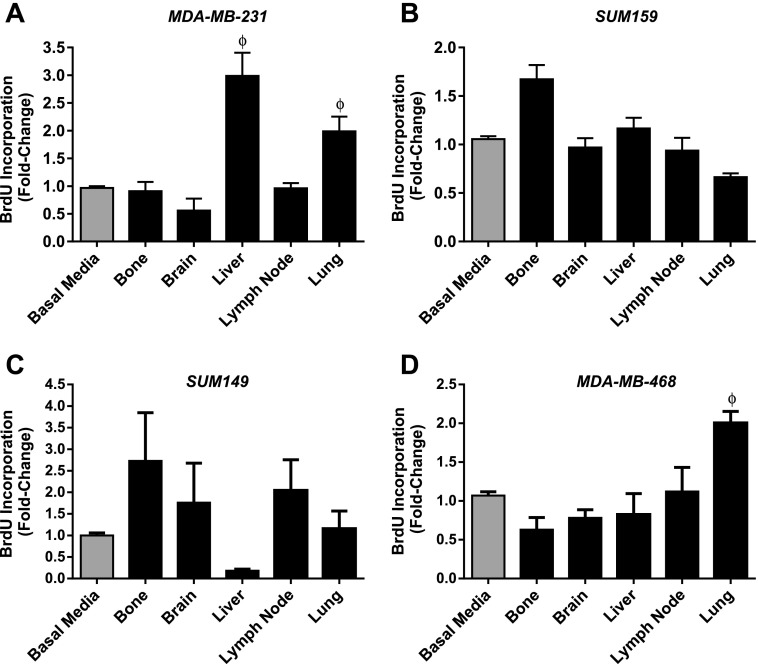
Analysis of cell proliferation using BrdU assays also demonstrated cell line-specific patterns in response to organ-CM (Figure 2), although with more variability than migration studies. MDA-MB-231 cells demonstrated increased proliferation in response to liver- and lung-CM (P < .05) (Figure 2A), whereas MDA-MB-468 cells demonstrated enhanced proliferation toward lung-CM relative to basal media (P < .05) (Figure 2D). Although SUM159 (Figure 2B) and SUM149 (Figure 2C) cell lines appeared to have differential patterns of response to various organ conditions, no statistically significant differences were observed. Taken together, MDA-MB-231, SUM159, SUM149, and MDA-MB-468 cells displayed cell line-specific and organ-specific patterns of migration and proliferation that corresponded to their in vivo metastatic behavior (Table 1).
Figure 2.
Human breast cancer cells demonstrate cell line-specific proliferative behavior in response to organ-CM. (A) MDA-MB-231, (B) SUM 159, (C) SUM 149, or (D) MDA-MB-468 cells (1.5 x 104 per well) were plated in triplicate (n = 3) in eight-well chamber slides and allowed to adhere for 24 hours. Cells were washed with PBS and incubated for 72 hours in serum-free media. The media were then changed to either basal media (DMEM/F12 + MITO+) or organ-CM (bone, brain, liver, LN, or lung). After 24 hours, cells were incubated with 5 µl/ml BrdU for 30 minutes before fixation and staining for BrdU incorporation through immunofluorescence. Positive cells were enumerated and determined as a percentage of total cells present (through nuclear staining with DAPI). Data are presented as means ± SEM (N = 3; fold change from negative control of basal media). ϕ, significantly different from basal media (ANOVA with Dunnett posttest; P < .05).
Lung-CM Contains Potential Mediators of Metastatic Behavior
We next wanted to identify soluble factors that may be influencing the observed organ-specific chemotactic and proliferative behavior of breast cancer cells. Lung is one of the most common sites of breast cancer metastasis and is a significant cause of morbidity and mortality, particularly in patients with aggressive basal-like and HER2+ subtypes of breast cancer [3]. Taken together with our observations that lung-CM consistently enhanced migration of all breast cancer cell lines tested and also enhanced proliferation in two of four cell lines tested, we decided to focus on investigating the composition of lung-CM.
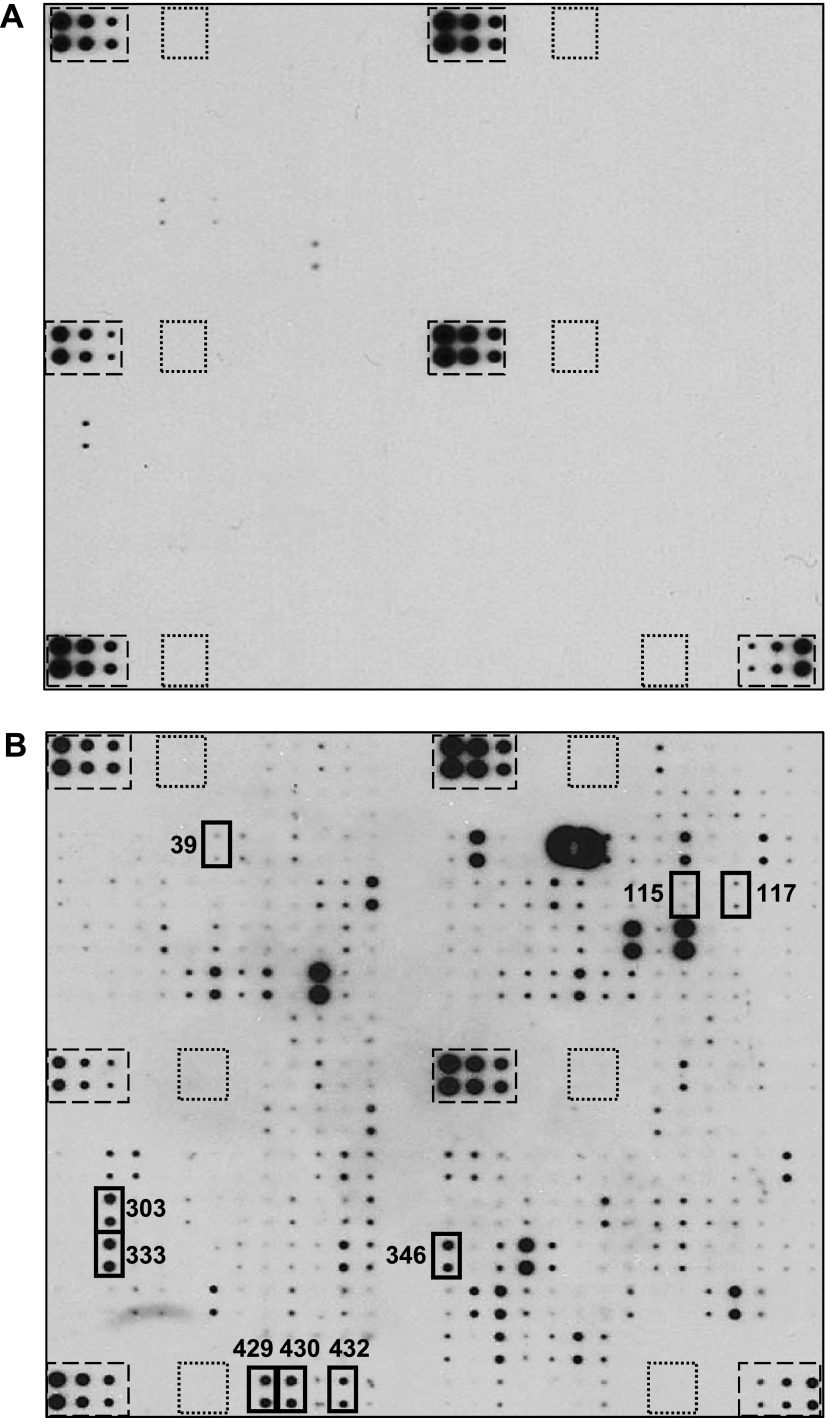
Protein arrays were exposed to lung-CM and basal media (Figure 3). After background subtraction and removal of any overlap present in basal media, densitometric analysis revealed that 70 proteins were present on triplicate arrays. These proteins belong to many general categories including chemokines, cytokines, growth factors, and soluble extracellular matrix (ECM) components that may play a role in metastatic behavior (Table W1). Queries using corresponding gene symbols and Ingenuity