Abstract
Single-cell functional proteomics assays can connect genomic information to biological function through quantitative and multiplex protein measurements. Tools for single-cell proteomics have developed rapidly over the past 5 years and are providing approaches for directly elucidating phosphoprotein signaling networks in cancer cells or for capturing high-resolution snapshots of immune system function in patients with various disease conditions. We discuss advances in single-cell proteomics platforms, with an emphasis on microchip methods. These methods can provide a direct correlation of morphological, functional and molecular signatures at the single-cell level. We also provide examples of how those platforms are being applied to both fundamental biology and clinical studies, focusing on immune-system monitoring and phosphoprotein signaling networks in cancer.
Cell-to-cell variation and single-cell functional proteomics analysis
Non-genetic cellular heterogeneity is a universal feature of any cell population [1,2]. Although this heterogeneity is often ascribed to some process (such as stochastic gene expression), it is also intrinsic to the finite nature of a single cell [3]. This heterogeneity is not without consequences; for example, it can contribute to the diversity of an immune response or to the emergence of therapeutic resistance in cancers. However, the detailed role of cellular heterogeneity in such processes is not always easy to capture. If some parameter is measured on a statistical number of 'identical' single cells, that parameter can almost always be used to stratify those cells into multiple populations. Whether the variance in the assayed parameter is biologically relevant may be debatable. Parameters for which the variance is thought to have high biological relevance are the levels of functional proteins. These include the signaling proteins (such as cytokines) that are secreted by immune cells, or the phosphorylated kinases and related effector proteins that comprise the heart of growth factor signaling networks within cells.
A single-cell functional proteomics assay is one that measures the quantity and functional state (such as phosphorylation) of a given protein or panel of proteins across many otherwise identical cells. A measurement of the average level of a protein requires many single-cell measurements. Such measurements, if compiled as a histogram of the frequency of observation versus the measured levels, reflect the fluctuations of that protein. Functional protein fluctuations can reflect changes in cellular activity, such as immune-cell activation or the activation or inhibition of protein signaling networks within, for example, tumor cells. However, the usefulness of fluctuations significantly expands with absolute quantification and increased numbers of proteins assayed per cell (multiplexing).
When multiple proteins are assayed from single cells, protein-protein correlations and anti-correlations are directly recorded. For cell-surface markers, such measurements provide a way to enumerate and sort highly defined cellular phenotypes. A multiplex analysis of secreted effector proteins from immune-cell phenotypes can provide a powerful view of immune-system function. For intracellular signaling networks, such as those associated with growth factor signaling, correlations and anti-correlations between phosphoproteins can indicate activating and inhibitory interactions, respectively. With increased multiplexing, such measurements increasingly resolve the structure of signaling networks. If the measurements are truly quantitative, it becomes possible to assess how perturbations to cells influence changes in the chemical potential of the measured proteins. This, in turn, allows the introduction of predictive models derived from physicochemical principles.
Single-cell functional proteomics can connect genomic information with biological context and biological function. For example, certain classes of genetically engineered immune cells are increasingly used for certain anti-cancer therapies. This clonal population of cells can show great functional heterogeneity [4,5]. That heterogeneity, which can be characterized by single-cell proteomics, arises from many epigenetic factors (biological context), such as exposure to specific cell types or to signaling proteins. This and other examples are discussed in detail below.
Here, we describe emerging technologies and their associated applications that are designed to characterize cellular heterogeneity by single-cell functional proteomics. We first provide an overview of the rapid development of single-cell proteomics tools that has occurred over the past half decade. We then discuss specific biological or clinical challenges that are either uniquely or most easily addressed by single-cell functional proteomics. These challenges include basic biology studies, such as the kinetics of T-cell activation, or the identification of effector proteins associated with cellular motility. Clinical applications include advanced immune monitoring of patients with a variety of disease conditions, ranging from HIV to cancer. Cancer biology applications include experiments aimed at resolving how targeted therapeutics alter the phosphoprotein signaling networks that are hyperactivated in many tumors. Each problem provides a venue for discussing platform advantages and limitations. We focus on multiplex microfluidics/nanotechnology-based platforms as these tools are proving uniquely suited for quantitative, single-cell functional proteomics.
Single-cell functional proteomics technologies
Single-cell functional proteomics tools range from flow cytometry to microfluidics-based platforms, many of which are listed and briefly characterized in Table 1. An ideal tool reports on the level of a given protein in copy numbers per cell, with a small uncertainty, a high level of sensitivity, and the capacity to analyze large numbers of cells quickly. The value of absolute quantification is that it enables direct comparisons across platforms, cell types, time points, clinical samples, and so on. However, many platforms enable quantification only in relative units, or allow for the identification of only the fraction of the cells that express a given protein. Other characteristics, such as the level of multiplexing, the types of proteins that can be assayed (such as cytoplasmic, membrane, or secreted), or the ability to integrate functional assays (such as cell motility) with proteomics assays, are also important attributes.
Table 1.
