Abstract
A strain of Escherichia coli with a 7-minute (340 kilobase pairs of DNA) deletion of the terminus region of the chromosome was isolated. This deletion was probably an IS10-promoted event and its extent was characterized by both genetic and DNA hybridization analyses. The most dramatic property of strains harboring this deletion was the absence of the sites that inhibit clockwise- and counterclockwise-traveling replication forks. These strains also grew slowly, produced many nonviable cells, were filamentous, and appeared to have an induced SOS system.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anilionis A., Ostapchuk P., Riley M. Identification of a second cryptic lambdoid prophage locus in the E. coli K12 chromosome. Mol Gen Genet. 1980;180(2):479–481. doi: 10.1007/BF00425865. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitner R. M., Kuempel P. L. P1 transduction map spanning the replication terminus of Escherichia coli K12. Mol Gen Genet. 1981;184(2):208–212. doi: 10.1007/BF00272906. [DOI] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché J. P., Gélugne J. P., Louarn J., Louarn J. M., Kaiser K. Relationships between the physical and genetic maps of a 470 x 10(3) base-pair region around the terminus of Escherichia coli K12 DNA replication. J Mol Biol. 1982 Jan 5;154(1):21–32. doi: 10.1016/0022-2836(82)90414-4. [DOI] [PubMed] [Google Scholar]
- Camakaris H., Pittard J. Regulation of tyrosine and phenylalanine biosynthesis in Escherichia coli K-12: properties of the tyrR gene product. J Bacteriol. 1973 Sep;115(3):1135–1144. doi: 10.1128/jb.115.3.1135-1144.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Cornish E. C., Davidson B. E., Pittard J. Cloning and characterization of Escherichia coli K-12 regulator gene tyrR. J Bacteriol. 1982 Dec;152(3):1276–1279. doi: 10.1128/jb.152.3.1276-1279.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diderichsen B., De Hauwer G. Improved mapping of the tyrS locus in Escherichia coli. Mol Gen Genet. 1980;178(3):647–650. doi: 10.1007/BF00337873. [DOI] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Cloning and physical analysis of the pyrF gene (coding for orotidine-5'-phosphate decarboxylase) from Escherichia coli K-12. Gene. 1983 Nov;25(1):39–48. doi: 10.1016/0378-1119(83)90165-8. [DOI] [PubMed] [Google Scholar]