Pathway Analysis revealed the presence of several proteins in the functional categories of metastasis, migration, neoplastic growth, and adhesion (Table W2). Of these, a further set of specific proteins of interest was identified (Table 2), including proteins related to lung metastasis mediators previously identified by Massagué and colleagues [epiregulin (EREG), OPN, and urokinase-type plasminogen activator (uPA)] [10,27–29] and proteins known to interact with CD44, a marker of aggressive breast cancer cells [basic fibroblast growth factor (bFGF), E-selectin (SELE), SELL, P-selectin (SELP), and OPN] [13,16,28,30–33].
Figure 3.
Lung-CM contain potential mediators of metastatic behavior. Lungs were harvested from healthy female nude mice, and lung-CM were generated and normalized as described in Materials and Methods section. RayBio Biotin Label-based Mouse Antibody Array I membranes (RayBiotech Inc; N = 3 per media condition) were exposed to dialyzed, biotin-labeled medium samples, washed, labeled with HRP-streptavidin, and visualized using chemiluminescence and film exposure. Representative arrays exposed to (A) basal media (DMEM/F12 + MITO+) and (B) lung-CM are shown. Boxes outlined with dashed lines indicate internal positive controls; boxes outlined with dotted lines indicate internal negative controls; and boxes outlined with solid bold lines highlight proteins of interest identified in lung-CM but not in basal media following the shortest (10-second) film exposure. Further details about proteins of interest are provided in Table 2.
Table 2.
Metastasis-Associated Proteins of Interest Identified by Protein Array Analysis of Lung-Conditioned Media*.
| Array Position (Figure 3) | Protein Name | Function/Association with Metastasis | References | ||
| 39 | bFGF, FGF2 |
|
[33] | ||
| 115 | EREG |
|
[10,29] | ||
| 117 | SELE |
|
[30] | ||
| 303 | SELL |
|
[31,32] | ||
| 333 | OPN |
|
[10,28] | ||
| 346 | SELP |
|
[31,32] | ||
| 429 | uPA |
|
[10,27] | ||
| 430 | VCAM1 |
|
[40] | ||
| 432 | VEGFA |
|
[41] |
Data are derived from analysis of three separate protein arrays (N = 3) of lung-CM versus basal media.
Depletion of SELL or OPN from Lung-CM Reduces Breast Cancer Cell Migration through CD44 Receptor-Ligand Interactions
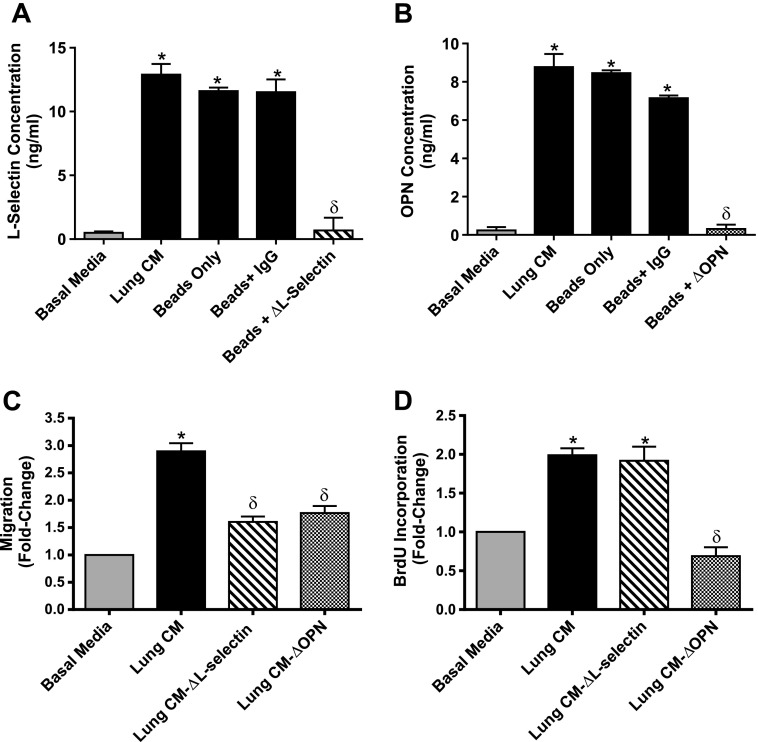
On the basis of our interest on the role of both OPN [28] and “stemlike” ALDHhiCD44+ cells [12,16,17] in breast cancer metastasis and the observation that all four cell lines investigated express cell_surface CD44 (Figure W4), we focused on investigating the functional consequences of two of the identified lung-derived CD44 ligands, SELL and OPN. Using an immunodepletion approach, SELL and OPN were successfully removed from lung-CM, down to equivalent levels of those observed in basal media (Figure 4, A and B). Relative to non-depleted lung-CM, depletion of SELL (ΔSELL) reduced the migration of MDA-MB-231 breast cancer cells, whereas depletion of OPN (ΔOPN) reduced both migration and proliferation (P < .05) (Figure 4, C and D).