Single-cell functional proteomics tools
| Technique | Numbers and types of poteins assayed | Through-put | Detection limit | Statistical accuracy and signal quantification | Notes and features | Literature |
|---|---|---|---|---|---|---|
| Flow cytometry methods | ||||||
| Fluorescence flow cytometry | Around 15 proteins (mostly membrane proteins, a few cytoplasmic proteins) | 104 cells s-1 | 500 copies per cell | 90% phenotyping accuracy; relative protein abundance | Standard for sorting and enumeration of cellular phenotypes. Secretion blocked and cells fixed for cytoplasmic proteins | [5,6] |
| Mass flow cytometry | Around 35 membrane and intracellular proteins, likely expandable | 103 cells s-1 | >103 copies per cell | Good cell counting statistics; relative protein abundance | Cells handled in bulk prior to analysis. Secretion blocked and cells fixed for cytoplasmic proteins | [8] |
| Surface methods | ||||||
| ELISpot | 1-3 secreted proteins | 6 spots per 105 cells | Quantitative for percentage active cells | Cells secrete proteins onto antibody coated surfaces; secretion activity correlated with cell location | [9] | |
| Microfluidics technologies | ||||||
| Image cytometry | 3-4 membrane or intracellular proteins and cell size | 103-104 cells per chip | 105 fluoro-phores per μm2 | Good cell counting statistics; relative protein abundance | Cells are fixed and stained (in bulk) with fluorescent antibodies; protein assay and cell location spatially correlated | [19,20] |
| Cell array | 1 intracellular protein | <103 cells per chip | Good cell counting statistics; relative protein abundance | Single cells separated and imaged on chip; continuous monitoring of cell physiology | [12-15] | |
| Micro-droplet | 1 membrane or intracellular protein | 102 μdrops s-1 | Not defined | Good cell sampling statistics | Cells entrained in microdroplets; microdroplet composition control permits screening cells | [16-18] |
| Micro-engraving | 3 secreted or 3 membrane proteins | 104-105 cells per chip | Not available | Very good cell number statistics; relative protein abundance | Cells isolated in miocrowells; surface immunoassays; proteins colorimetrically detected; secretome kinetics from single cells; proteomeic and functional assays from same cell | [21-23,37] |
| Single cell barcode chips | About 20 secreted, membrane, or cytoplasmic proteins, expandable | 103-105 cells per chip | 102 copies | Good cell counting statistics, absolute quantification, 10% measurement error per protein per cell | Cells isolated in microchambers, miniature antibody arrays yield spatial separation of specific protein assays; proteomeic and functional assays from same cell; single cells or defined small cell populations accessed. | [4,5,26-30] |
Single-cell functional proteomics tools may be classified into three classes (Table 1). The first class comprises cytometry methods (Figure 1b illustrates flow cytometry), which have evolved over 40 years. The basic idea is to label specific cellular proteins. The cells are then suspended in bulk, and then analyzed, one by one, for the presence of the label. For fluorescence flow cytometry (FFC) (or fluorescence activated cell sorting (FACS)), cellular proteins are labeled with fluorescent antibodies [6,7]. The degree of multiplexing is limited to around 15 by the availability of spectrally distinct fluorophores. The recently developed mass cytometry [8] expands multiplexing to more than 30 by using transition metal mass labels, instead of fluorophores, followed by mass spectrometric analysis of individual cells. For these tools, most assayed proteins are cell surface markers, rather than functional proteins. Intracellular staining (ICS) [9], which requires blocking protein secretion and fixing the cells, can be coupled with cytometry to interrogate for the relative levels of functional proteins such as cytokines or phospho-kinases. Cytometry methods (particularly FFC and FACS) readily handle large numbers of cells, and so can be used to identify (and sort) relatively rare cell types. Cytometry tools capable of a high degree of multiplexing are very powerful, but are also expensive, and usually require a staffed facility for their operation.
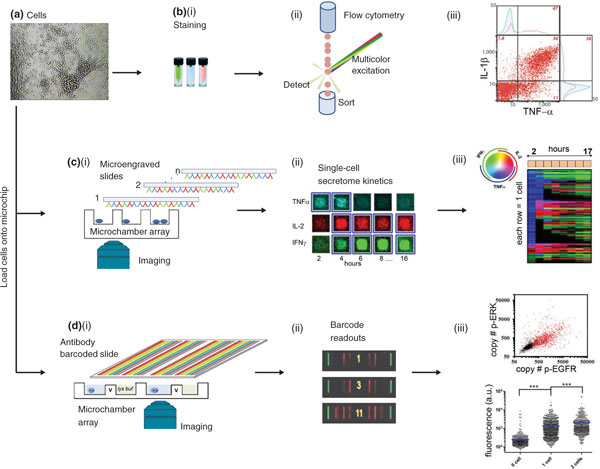
Figure 1.
Selected tools for single-cell functional proteomics. Three technology platforms are illustrated, along with data that highlight the unique strengths of each platform. (a) All platforms start with a single-cell suspension. (b)(i) Intracellular staining (ICS) flow cytometry for assaying for secreted (functional) proteins requires blocking cell secretion during an incubation step, fixing the cells, and then permeabilizing the cells to enable antibody staining. (b)(ii) Proteins are colorimetrically detected by streaming the cells, one at a time, through multicolor laser excitation. (b)(iii) A flow cytometry scatter plot showing the correlation of two effector proteins detected from stimulated CD8+ T cells. This plot reflects the excellent statistics achievable using this technique (adapted from [5] with permission). (c)(i) Microengraving assays start by isolating single cells into microwells, several thousand of which are patterned onto a single chip. A glass substrate that is microengraved with various capture antibodies covers the microwells. The substrate can be replaced at various times to reveal protein-specific secretion kinetics. The phenotype of the cells can also be determined by imaging, using fluorophore-labeled antibodies against specific cell-surface markers. (c)(ii) Secreted protein levels are measured by developing the microengraved slides with fluorophore-labeled, secondary antibodies and correlating the fluorescence signal with the microchamber address. (c)(iii) Assembled traces reveal the secretion kinetics for three proteins from a specific T-cell phenotype. The color coding key is provided in the colored circle at top left. Adapted from [37] with permission. (d)(i) Single-cell barcode chip (SCBC) assays also begin by isolating cells within small-volume microchambers. Flexibility of microfluidics design enables individual cells to be lyzed for analysis of cytoplasmic proteins and membrane and secreted proteins. Proteins are captured on miniature antibody barcode arrays. A full barcode representing the panel of proteins to be assayed is incorporated into each microchamber. (d)(ii) SCBC assays yield data on single cells and on small cell populations. Three developed barcodes are shown; the yellow number indicates the numbers of cells in the associated microchamber. (d)(ii) Statistical analysis of single-cell data collected from model brain cancer cells. Top: scatter plot showing the correlation of two phosphoproteins. The black or red dots represent data from microchambers containing 0 or 1 cells, respectively. Bottom: scatter plots show the statistical uniqueness of the 0-cell, 1-cell, and 2-cell datasets for p-EGFR. a.u., arbitrary units. Adapted from [28] with permission.