- Fouts K. E., Barbour S. D. Insertion of transposons through the major cotransduction gap of Escherichia coli K-12. J Bacteriol. 1982 Jan;149(1):106–113. doi: 10.1128/jb.149.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest J. R., Roberts R. E. Cloning, mapping, and expression of the fumarase gene of Escherichia coli K-12. J Bacteriol. 1983 Feb;153(2):588–596. doi: 10.1128/jb.153.2.588-596.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Engström P., Wolf-Watz H., Iino T., Hazelbauer G. L. Cloning of trg, a gene for a sensory transducer in Escherichia coli. J Bacteriol. 1982 Oct;152(1):372–383. doi: 10.1128/jb.152.1.372-383.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Harayama S. Mutants in transmission of chemotactic signals from two independent receptors of E. coli. Cell. 1979 Mar;16(3):617–625. doi: 10.1016/0092-8674(79)90035-7. [DOI] [PubMed] [Google Scholar]
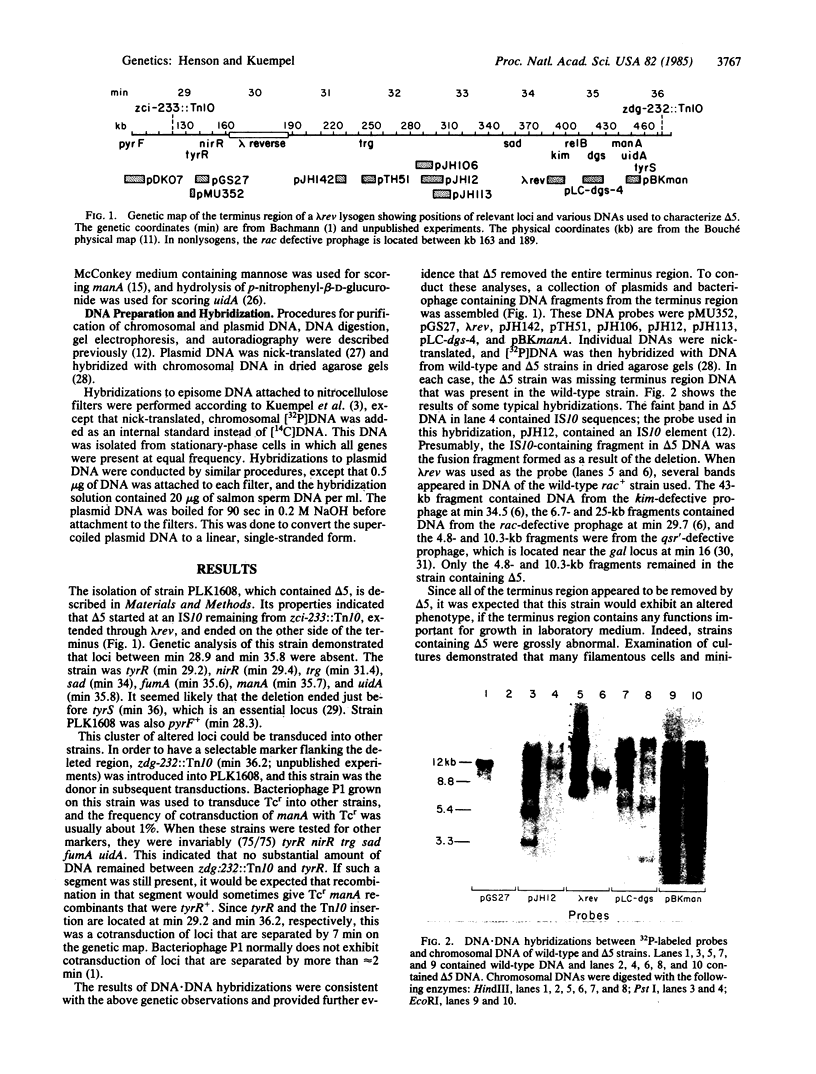
- Henson J. M., Kopp B., Kuempel P. L. Deletion of 60 kilobase pairs of DNA from the terC region of the chromosome of Escherichia coli. Mol Gen Genet. 1984;193(2):263–268. doi: 10.1007/BF00330678. [DOI] [PubMed] [Google Scholar]
- Henson J. M., Kuempel P. L. The use of transposon insertion zdc-235::Tn10 (min 32) to clone and delete DNA from the terminus region of Escherichia coli. Mol Gen Genet. 1983;189(3):506–512. doi: 10.1007/BF00325918. [DOI] [PubMed] [Google Scholar]
- Kaiser K. The origin of Q-independent derivatives of phage lambda. Mol Gen Genet. 1980;179(3):547–554. doi: 10.1007/BF00271744. [DOI] [PubMed] [Google Scholar]
- Kuempel P. L., Duerr S. A. Chromosome replication in Escherichia coli is inhibited in the terminus region near the rac locus. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):563–567. doi: 10.1101/sqb.1979.043.01.062. [DOI] [PubMed] [Google Scholar]