Figure 4.
Depletion of SELL and OPN from lung-CM reduces breast cancer cell migration. (A and B) Lungs were harvested from healthy female nude mice, and lung-CM were generated, normalized, and immunodepleted of SELL (A) or OPN (B) as described in Materials and Methods section. The resulting SELL or OPN levels were assessed by ELISA in undepleted lung-CM, lung-CM exposed to Dynabeads Protein G only, lung-CM exposed to beads plus a nonspecific anti-mouse IgG antibody, and lung-CM depleted of SELL (A) or OPN (B) using beads plus specific anti-mouse antibodies (“ΔSELL” or “ΔOPN”). (A and B) Data are presented as means ± SEM (N = 3). (C) MDA-MB-231 human breast cancer cells (5 x 104 per well) were plated in triplicate (n = 3) on top of gelatin-coated transwells (8-µm pore size) before placement into basal media (DMEM/F12 + MITO+), undepleted lung-CM, or lung-CM depleted of SELL or OPN. Migration was allowed to occur for more than 18 hours at 37°C (5% CO2). Transwells were then fixed with glutaraldehyde, stained with Harris' hematoxylin, and developed with ammonium hydroxide. Five HPFs of view were captured per transwell, and migrated cells were enumerated using ImageJ software (NIH). (D) MDA-MB-231 cells (1.5 x 104 per well) were plated in triplicate (n = 3) in eight-well chamber slides and allowed to adhere for 24 hours. Cells were washed with PBS and then incubated for 72 hours in serum-free media. The media were then changed to basal media (DMEM/F12 + MITO+), undepleted lung-CM, or lung-CM depleted of SELL or OPN. After 24 hours, cells were incubated with 5 µl/ml BrdU for 30 minutes before fixation and staining for BrdU incorporation through immunofluorescence. Positive cells were enumerated and determined as a percentage of total cells present (through nuclear staining with DAPI). (C and D) Data are presented as means ± SEM (N = 3; fold change from negative control of basal media). *, significantly different from basal media; δ, significantly different from undepleted lung-CM (ANOVA with Tukey posttest; P < .05).
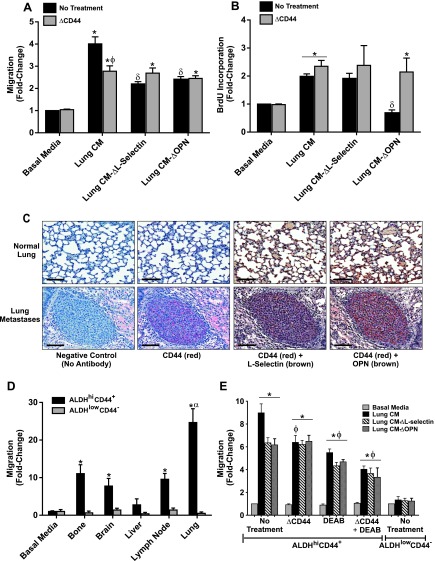
We next wanted to investigate whether the observed functional effect of lung-derived OPN and SELL was occurring through CD44 receptor-ligand interactions with breast cancer cells. Pretreatment of MDA-MB-231 cells with a CD44-blocking antibody before exposure to nondepleted lung-CM resulted in decreased migration, down to the level of migration seen in the presence of lung-CM depleted of SELL or OPN (P < .05) (Figure 5A). In addition, blocking of CD44 function in MDA-MB-231 cells in the presence of ΔSELL- or ΔOPN-depleted media resulted in no further reduction in migratory response, suggesting that both SELL-mediated and OPN-mediated migrations toward lung-CM are occurring through CD44 receptor-ligand interactions (Figure 5A). In contrast, blocking of CD44 function did not influence the growth of MDA-MB-231 cells in either nondepleted or depleted lung-CM (Figure 5B).
Figure 5.