Surface methods (Table 1) for single cell functional proteomics include the established and relatively simple and inexpensive ELISpot technique for detecting protein secretion from live cells [9]. Cells are immobilized on an antibody-coated surface, and sandwich-type immunoassays are utilized to detect secreted proteins in the vicinity of individual cells.
Microfluidics technologies constitute the third class of tools. Common advantages of microfluidics tools are that they can often be cheaply manufactured in large quantities, they can handle very small numbers of cells and require only tiny quantities of expensive reagents, and they may often be customized to allow for on chip incubation, cell lysis, and so on. For single cell proteomics, microfluidics platforms fall into two groups - those in which the cells are stained to identify specific proteins, and those for which proteins are released from the cells and measured using surface immunoassays. The first group includes the image cytometry, cell-array, and micro-droplet techniques. Early variations of such tools detected proteins from single cells by imaging stained cells, or by flowing the labeled cells or cell-encapsulation droplets through a microfluidic channel designed to allow fluorescence detection. These were basically microchip versions of FFC or FACS [10]. More recent approaches have significantly diverged to take advantage of some of the unique aspects of microfluidics. For example, cells can be spatially segregated into large arrays (cell arrays [11-15]), or they can be entrained within arrays of drops [16-18]. Such manipulations are followed by immunostaining of membrane proteins, followed by automated imaging to quantify single-cell fluorescence signals. These approaches can offer control over the cell environment before analysis, which make them attractive screening tools. One disadvantage of these and other cell-staining approaches [19,20] is that they have limited multiplexing capacity.
The most advanced microfluidic single-cell proteomics tools use surface-immobilized antibodies for separating protein detection from cell manipulation (Figure 1c,d). This approach has several advantages: it can yield increased multiplexing capacity, it can be extended to assays for secreted, cytoplasmic, and membrane proteins, and measurements of cellular functions can be integrated with the proteomics assays. The experimental challenge is that a given cell may only produce between a few hundred and a few thousand copies of a protein of interest - such numbers are typical for many phosphoproteins or secreted signaling proteins. The solution is to enclose the cell within a microenvironment with a volume of about 1 nl. In this way, the resultant protein concentration can be sufficiently high to allow detection with standard immunoassays. The beauty of microfabrication is that such tiny volume assays can be repeated many times, in parallel, on a single microchip. The microengraving approach developed by Love's group (Figure 1c) [21-23] uses small volume microwells in an array format to isolate and culture single cells. A 'microengraved' (antibody-coated) substrate is used to cap the microwell array and to capture secreted proteins. Proteins are detected using sandwich-type ELISA immunoassays. Different fluorophores colorimetrically distinguish between different detection antibodies to allow the simultaneous detection of about three secreted proteins. The microengraved substrate can be replaced multiple times in situ, thus enabling kinetic studies (Figure 1c(ii,iii)) at the single-cell level. The multiplexing capacity of the microengraving method can be increased using fluorophore-labeled antibody staining of membrane proteins; fluorescence imaging of the captured cells yields information on membrane protein levels (to identify cellular phenotypes), and the microengraved substrate assays for secreted proteins (to assess cellular function).
A related approach is the single-cell barcode chips (SCBCs). The basic concept is to pattern a many-element capture antibody array in each single-cell microwell so that different proteins are detected at different designated array spots. The key enabling technology of SCBCs is the miniature antibody arrays. A related challenge is that antibody arrays are not stable to the physical conditions of microfluidics device fabrication. The solution has been to couple the technique of DNA-encoded antibody libraries (DEAL) [24] with microfluidics-based flow patterning. Specifically, an elastomer film is molded so that it contains a series of long, serpentine channels. It is adhered to the top of a glass slide. Solutions containing a different single-stranded DNA (ssDNA) oligomer are flowed through each channel. Those solutions evaporate. The molded elastomer is then removed, leaving a series of 10 to 20 µm wide stripes of different ssDNA oligomers across the glass substrate. A second elastomer layer, patterned with between 300 and 10,000 microchambers for single-cell assays, is adhered to the glass slide. The design is such that each microchamber contains a full complement of ssDNA stripes. Just before use, these miniature ssDNA arrays are converted into antibody arrays using a cocktail of complementary ssDNA-labeled antibodies. The resultant antibody array (the barcode) [25] provides the detection technology for SCBCs (Figure 1d(ii)) [4,26]. Up to 20 functional proteins have been assayed per cell [5], and the limit is probably around 100. Specific SCBC designs enable cell lysis, thus allowing cytoplasmic, membrane, and secreted proteins to be assayed from the same single cell. SCBC assays can yield absolute protein level quantification [27] and access to discrete cell populations (one cell, two cells, three cells and so on) [28] (Figure 1d(iii)). Both the SCBC and microengraving platforms can be integrated with multicolor FACS to enable the integration of phenotype analysis with functional proteomics [28]. Quantitative data comparison across different SCBC assays [29] allows clinical studies or investigations in which statistical cell behaviors are compared across a perturbation series.
Most microfluidics tools enable the single cells to be imaged. When integrated with proteomics measurements, this can enable several interesting assays, such as correlating cell motility or cell-cell interactions [28,30] with specific protein levels. Unlike flow cytometry analyses, cells can remain in their native morphology so that cell size, spreading, or motility can be correlated with proteomic signature for each cell assayed [30]. The ability of a cytotoxic T cell to kill the target cell can be directly visualized under an optical microscope. Once conducted in a microengraving device, this allows direct comparison of cytolic activity with the protein profile of the same T cell [31]. Finally, cells can be recovered from these types of assays for additional analysis, or for establishing clonal cell lines with desired properties [23]. This direct coupling of many different assays on the same single cells is, so far, a unique attribute of these microfluidics platforms.