- Kuempel P. L., Duerr S. A., Seeley N. R. Terminus region of the chromosome in Escherichia coli inhibits replication forks. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3927–3931. doi: 10.1073/pnas.74.9.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuempel P. L., Maglothin P., Prescott D. M. Bidirectional termination of chromosome replication in Escherichia coli. Mol Gen Genet. 1973 Sep 5;125(1):1–8. doi: 10.1007/BF00292981. [DOI] [PubMed] [Google Scholar]
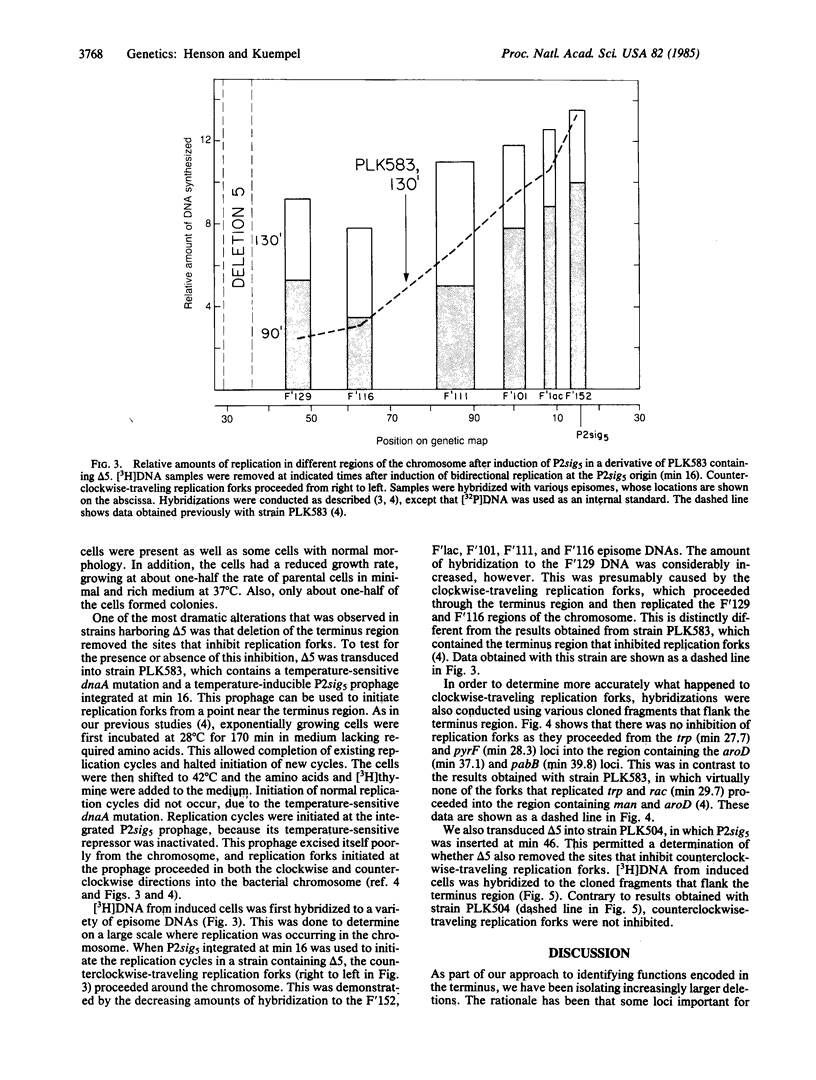
- Lambden P. R., Guest J. R. Mutants of Escherichia coli K12 unable to use fumarate as an anaerobic electron acceptor. J Gen Microbiol. 1976 Dec;97(2):145–160. doi: 10.1099/00221287-97-2-145. [DOI] [PubMed] [Google Scholar]
- Louarn J., Patte J., Louarn J. M. Evidence for a fixed termination site of chromosome replication in Escherichia coli K12. J Mol Biol. 1977 Sep 25;115(3):295–314. doi: 10.1016/0022-2836(77)90156-5. [DOI] [PubMed] [Google Scholar]
- Novel G., Novel M. Mutants d'Escherichia coli K 12 affectés pour leur croissance sur méthyl-beta-D-glucuronide: localisation of gène de structure de la beta-D-glucuronidase (uid A. Mol Gen Genet. 1973;120(4):319–335. [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Ross D. G., Swan J., Kleckner N. Physical structures of Tn10-promoted deletions and inversions: role of 1400 bp inverted repetitions. Cell. 1979 Apr;16(4):721–731. doi: 10.1016/0092-8674(79)90088-6. [DOI] [PubMed] [Google Scholar]
- Shaw D. J., Guest J. R. Amplification and product identification of the fnr gene of Escherichia coli. J Gen Microbiol. 1982 Oct;128(10):2221–2228. doi: 10.1099/00221287-128-10-2221. [DOI] [PubMed] [Google Scholar]
- Skinner M. A., Cooper R. A. An Escherichia coli mutant defective in the NAD-dependent succinate semialdehyde dehydrogenase. Arch Microbiol. 1982 Sep;132(3):270–275. doi: 10.1007/BF00407964. [DOI] [PubMed] [Google Scholar]
- Tsao S. G., Brunk C. F., Pearlman R. E. Hybridization of nucleic acids directly in agarose gels. Anal Biochem. 1983 Jun;131(2):365–372. doi: 10.1016/0003-2697(83)90185-9. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]