Soluble lung-derived factors mediate breast cancer cell migration through CD44 receptor-ligand interactions. (A and B) To assess the combined role of CD44 in SELL- and OPN-mediated breast cancer migration and growth, MDA-MB-231 cells were preincubated at RT for 30 minutes in the presence (ΔCD44) or absence of 10 µg/ml rat anti-human CD44 antibody before being subjected to transwell migration (A) or BrdU incorporation (B) assays using lung-CM as described for Figure 4. (C) MDA-MB-231 human breast cancer cells were injected into the tail vein of female nude mice using an established model of experimental metastasis (1 x 106 cells per mouse; n = 5 mice). At 8 weeks post-injection, mice were killed and assessed for metastatic burden in the lung using histopathology. Serial tissue sections were subjected to immunohistochemical analysis of CD44 (red staining; AP-Red), SELL (brown staining; DAB), and/or OPN (brown staining; DAB) in normal lung tissue (top panels) and lung metastases (bottom panels). Representative sections are shown; all scale bars, 100 µm. (D and E) ALDHhiCD44+ (black bars) or ALDHlowCD44- (gray bars) cell populations were isolated from the MDA-MB-231 human breast cancer cell line using FACS as described in Figure W3 and subjected to transwell migration assays using organ-CM (bone, brain, liver, LN, and lung) as described for Figure 1 (D) or preincubated at RT for 30 minutes in the presence (ΔCD44) or absence of 10 µg/ml rat anti-human CD44 antibody and/or 100 µM of the ALDH inhibitor DEAB (100 µM) before being subjected to transwell migration assays using lung-CM as described for Figure 4 (E). All graphical data are presented as means ± SEM (N = 3; fold change from negative control of basal media). *, significantly different from basal media; δ, significantly different from undepleted lung-CM; ϕ, significantly different from untreated MDA-MB-231 cells in the same respective lung-CM conditions (ANOVA with Tukey posttest; P < .05).
To validate our ex vivo observations, we also investigated whether CD44 receptor-ligand interactions might occur during in vivo lung metastasis. Using an experimental breast cancer metastasis model, we observed that normal mouse lung stained strongly for OPN and SELL (Figure 5C, top right panels) but not for CD44 (Figure 5C, top left panels). In contrast, breast cancer lung metastases stained positive for CD44 and colocalized with OPN (and to a lesser extent SELL) (Figure 5C, bottom panels), suggesting that CD44 receptor-ligand interactions could also occur during in vivo lung metastasis.
Finally, we have previously shown that breast cancer cells with a “stemlike” ALDHhiCD44+ phenotype exhibit an overall increase in metastatic behavior in vivo, particularly to the lung [16]. To determine if this behavior may be due to CD44 ligand-receptor interactions, we isolated ALDHhiCD44+ and ALDHlowCD44- subsets from the MDA-MB-231 cell line through FACS (Figure W5) and exposed them to different organ-CM. Similar to cell line-specific in vivo and whole-population ex vivo observations (Figure 1 and Table 1), ALDHhiCD44+ cells from the MDA-MB-231 cell line demonstrated increased migration toward LN-, lung-, bone-, and brain-CM (P < .05) (Figure 5D). These organ-specific increases in migration were not observed for ALDHlowCD44- cells. Notably, ALDHhiCD44+ cells showed the greatest migration toward lung-CM relative to all other organ-CM (P < .05) (Figure 5D), and we hypothesized that this was due to CD44 receptor-ligand interactions with lung-derived SELL and/or OPN. Similar to whole-population observations (Figure 5A), pretreatment of ALDHhiCD44+ cells with CD44-blocking antibody before exposure to nondepleted lung-CM resulted in decreased migration, down to the level of migration in lung-CM depleted of SELL or OPN (P < .05) (Figure 5E). In addition, blocking of CD44 function in ALDHhiCD44+ cells in the presence of ΔSELL- or ΔOPN-depleted media resulted in no further reduction in migratory response, suggesting that both SELL-mediated and OPN-mediated migrations toward lung-CM are occurring through CD44 receptor-ligand interactions in ALDHhiCD44+ cells (Figure 5E). Interestingly, when cells were pretreated with the ALDH inhibitor DEAB, migration of ALDHhiCD44+ cells also decreased in response to nondepleted or ΔSELL/ΔOPN-depleted media (P < .05). Furthermore, combined blockade of both CD44 and ALDH reduced migration of ALDHhiCD44+ cells down to the level of ALDHlowCD44- cells (Figure 5E).
Discussion
The majority of breast cancer deaths occur as a result of metastatic disease rather than from the effects of the primary tumor. Although clinical studies have shown that breast cancer preferentially metastasizes to lung, liver, bone, LN, and brain [3], it remains unclear whether properties of cancer cells (“seeds”), properties of organ microenvironments (“soil”), or a combination of both is responsible for this observed organ tropism. Previous studies by Massagué and colleagues have identified particular genes expressed by breast cancer cells that mediate organ-specific metastasis [4,8–10]. However, investigation of the factors contributed by specific metastatic microenvironments has received much less attention. We believe that this is largely due to the technical challenges of studying the organ microenvironment in vitro, juxtaposed with the challenges of identifying and studying specific molecular factors in detail using complex in vivo animal models of metastasis [34]. Although interactions between breast cancer cells and the bone microenvironment have been extensively studied using two-dimensional and three-dimensional coculture models [35], studies investigating the influence of other organ microenvironments have been limited and have focused mainly on murine mammary cancer cells [36–39].
In the current study, we established a comprehensive ex vivo model system for investigating the influence of organ-specific soluble factors on the metastatic behavior of human breast cancer cells. This model system facilitates investigation of individual metastatic behaviors of human breast cancer cells in soluble organ conditions that mirror common clinical sites of breast cancer metastasis, as well as allows for identification of organ-derived factors that may provide a basis for cross talk between disseminated tumor cells and their metastatic microenvironments. Our results indicate that human breast cancer cells with varying genetic backgrounds exhibit cell line-specific and organ-specific migration and growth patterns in different organ-CM. Importantly, these patterns reflect the known metastatic dissemination patterns of these cell lines in vivo, thus providing validation for our newly developed ex vivo model system. The breast cancer cell lines used in this study have been previously classified as being either an ER-/PR-/HER2+ subtype (SUM159 and SUM149) [19] or a triple-negative ER-/PR-/HER2- subtype (MDA-MB-231 and MDA-MB-468) [18,20]. Patients with breast cancer with these aggressive molecular subtypes tend to display a high propensity for lung metastasis that results in significant morbidity and mortality [3]. Notably, these clinical data support the results derived from our ex vivo model system because exposure to lung-CM increased migration of all breast cancer cell lines tested and increased proliferation in two of four cell lines.