Applications to immune monitoring and function
Immune cells are classified along the hematopoietic lineage, starting with myeloid and lymphoid lineages. A triumph of immune system biology has been the identification of cell surface markers that allow, by FACS, the enumeration and sorting of specific immune-cell phenotypes from blood or tissues. For example, a cytotoxic T cell is defined by the cell surface markers Cluster of Differentiation (CD)3, CD45, and CD8, with additional markers specifying the antigen specificity of the T cell receptor (TCR) or providing further phenotypic classification, such as effector memory. However, functional information requires assays of secreted effector proteins (such as cytokines and cytotoxic granules) that mediate the tasks of target killing, self-renewal, recruitment of other immune cell types, and inflammation. Because of the variety of potential pathogen targets, cellular immunity is functionally heterogeneous. Recent studies using different single-cell proteomics platforms have begun to capture and characterize this heterogeneity.
The function of an immune cell is largely delineated by a range of proteins it produces. Early efforts to profile multiple immune effector functions of single immune cell function used ICS FFC. Betts et al. [32] measured five functions (degranulation and levels of interferon (IFN)-γ, macrophage inflammatory protein (MIP)-1b, tumor necrosis factor (TNF)-α, and interleukin (IL)-2) from single HIV-specific CD8+ T cells collected from chronically HIV-infected individuals and people whose HIV infection has not progressed over a long term (called non-progressors or elite controllers). The number of effector functions displayed in T cells from people with chronic HIV was limited relative to those from non-progressors, and the number of functions ('polyfunctionality') was inversely correlated with viral load. Another example of the use of ICS FFC was by Darrah et al. [33], who showed that the degree of protection against Leishmania major infection in mice is predicted by the frequency of CD4+ T cells simultaneously producing IFN-γ, IL-2, and TNF-α. More recent studies have used ICS mass cytometry. For example, Newell and coworkers [34] used this approach to assay 17 membrane protein markers, 6 intracellular cytokines and 2 cytotoxic granules from stimulated CD8+ T cells from healthy patients. They found that the cytokine secretion profiles were almost statistically distributed across the individual cells, but there were distinct niches occupied by virus-specific cells.
Microfluidic functional proteomics has been used for longitudinal monitoring of patients undergoing adoptive cell transfer (ACT) trials, a form of immunotherapy for metastatic melanoma. Ma and coworkers [4] used SCBCs to compare the functional diversity of tumor antigen (MART-1)-specific CD8+ T cells collected from the blood of a melanoma cancer patient with CD8+ T cells collected from healthy donors. At the time of collection, the patient was participating in an ACT trial that used TCR-engineered T cells specific for the MART-1 melanosomal antigen [35]. In this therapy, the TCR-engineered T cells are expanded ex vivo and infused into the patient with the aim that the T cells will drive an anti-tumor immune response. Ma's team assayed a panel of 12 secreted proteins and found a large (albeit not statistically random) range of functional phenotypes within a tightly defined T-cell phenotype [4]. A follow-up kinetic study [5] helped define some of this functional diversity (Figure 2). The authors [5] studied three melanoma cancer patients participating in the same ACT trial and combined 19-plex SCBC functional (secreted) protein assays with 10-color FACS to measure the functional evolution of specific T-cell phenotypes at 5 to 10 time points over a 90-day trial (Figure 2a). These measurements led to several conclusions. First, for a given patient and T-cell phenotype, if all single-cell data from all time points were co-analyzed, a level of functional coordination was resolved, meaning that the T cells could be loosely classified according to biological behaviors, such as anti-tumor or pro-inflammatory. Second, the most polyfunctional cells dominated the immune response (Figure 2b). Roughly 10% of the cells of a given phenotype secreted five or more different proteins. For any one of those proteins, those highly functional cells secreted, on average, 100-fold more protein copies than the less polyfunctional cells. Thus, for a given phenotype, 10% of the cells dominated the overall immune response by 10-fold. This led to the defining of a polyfunctionality strength index (Figure 2c). Interestingly, although the cellular population dynamics or phenotype changes (such as naïve or central memory) over the course of the trial did not yield clear clinical correlates, the polyfunctionality kinetics did correlate with clinical observations, providing feedback for potentially improving the ACT trial design. This collective work over the past decade has refined the notion that the quality of a T-cell immune response is best captured by the functional performance of the T cells, rather than their quantity [36].
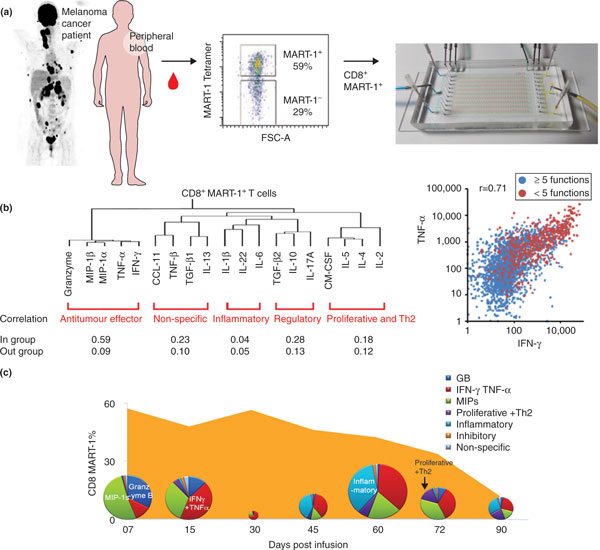
Figure 2.