In addition to the usefulness of our ex vivo model system for functional studies of organ tropism, the findings presented here also highlight the lung as an important source of soluble factors that can mediate metastasis, migration, neoplastic growth, and adhesion. Ingenuity Pathway Analysis identified recurring molecules across these categories including bFGF, EREG, selectins, OPN, uPA, vascular cell adhesion molecule 1 (VCAM-1), and vascular endothelial growth factor A (VEGFA). Due to its heparin-binding ability, bFGF can become concentrated in CD44-associated proteoglycans, and bursts of bFGF can be released on ECM remodeling, causing angiogenesis and an opportunity for tumor cells to enter the vasculature [33]. The SELE, SELP, and SELL are adhesion molecules found in both surface-bound and soluble forms from different cell types (endothelial, platelet, and leukocytes, respectively), and all three selectins act as CD44 ligands [30–32]. SELE has been implicated in the successful transendothelial migration of tumor cells [30], whereas SELLs and SELPs have been associated with formation of permissive metastatic microenvironments in lung [32]. VCAM-1 is an adhesion molecule that exists in both soluble and surface-bound forms, and high plasma levels of VCAM-1 are associated with clinical breast cancer progression to lung [40]. VEGFA is a soluble growth factor and a key regulator of angiogenesis that has also been shown to exert a chemoattractive effect on many types of cancer cells including breast cancer cells [41].
Importantly, our protein array studies also revealed some complementary lung-derived factors that correspond to tumor cell-derived factors identified by Massagué's group as being mediators of lung metastasis, including EREG, OPN, and uPA [10]. EREG is a member of the EGF family of growth factors and has been shown to exert mitogenic effects in addition to being a key component of vascular remodeling in metastasis [29]. OPN is a secreted phosphoprotein that acts as both a chemoattractant and a matrix protein and has been implicated in promotion of breast cancer cell adhesion, migration, invasion, and metastasis through interactions with integrins and CD44 [28]. Finally, uPA is a secreted serine protease that interacts with cyclooxygenase 2 (COX-2) (a tumor-derived factor identified by Massagué and colleagues) to mediate breast cancer motility and invasion [27].
We were particularly intrigued by the observation that several of the factors identified in lung-CM were CD44 ligands because CD44 has a well-established role in metastasis [33]. We thus decided to investigate CD44 ligand-receptor interactions as an underlying mechanism of the observed breast cancer cell migratory and proliferative responses to lung-CM, using OPN and SELL as proof-of-principle ligands. We observed that immunodepletion of SELL from lung-CM resulted in decreased migration of MDA-MB-231 cells, whereas depletion of OPN decreased both migration and proliferation. Although the migration effects could be abrogated by pretreatment with a CD44 antibody, blocking CD44 function did not influence breast cancer cell proliferation. This indicates that, although migration in response to soluble lung-derived factors can be mediated by CD44 ligand-receptor interactions, proliferative responses may be occurring through CD44-independent mechanisms such as OPN-integrin interactions [28] or additional mitogenic mechanisms that remain to be uncovered.
It is worthwhile noting that, although all four cell lines tested (MDA-MB-231, SUM159, SUM-149, and MDA-MB-468) were observed to express consistently high levels of the standard form of CD44 that can interact with soluble lung-derived ligands to promote a significant chemotactic response to lung-CM, we observed some cell line-specific differences with regards to the degree of chemotactic response. Although the underlying molecular mechanisms of this are unclear at this time, there are several possibilities that might help to explain these differences. Although we only examined expression of the standard form of CD44, there are a number of different CD44 isoforms (both metastasis promoting and metastasis inhibiting) [42] that may be playing a role in the observed cell line-specific responses to lung-CM, and the differential expression patterns of these in the different breast cancer cell lines will be examined in future studies. Interestingly, the two cell lines that showed the greatest chemotactic responses toward lung-CM (SUM149 and MDA-MB-468) also express very high levels of the epidermal growth factor receptor [19,43], which has been shown to cross-talk with OPN/integrin and OPN/CD44 pathways at the molecular level to promote migratory and metastatic behavior [28]. The MDA-MB-468 cell line also expresses higher endogenous levels of OPN than the MDA-MB-231 cell line [44] that may be influencing the chemotactic response to lung-CM in an autocrine fashion, although the relative contributions and roles of tumor-derived versus microenvironment-derived OPN remain poorly understood [28]. Ongoing studies are aimed at further delineating the differential tumor-derived molecular factors that can interact with lung-derived soluble factors to mediate breast cancer migration and growth.
In addition to the role of CD44 in metastasis, recent studies have also highlighted its role in normal stem cell mobilization and homing as well as establishment of the premetastatic niche [45]. CD44 receptor-ligand interactions may be especially important in the context of “stemlike” tumor cells in metastasis, as one of the key putative characteristics of breast cancer tumor-initiating cells is that of CD44 positivity [13]. Indeed, we observed that isolated breast cancer cells with an ALDHhiCD44+ phenotype responded in an organ-specific chemotactic manner, whereas ALDHlowCD44- cells did not. This suggests that ALDHhiCD44+ breast cancer cells may be driving the organotropic behavior of whole cell populations. Furthermore, our previous observation that ALDHhiCD44+ breast cancer cells exhibit an overall increase in metastatic behavior in vivo with a particular propensity for the lung [16] is supported by the findings of the current study, where we observed that ALDHhiCD44+ cells displayed preferential migration toward lung-CM relative to all other organs. Similar to our results in whole breast cancer cell populations, we observed that migration of ALDHhiCD44+ cells could be mediated by CD44 receptor-ligand interactions. However, we further observed that pretreatment with the ALDH inhibitor DEAB also decreased migration of ALDHhiCD44+ cells in response to nondepleted or ΔSELL/ΔOPN-depleted lung-CM, and that combined blockade of both CD44 and ALDH reduced migration of ALDHhiCD44+ cells down to the level of ALDHlowCD44- cells. This suggests an additive role for both CD44 and ALDH pathways in mediating migration toward soluble lung-derived factors. This was somewhat unexpected, as previous studies have mainly implicated ALDH in retinoic acid synthesis, cellular self-protection, differentiation, and/or proliferation, rather than in processes such as migration [46]. Furthermore, the ALDH-mediated migration effect was only observed in lung-CM but not in basal media, supporting the idea that interactions with soluble lung-derived factors may be influencing this behavior. To the best of our knowledge, none of the 70 proteins identified in lung-CM have previously been shown to interact with ALDH; however, this is currently under further investigation.
In summary, this study reports the establishment and validation of a comprehensive ex vivo model system for investigating the influence of organ-specific soluble factors on metastatic behavior of human breast cancer cells. Our results indicate that multiple human breast cancer cells exhibit organ-specific and cell line-specific migration and growth patterns toward specific organ conditions in a manner that reflects their metastatic behavior in vivo. Our results also highlight the lung as an important source of soluble factors that mediate metastatic behavior and suggest for the first time that interactions between subpopulations of CD44-expressing breast cancer cells (including ALDHhiCD44+ cells) and soluble factors present in the lung microenvironment may play an important role in determining organotropic metastatic behavior. These findings lay the groundwork for a number of future studies, including investigation of the role of other soluble CD44 ligands in lung metastasis, the complementary involvement of lung-derived insoluble/ECM factors in mediating breast cancer cell behavior (particularly colonization and proliferation), and molecular interrogation of CM derived from other organs. Taken together, further elucidation of the cross talk between “seed” and “soil” during organ-specific metastasis and the translation of this knowledge into the clinic will have important implications for the management and treatment of breast cancer.
Supplemental Materials and Methods
Flow Cytometry Characterization of Primary Cell Isolates and Human Breast Cancer Cells
Isolated LNSC and BMSC populations were characterized by flow cytometry on the basis of previous studies [1, 2] (Figure W1). LNSCs (1 x 105) were incubated with FITC-conjugated anti-CD45 and phycoerythrin (PE)-conjugated anti-gp38 antibodies (eBioscience, San Diego, CA), followed by incubation with 7-aminoactinomycin D (7-AAD; BD Biosciences). BMSCs (1 x 105) were labeled with anti-Sca-1, anti-CD105, anti-CD73, anti-CD29, or anti-CD44 antibodies (R&D Systems Inc, Minneapolis, MN). All samples were then labeled with a goat anti-rat PE-conjugated secondary antibody (R&D Systems), followed by incubation with 7-AAD.
Human breast cancer cell lines (MDA-MB-231, SUM159, SUM149, and MDA-MB-468; 1 x 105 cells) were characterized for CD44 expression by flow cytometry using a PE-conjugated anti-CD44 antibody (clone IM7; BD Biosciences) (Figure W2).
For all flow cytometry experiments, appropriate IgG isotype controls were used to assess nonspecific staining. Cells (1 x 105 viable events) were analyzed using an EPICS XL-MCL flow cytometer (Beckman Coulter, Fullerton, CA). Methodology for the FACS analysis (Figure W3) is provided in the main body of the manuscript.
Fluorescence-Activated Cell Sorting
Cell subpopulations (ALDHhiCD44+CD24- and ALDHlow-CD44low/-CD24+) were isolated from the MDA-MB-231 cell line as described previously [3, 4]. Briefly, cells were concurrently labeled with 7-AAD, ALDEFLUOR assay kit (STEMCELL Technologies, Vancouver, British Columbia) and fluorescently conjugated antibodies including anti-CD44 (clone IM7) conjugated to allophycocyanin and anti-CD24 (clone ML5) conjugated to PE (BD Biosciences). ALDH activity was used as the primary sort criteria (top ∼20% = ALDHhi; bottom ∼20% = ALDHlow) and CD44+CD24- phenotype as the secondary sort criteria (top ∼10% gated on ALDHhi; bottom ∼10% gated on ALDHlow). Cell viability was assessed by 7-AAD staining during cell sorting and confirmed by trypan blue exclusion postsorting. FACS-isolated cells were used immediately for in vitro migration assays.
Acknowledgments
We thank Benjamin Hedley and Kristin Chadwick for their invaluable advice and technical help with the flow cytometry/FACS experiments, Joseph Andrews for bioinformatics expertise in protein array data analysis, Carl Postenka for assistance with histopathology, and Ann Chambers and Danny Welch for helpful discussions during manuscript preparation.