Integrated FACS/SCBC phenotypic/functional proteomic analysis of tumor-antigen-specific T-cell populations collected from a melanoma cancer patient participating in an ACT trial. (a) Measurement protocol. MART-1 tumor-antigen-specific CD8+ T cells are separated from the blood of the patient using 10-parameter FACS sorting and then loaded onto an SCBC for assaying 19 secreted effector proteins. (b) Analysis of SCBC data. Unsupervised clustering of the single-cell proteomic data (tree, left) reveals coordinated behaviors that reflect specific immune functions. Correlation coefficients, calculated from single cell assays, are provided for proteins within the specified groupings (In group) and outside those groupings (Out group). The scatter plot (right) shows correlations between two anti-tumor effector proteins (IFN-γ and TNF-α) and also shows that the roughly 10% of the cell population that secretes five or more different proteins are also about 100-fold more active for any given protein, and so dominate the immune response for that phenotype. (c) The population kinetics of the TCR-engineered MART-1+ CD8+ T cells, as a percentage of CD3+ T cells (orange solid curve), along with the polyfunctional index (pie chart areas) for tracking population of the MART-1+ CD8+ T cells secreting five or more proteins. The pie chart composition reflects the relative abundances of those proteins. GB refers to the protein Granzyme B. The dynamics of the polyfunctional cells showed much stronger correlations with the observed clinical responses in the patients. Adapted from [5] with permission.
Microfluidics platforms offer the unique capacity for coupling cell adhesion, spreading, and migration assays with multiplex functional proteomics from the same single cells. This is because cells can be incubated and observed within the same microenvironment in which the protein assays are executed. Such assays have relevance for understanding cancer cell behaviors. Cell migration, for example, can be influenced by certain of the cytokines more commonly associated with immune cells. Lu et al. [30] used an SCBC-type antibody array coupled with custom-designed microchip (Figure 3), and identified a few cytokines (IL-6, IL-8, and monocyte chemotactic protein (MCP)-1) that correlated with cell motility.
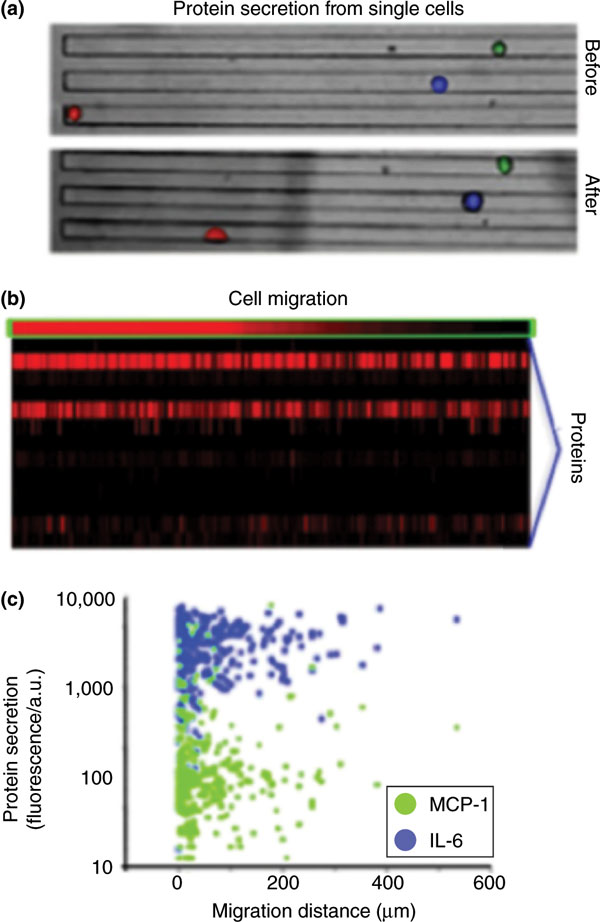
Figure 3.
Multiplexed proteomics for co-measurement of cell migration and cytokine secretion of the same A549 (model lung carcinoma) cancer cells. (a) Light field images showing migration of three single cancer cells within microfluidic channels collected at 0 (before) and 24 (after) hours. (b) Heatmap: each column is a single-cell assay; each row is an assayed parameter. Cell migration distance (top row) is shown with the entire protein secretion profile (lower 14 rows). Approximately 1,000 single cells were assayed. (c) Scatter plots showing how the levels of three proteins (MCP-1 and IL-6) varied with cell migration distance. a.u., arbitrary units. Adapted from [30] with permission.
Love and colleagues used microengraving to carry out two sets of studies that coupled functional behaviors with functional proteomics on single T cells [31,37]. In the first [31], they measured cytolytic activity of CD8+ T cells by performing live-cell imaging of these cells cultured together with single target cells in a microengraving device. This allowed the killing ability of individual T cells to be directly correlated with the production of multiple cytokines, and it revealed a discordance between cytokine secretion and cytolysis. The authors [31] found that the majority of in vivo primed, circulating HIV-specific CD8+ T cells were discordant for cytolysis and secretion of cytokines, notably IFN-γ, when encountering cognate antigen presented on defined numbers of cells. In their second study [37], they investigated the kinetics of cytokine production using serial analyses of single primary human T cells under various conditions (Figure 1c). They showed that for multifunctional T helper 1-skewed cytokine responses (IFN-γ, IL-2, and TNF-α), cells predominantly release those cytokines sequentially, rather than simultaneously. These kinetic trajectories were associated with states of cell differentiation, suggesting that transient programmatic activities of many individual T cells contribute to sustained, population-level responses.
The value of absolute quantification was demonstrated by Shin et al. [38], who used a 12-plex SCBC assay to investigate how the secretome of lipopolysaccharide-stimulated macrophage cells responded to neutralizing antibody perturbations. They reported on the use of statistical-physics-derived models as a means for correctly predicting how specific secreted protein levels would vary with the perturbations. We cover related concepts below in our discussions of phosphoprotein signaling networks.