Abbreviations
- ALDH
aldehyde dehydrogenase
- ANOVA
analysis of variance
- AP
alkaline phosphatase
- bFGF
basic fibroblast growth factor
- BM
bone marrow
- BMSC
bone marrow stromal cell
- BrdU
bromodeoxyuridine
- CD
cluster of differentiation
- CM
conditioned media
- COX-2
cyclooxygenase 2
- DAPI
4′,6-diamidino-2-phenylindole
- DEAB
diethylaminobenzaldehyde
- DMEM
Dulbecco's modified Eagle's medium
- ECM
extracellular matrix
- ER
estrogen receptor
- EREG
epiregulin
- FITC
fluorescein isothiocyanate
- HPF
high-powered field
- LN
lymph node
- LNSC
lymph node stromal cell
- NIH
National Institutes of Health
- OPN
osteopontin
- PR
progesterone receptor
- RT
room temperature
- SELE
E-selectin
- SELL
L-selectin
- SELP
P-selectin
- uPA
urokinase-type plasminogen activator
- VCAM-1
vascular cell adhesion molecule 1
- VEGFA
vascular endothelial growth factor A
Footnotes
This work was supported by grants from the Canadian Breast Cancer Foundation-Ontario Region, the Canada Foundation for Innovation (No. 13199), and donor support from John and Donna Bristol through the London Health Sciences Foundation (to A.L.A.). Studentship and fellowship support were provided by an Ontario Graduate Scholarship (Province of Ontario, to J.E.C.), the Canadian Institutes of Health Research (CIHR)-Strategic Training Program (to J.E.C. and B.C-Y.), and the Pamela Greenaway-Kohlmeier Translational Breast Cancer Research Unit at the London Regional Cancer Program (to J.E.C., Y.X., and A.K.C.). A.L.A. is supported by a CIHR New Investigator Award and an Early Researcher Award from the Ontario Ministry of Research and Innovation.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 to W5 and are available online at www.neoplasia.com.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 3.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28:3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 5.Ewing J. Neoplastic Diseases. A Treatise on Tumors. London, United Kingdom: WB Saunders Co; 1928. pp. 77–89. [Google Scholar]
- 6.Paget S. The distribution of secondary growths in cancer of the breast. The Lancet. 1889;1:99–101. [PubMed] [Google Scholar]
- 7.Weiss L. Comments on hematogenous metastatic patterns in humans as revealed by autopsy. Clin Exp Metastasis. 1992;10:191–199. doi: 10.1007/BF00132751. [DOI] [PubMed] [Google Scholar]
- 8.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 10.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss L. Metastatic inefficiency. Adv Cancer Res. 1990;54:159–211. doi: 10.1016/s0065-230x(08)60811-8. [DOI] [PubMed] [Google Scholar]
- 12.Croker AK, Allan AL. Cancer stem cells: implications for the progression and treatment of metastatic disease. J Cell Mol Med. 2008;12:374–390. doi: 10.1111/j.1582-4934.2007.00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2009;13:2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croker AK, Allan AL. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44+ human breast cancer cells. Breast Cancer Res Treat. 2012;133:75–87. doi: 10.1007/s10549-011-1692-y. [DOI] [PubMed] [Google Scholar]
- 18.Cailleau R, Olivé M, Cruciger QV. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In Vitro. 1978;14:911–915. doi: 10.1007/BF02616120. [DOI] [PubMed] [Google Scholar]
- 19.Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi OP, Ethier SP. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Brit J Cancer. 1999;81:1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price JE, Polyzos A, Zhang RD, Daniels LM. Tumorigenicity and metastasis of human breast carcinoma cell lines in nude mice. Cancer Res. 1990;50:717–721. [PubMed] [Google Scholar]
- 21.Zhou YW, Aritake S, Tri Endharti A, Wu J, Hayakawa A, Nakashima I, Suzuki H. Murine lymph node-derived stromal cells effectively support survival but induce no activation/proliferation of peripheral resting T cells in vitro. Immunology. 2003;109:496–503. doi: 10.1046/j.1365-2567.2003.01693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Kuroda S, Shichinohe H, Ikeda J, Seki T, Hida K, Tada M, Sawada K, Iwasaki Y. Migration and differentiation of nuclear fluorescence-labeled bone marrow stromal cells after transplantation into cerebral infarct and spinal cord injury in mice. Neuropathology. 2003;23:169–180. doi: 10.1046/j.1440-1789.2003.00496.x. [DOI] [PubMed] [Google Scholar]
- 23.Khan SA, Cook AC, Kappil M, Günthert U, Chambers AF, Tuck AB, Denhardt DT. Enhanced cell surface CD44 variant (v6, v9) expression by osteopontin in breast cancer epithelial cells facilitates tumor cell migration: novel post-transcriptional, post-translational regulation. Clin Exp Metastasis. 2005;22:663–673. doi: 10.1007/s10585-006-9007-0. [DOI] [PubMed] [Google Scholar]
- 24.Hammerschmidt SI, Ahrendt M, Bode U, Wahl B, Kremmer E, Förster R, Pabst O. Stromal mesenteric lymph node cells are essential for the generation of gut-homing T cells in vivo. J Exp Med. 2008;205:2483–2490. doi: 10.1084/jem.20080039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 26.Baddoo M, Hill K, Wilkinson R, Gaupp D, Hughes C, Kopen GC, Phinney DG. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem. 2003;89:1235–1249. doi: 10.1002/jcb.10594. [DOI] [PubMed] [Google Scholar]
- 27.Singh B, Berry JA, Shoher A, Ramakrishnan V, Lucci A. COX-2 overexpression increases motility and invasion of breast cancer cells. Int J Oncol. 2005;26:1393–1399. [PubMed] [Google Scholar]
- 28.Tuck AB, Chambers AF, Allan AL. Osteopontin overexpression in breast cancer: knowledge gained and possible implications for clinical management. J Cell Biochem. 2007;102:859–868. doi: 10.1002/jcb.21520. [DOI] [PubMed] [Google Scholar]
- 29.Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C, Gomis RR, Manova-Todorova K, Massagué J. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 30.Hanley WD, Burdick MM, Konstantopoulos K, Sackstein R. CD44 on LS174T colon carcinoma cells possesses E-selectin ligand activity. Cancer Res. 2005;65:5812–5817. doi: 10.1158/0008-5472.CAN-04-4557. [DOI] [PubMed] [Google Scholar]
- 31.Hanley WD, Napier SL, Burdick MM, Schnaar RL, Sackstein R, Konstantopoulos K. Variant isoforms of CD44 are P- and L-selectin ligands on colon carcinoma cells. FASEB J. 2006;20:337–339. doi: 10.1096/fj.05-4574fje. [DOI] [PubMed] [Google Scholar]