Applications to intracellular signaling networks
For many cancers, genomic surveys are revealing a rich molecular landscape of altered signal transduction cascades that often cluster along a set of druggable core pathways. In fact, these pathways contain many of the targets of the newer generations of targeted cancer therapies [39]. However, the translation of genomic data into effective clinical treatments has not been straightforward. This is at least partly because non-genetic cell-to-cell variability is profound in drug responses and resistance development, yet it cannot be readily captured from genome sequencing data. A recent editorial [40] has pointed out that capturing the functional protein signaling networks may prove valuable for this purpose, because it is those 'signaling proteins, not the genes per se, that are responsible for the phenotypes of tumors and for the emergence of therapeutic resistance'. Single-cell proteomics provides the most direct approach for elucidating signaling network structure and coordination, and for interrogating how that coordination is disrupted by drugs. It thus may provide a powerful tool for translating genomic information into effective clinical practices for many highly challenging types of cancer [41].
An early single-cell study of phosphoprotein signaling [42] used ICS FFC to assay, in various cancer cells, the cytokine responses of six phosphoproteins, mostly from the signal transducers and activators of transcription (STAT) family. Signaling network heterogeneity and network remodeling was observed in both normal cells in a hematopoietic compartment [43] and cancerous cells such as acute myeloid leukemia [42], suggesting that cells could be classified according to functional phenotype. There have been other highly multiplex studies of phosphoprotein signaling networks using flow (or mass) cytometry [44] or image cytometry [19] over the past decade, and more recent work using SCBC platforms [27,28,45]. Such a sparse literature (especially compared with the routine use of cytometry techniques for cellular phenotyping) highlights the difficulty of these assays, even though the specific studies have illustrated their value. We now turn to discussion of this value, within the specific context of cancer pathways.
Cancer pathway models are essentially maps of the protein-protein interactions that describe the flow from a cell signaling trigger (ligand-receptor binding) to functional behaviors, such as cell division or apoptosis. These pathways are often assembled from diverse datasets (high-throughput data on cell populations, integrated with small interfering RNA perturbations, knockout models, and so on) to yield maps in which the nodes are functional proteins and the edges are inhibitory or activating interactions. These models generally assume linear relationships between upstream effector proteins, ATP, and nutrient levels and activation downstream. However, most signaling cascades behave as excitable devices with thresholds, enabling them to integrate diverse temporal and spatial inputs to produce specific signaling responses [46]. Single-cell proteomics discerns much of this detail, and, if truly quantitative, can yield simplifying approaches towards understanding how such pathways function (Figure 4).
Figure 4.
Phosphoprotein signaling networks from multiplex, quantitative single-cell proteomics. All data represented are uniquely measured at the single-cell level. (a) A Monte-Carlo simulation of fluctuations that represent the copy numbers per cell of an activated (such as phosphorylated) form of a protein, as that protein is involved in increasing numbers of regulatory processes. On the right are the experimentally measured fluctuations of HIF-1α from model GBM cancer cells as these cells are exposed to different O2 partial pressures. The increasingly important role of HIF-1α under hypoxic conditions is evident. Reproduced from [45]. (b) Scatter plot showing protein-protein correlations for two phosphoproteins. The black and red dots represent measurements from 0-cell and 1-cell SCBC microchambers, respectively. Reproduced from [28]. (c) A protein-protein correlation network for model GBM cancer cells following epidermal growth factor (EGF) stimulation (top), and following EGF stimulation + erlotinib (anti-EGF receptor) inhibition (bottom). The weight of the network edges reflects the correlation strength, and a red edge indicates an anti-correlation. Reproduced from [27]. (d) Collective signaling modes, as determined by the eigenvectors of the single-cell protein-protein covariance matrix. Shown are the eigenvectors associated with mTORC1 signaling in model GBM cells, as pO2 is varied. The composition of the green, red, and blue eigenvectors (top plot) is given in the pie charts below for each value of pO2 investigated. The amplitude of the mTORC1 associated eigenvectors shows a minimum between 1.5% and 2% pO2, indicating the loss (and undruggability) of that signaling within this narrow window of pO2 values. Note that HIF-1α is strongly associated with mTORC1 signaling above 2% pO2, but not below 2% pO2, indicating a switch in the structure of the signaling network. The cells studied were model GBM cell lines containing the EDFR variant III (vIII) oncogene (U87 EGFRvIII; panels a, b, d) or the EGRFvIII oncogene plus loss of the phosphatase and tensin homolog (PTEN) tumor suppressor gene (EGFRvIII PTEN). Reproduced from [45] with permission.
Population heterogeneity can arise from factors such as the stochastic nature of intracellular events controlled by low-copy-number transcription factors [47] or through cell-cell interactions [48,49]. The net result is often high-amplitude fluctuations at the single-cell level but stable distributions across a population [50]. The concept of a stable population existing in the presence of random fluctuations is reminiscent of many physical systems that are successfully understood using statistical physics. Thus, tools derived from that field can probably be applied to using fluctuations to determine the nature of signaling networks. This approach contrasts with traditional biology thinking, which might seek to classify the population into functional phenotypes.
Wei and coworkers [45] reported simulations to account for how an increasing signaling activity of a hypothetical protein would be reflected in the fluctuations of the activated state of that protein (Figure 4a). They used a mean field theory, which treated the increasing signaling activity of the hypothetical protein as arising from the statistically averaged (mean field) influences of effector proteins. As the activity increases, the fluctuations shift to higher average copy numbers and are increasingly dispersed. The simulations captured how the experimentally measured fluctuations of hypoxia inducible factor (HIF)-1α in single glioblastoma multiforme (GBM) cancer cells evolve as the cells were exposed to increasingly hypoxic conditions. HIF-1α is, in fact, steadily activated as the cells transition from normoxia to hypoxia [51]. This conclusion can be drawn by simply inspecting the HIF-1α fluctuations.
Quantitative, multiplexed assays can also provide protein-protein correlations. This means that one can use statistical models that explicitly account for protein-protein interactions (Figure 4b,c) and begin defining the state of the signaling network. Shin et al. [38] developed a quantitative Le Chatelier principle that relates how the changes in average signaling protein levels following a weak perturbation to a cell correlate to the changes in the chemical potentials of those proteins. The Le Chatelier principle states that a stable system will respond to a weak perturbation so as to restore that stability. The theory is summarized by the matrix equation . Here, is a column vector with P components representing the average protein levels of the P assayed proteins; β is 1/kBT, where kB is Boltzmann's constant and T is temperature; Σ is a P × P matrix where each element is the experimentally measured covariance of a specific protein Pi with another specific protein Pj; and Δµ is a column vector whose P components describe the change in the chemical potentials of the P proteins, due to a change in external conditions (the perturbation). If the predicted changes in protein levels match experiment, the implication is that the signaling network is described by a stable state and responds to a weak perturbation so as to restore that state. If the calculation does not match experiment, then either the perturbation is strong or the signaling network is not stable. The theoretical tools were coupled with single-cell proteomics assays of mammalian target of rapamycin (mTOR) complex1 (C1) and HIF-1α signaling in model GBM cancer cells, to capture the response of these networks to the transition from normoxia (21% O2 partial pressure (pO2)) to hypoxia (1% pO2) (Figure 4d). mTORC1 signaling was identified as one stable state above 2% pO2 and as a different stable state between 1.5% and 1% pO2, with a switch between those two states near 2 to 1.5% pO2. Within this narrow window of pO2, the models predicted that mTORC1 would be unresponsive to inhibitors, but that it could be drugged at higher or lower pO2. These surprising predictions were found to be correct in both cell lines and tumor models [45].
These results have several implications. First, single-cell proteomics, coupled with approaches derived from statistical physics, can yield detailed (and often surprising) predictions, which can be experimentally validated. Traditional biology experiments on bulk cell cultures or disease models rarely yield such detailed predictions. Furthermore, cellular heterogeneity was not assessed to capture functional phenotypes. Instead the fluctuations were analyzed to identify a stable state or to point to where that state was unstable. Although this general concept is not new, the experimental challenge has been to find approaches that can accurately sample the relevant fluctuations. Related examples have drawn from model systems using, for example, green fluorescent protein reporter genes to provide signatures of protein fluctuations. In one such case [50], time-lapse live-cell microscopy was used to capture specific promoter activity fluctuations in fibroblast cells. The authors [50] identified switching rates between two stable states within the cells. A major advantage of the multiplexed platforms, such as SCBCs, is that fluctuations of broadly sampled signaling networks from primary cells can be measured, allowing predictive applications to non-model systems, with extensions to clinically relevant problems.
Extending such assays to discrete cell populations (unique to microfluidic/nanotechnology platforms) allows the investigation of cell-cell interactions. A few studies have explored the inhibitory or activating nature of such interactions using a combination of protein assays and/or functional observations [52-55]. A recent study [28] correlated the levels of a panel of phospho- (and effector) signaling proteins in model GBM cells, with cell-cell distances in two-cell assays. This indicated that a detailed knowledge of pairwise cell interaction functions could be used to predict specific properties of larger cell populations. Such experiments again draw from concepts derived from statistical physics [56], and may eventually allow complex phenomena within tissue microenvironments to be understood.
Looking forward
The advance of methods for single-cell functional proteomics has been rapid, and the majority of tools discussed here did not exist 5 years ago. These platforms offer unique and emerging opportunities. The coupling of functional and proteomic assays at the single-cell level is one such advantage. Most microfluidic proteomics platforms, however, cannot yet match the statistics and throughput of cytometry tools. However, as these technologies evolve, the range of potential applications will continue to expand, as will the thinking regarding how the resultant datasets can be interpreted. It is likely that, in the near future, microchip platforms will enable as many as 100 proteins to be assayed from single cells, and platforms that enable 10- to 20-plex assays will become routine biological and clinical tools. However, beyond about 100 proteins, all (microchip or cytometry) single-cell proteomics approaches will ultimately be limited by antibodies or other capture agents. Thus, an important underlying challenge is the production of high-performance and robust protein capture agents at low cost. A second outstanding challenge is the development of a capture-agent-independent approach that allows discovery.
One area that has not been covered here is that of mass spectrometry. However, that field has seen remarkable advances over the past few years, and single-cell proteomics may be on the horizon. Targeted proteomics using mass spectrometry has evolved to the extent that small cell numbers, or even single cells, can be analyzed for highly abundant proteins. Protein processing with immobilized enzymes [57] or novel column chromatography methods [58] may eventually allow mass spectrometry to be a single-cell proteomics discovery tool. Finally, the idea that single-cell functional proteomics can provide a conduit to the predictive world of statistical physics is exciting, but the benefits (and limitations) of this type of thinking are largely untapped. It is certain, however, that as measurement quantification, multiplexing capacity, statistical sampling, and sensitivity all improve, so will the power of the models that can use these data to resolve what are otherwise complex biological problems.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Wei Wei, Email: wwei@caltech.edu.
Young Shik Shin, Email: ysshin@caltech.edu.
Chao Ma, Email: machaoga@gmail.com.
Jun Wang, Email: wangj@caltech.edu.
Meltem Elitas, Email: meltem.elitas@yale.edu.
Rong Fan, Email: rong.fan@yale.edu.
James R Heath, Email: heath@caltech.edu.
Acknowledgments
Some of the work reviewed here was supported by the National Cancer Institute (5U54 CA119347 and R01 CA170689-01 to JRH) and the National Institutes of Health (NIH 1 U01 CA164252-01 to RF and U54 CA143798 to RF).