- 32.Laubli H, Borsig L. Selectins as mediators of lung metastasis. Cancer Microenviron. 2010;3:97–105. doi: 10.1007/s12307-010-0043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naor D, Nedvetzki S, Golan I, Melnik L, Faitelson Y. CD44 in cancer. Crit Rev Clin Lab Sci. 2002;39:527–579. doi: 10.1080/10408360290795574. [DOI] [PubMed] [Google Scholar]
- 34.Chu JE, Allan AL. The role of cancer stem cells in the organ tropism of breast cancer metastasis: a mechanistic balance between the “seed” and the “soil”? Int J Breast Cancer. 2012;2012:209748. doi: 10.1155/2012/209748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mastro AM, Vogler EA. A three-dimensional osteogenic tissue model for the study of metastatic tumor cell interactions with bone. Cancer Res. 2009;69:4097–4100. doi: 10.1158/0008-5472.CAN-08-4437. [DOI] [PubMed] [Google Scholar]
- 36.Horak E, Darling DL, Tarin D. Analysis of organ-specific effects on metastatic tumor formation by studies in vitro. J Natl Cancer Inst. 1986;76:913–922. [PubMed] [Google Scholar]
- 37.Martin MD, Fingleton B, Lynch CC, Wells S, McIntyre JO, Piston DW, Matrisian LM. Establishment and quantitative imaging of a 3D lung organotypic model of mammary tumor outgrowth. Clin Exp Metastasis. 2008;25:877–885. doi: 10.1007/s10585-008-9206-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bal de Kier Joffé E, Alonso DF, Puricelli L. Soluble factors released by the target organ enhance the urokinase-type plasminogen activator activity of metastatic tumor cells. Clin Exp Metastasis. 1991;9:51–56. doi: 10.1007/BF01831709. [DOI] [PubMed] [Google Scholar]
- 39.Puricelli L, Gómez DE, Vidal MC, Eiján AM, Spinelli O, Alonso DF, de Lustig ES, Bal de Kier Joffé E. Effect of host-organ environment on the in vivo and in vitro behavior of a murine mammary adenocarcinoma. Tumour Biol. 1994;15:284–293. doi: 10.1159/000217903. [DOI] [PubMed] [Google Scholar]
- 40.Chen Q, Zhang XH, Massagué J. Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell. 2011;20:538–549. doi: 10.1016/j.ccr.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsson E, Honeth G, Bendahl PO, Saal LH, Gruvberger-Saal S, Ringnér M, Vallon-Christersson J, Jönsson G, Holm K, Lövgren K, et al. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Cancer. 2011;11:418. doi: 10.1186/1471-2407-11-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mamot C, Drummond DC, Greiser U, Hong K, Kirpotin DB, Marks JD, Park JW. Epidermal growth factor receptor (EGFR)-targeted immunoliposomes mediate specific and efficient drug delivery to EGFR- and EGFRvIII-overexpressing tumor cells. Cancer Res. 2003;63:3154–3161. [PubMed] [Google Scholar]
- 44.Ortiz-Martínez F, Perez-Balaguer A, Ciprián D, Andrés L, Ponce J, Adrover E, Sánchez-Payá J, Aranda FI, Lerma E, Peiró G. Association of increased osteopontin and splice variant-c mRNA expression with HER2 and triple-negative/basal-like breast carcinomas subtypes and recurrence. Hum Pathol. 2014;45:504–512. doi: 10.1016/j.humpath.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 45.Williams K, Motiani K, Giridhar PV, Kasper S. CD44 integrates signaling in normal stem cell, cancer stem cell and (pre)metastatic niches. Exp Biol Med (Maywood) 2013;238:324–338. doi: 10.1177/1535370213480714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7:292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- 47.Adams LS, Kanaya N, Phung S, Liu Z, Chen S. Whole blueberry powder modulates the growth and metastasis of MDA-MB-231 triple negative breast tumors in nude mice. J Nutr. 2011;141:1805–1812. doi: 10.3945/jn.111.140178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lowes LE, Goodale D, Keeney M, Allan AL. Image cytometry analysis of circulating tumor cells. Methods Cell Biol. 2011;102:261–290. doi: 10.1016/B978-0-12-374912-3.00010-9. [DOI] [PubMed] [Google Scholar]
- 49.Johnson RW, Nguyen MP, Padalecki SS, Grubbs BG, Merkel AR, Oyajobi BO, Matrisian LM, Mundy GR, Sterling JA. TGF-β promotion of Gli2-induced expression of parathyroid hormone-related protein, an important osteolytic factor in bone metastasis, is independent of canonical Hedgehog signaling. Cancer Res. 2011;71:822–831. doi: 10.1158/0008-5472.CAN-10-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGowan PM, Simedrea C, Ribot EJ, Foster PJ, Palmieri D, Steeg PS, Allan AL, Chambers AF. Notch1 inhibition alters the CD44hi/CD24lo population and reduces the formation of brain metastases from breast cancer. Mol Cancer Res. 2011;9:834–844. doi: 10.1158/1541-7786.MCR-10-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flanagan L, Van Weelden K, Ammerman C, Ethier SP, Welsh J. SUM-159PT cells: a novel estrogen independent human breast cancer model system. Breast Cancer Res Treat. 1999;58:193–204. doi: 10.1023/a:1006331716981. [DOI] [PubMed] [Google Scholar]
- 52.Singh B, Cook KR, Martin C, Huang EH, Mosalpuria K, Krishnamurthy S, Cristofanilli M, Lucci A. Evaluation of a CXCR4 antagonist in a xenograft mouse model of inflammatory breast cancer. Clin Exp Metastasis. 2010;27:233–240. doi: 10.1007/s10585-010-9321-4. [DOI] [PubMed] [Google Scholar]
- 53.Allan AL, George R, Vantyghem SA, Lee MW, Hodgson NC, Engel CJ, Holliday RL, Girvan DP, Scott LA, Postenka CO, et al. Role of the integrin-binding protein osteopontin in lymphatic metastasis of breast cancer. Am J Pathol. 2006;169:233–246. doi: 10.2353/ajpath.2006.051152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- 1.Hammerschmidt SI, Ahrendt M, Bode U, Wahl B, Kremmer E, Förster R, Pabst O. Stromal mesenteric lymph node cells are essential for the generation of gut-homing T cells in vivo. J Exp Med. 2008;205:2483–2490. doi: 10.1084/jem.20080039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 3.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2009;13:2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Croker AK, Allan AL. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44+ human breast cancer cells. Breast Cancer Res Treat. 2012;133:75–87. doi: 10.1007/s10549-011-1692-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.