References
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;5:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference? Cell. 2010;5:559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TL. Thermodynamics of Small Systems Parts I and II. New York: Dover; 1994. (Originally published 1963, 1964) [Google Scholar]
- Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu C-C, Kwong GA, Radu CG, Ribas A, Heath JR. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat Med. 2011;5:738–743. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Cheung AF, Chodon T, Koya RC, Wu Z, Ng C, Avramis E, Cochran AJ, Witte ON, Baltimore D, Chmielowski B, Economou JS, Comin-Anduix B, Ribas A, Heath JR. Multifunctional T cell analyses to study response and progression in adoptive cell transfer immunotherapy. Cancer Discov. 2013;5:418–429. doi: 10.1158/2159-8290.CD-12-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzenberg LA, Parks D, Sahaf B, Perez O, Roederer M, Herzenberg LA. The history of the fluorescence activated cell sorter and flow cytometry: a view from Stanford. Clin Chem. 2002;5:1819–1827. [PubMed] [Google Scholar]
- De Rosa SC, Herzenberg LA, Roederer M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med. 2001;5:245–248. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir ED, Krutzik PO, Finck R, Bruggner R V, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe'er D, Tanner SD, Nolan GP. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;5:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoreaux L, Roederer M, Koup R. Intracellular cytokine optimization and standard operating procedure. Nat Protoc. 2006;5:1507–1516. doi: 10.1038/nprot.2006.268. [DOI] [PubMed] [Google Scholar]
- Fu AY, Spence C, Scherer A, Arnold FH, Quake SR. A microfabricated fluorescence-activated cell sorter. Nat Biotechnol. 1999;5:1109–1111. doi: 10.1038/15095. [DOI] [PubMed] [Google Scholar]
- Moodie Z, Price L, Gouttefangeas C, Mander A, Janetzki S, Löwer M, Welters MJP, Ottensmeier C, Van der Burg SH, Britten CM. Response definition criteria for ELISPOT assays revisited. Cancer Immunol Immunother. 2010;5:1489–1501. doi: 10.1007/s00262-010-0875-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo D, Aghdam N, Lee LP. Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays. Anal Chem. 2006;5:4925–4930. doi: 10.1021/ac060541s. [DOI] [PubMed] [Google Scholar]
- Di Carlo D, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip. 2006;5:1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- Faley SL, Copland M, Wlodkowic D, Kolch W, Seale KT, Wikswo JP, Cooper JM. Microfluidic single cell arrays to interrogate signalling dynamics of individual, patient-derived hematopoietic stem cells. Lab Chip. 2009;5:2659–2664. doi: 10.1039/b902083g. [DOI] [PubMed] [Google Scholar]
- Faley SL, Copland M, Reboud J, Cooper JM. Cell chip array for microfluidic proteomics enabling rapid in situ assessment of intracellular protein phosphorylation. Biomicrofluidics. 2011;5:24106. doi: 10.1063/1.3587095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Droplet microfluidic technology for single-cell high-throughput screening. Proc Natl Acad Sci USA. 2009;5:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner A, Srisa-Art M, Holt D, Abell C, Hollfelder F, deMello AJ, Edel JB. Quantitative detection of protein expression in single cells using droplet microfluidics. Chem Comm. 2007;5:1218–1220. doi: 10.1039/b618570c. [DOI] [PubMed] [Google Scholar]
- Martino C, Zagnoni M, Sandison ME, Chanasakulniyom M, Pitt AR, Cooper JM. Intracellular protein determination using droplet-based immunoassays. Anal Chem. 2011;5:5361–5368. doi: 10.1021/ac200876q. [DOI] [PubMed] [Google Scholar]
- Sun J, Masterman-Smith MD, Graham NA, Jiao J, Mottahedeh J, Laks DR, Ohashi M, DeJesus J, Kamei K, Lee K-B, Wang H, Yu ZTF, Lu Y-T, Hou S, Li K, Liu M, Zhang N, Wang S, Angenieux B, Panosyan E, Samuels ER, Park J, Williams D, Konkankit V, Nathanson D, Van Dam RM, Phelps ME, Wu H, Liau LM, Mischel PS. et al. A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Canc Res. 2010;5:6128–6138. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wu Y, Lee S-K, Fan R. High-content single-cell analysis on-chip using a laser microarray scanner. Lab Chip. 2012;5:5025–5033. doi: 10.1039/c2lc40309a. [DOI] [PubMed] [Google Scholar]
- Love JC, Ronan JL, Grotenbreg GM, Van der Veen AG, Ploegh HL. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat Biotechnol. 2006;5:703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC. Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving. Lab Chip. 2010;5:1391–1400. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan N, Kwon DS, Law KM, Ogunniyi AO, Anahtar MN, Richter JM, Walker BD, Love JC. Rapid, efficient functional characterization and recovery of HIV-specific human CD8+ T cells using microengraving. Proc Nat Acad Sci USA. 2012;5:3885–3890. doi: 10.1073/pnas.1111205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. DNA-encoded antibody libraries: a unified platform for multiplexed cell sorting and detection of genes and proteins. J Am Chem Soc. 2007;5:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R, Vermesh O, Srivastava A, Yen BKH, Qin L, Ahmad H, Kwong GA, Liu C-C, Gould J, Hood L, Heath JR. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nat Biotechnol. 2008;5:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YS, Ahmad H, Shi Q, Kim H, Pascal TA, Fan R, Goddard WA, Heath JR. Chemistries for patterning robust DNA microbarcodes enable multiplex assays of cytoplasm proteins from single cancer cells. ChemPhysChem. 2010;5:3063–3069. doi: 10.1002/cphc.201000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Qin L, Wei W, Geng F, Fan R, Shin YS, Guo D, Hood L, Mischel PS, Heath JR. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc Nat Acad Sci USA. 2012;5:419–424. doi: 10.1073/pnas.1110865109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tham D, Wei W, Shin YS, Ma C, Ahmad H, Shi Q, Yu J, Levine RD, Heath JR. Quantitating cell-cell interaction functions with applications to glioblastoma multiforme cancer cells. Nano Lett. 2012;5:6101–6106. doi: 10.1021/nl302748q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad H, Sutherland A, Shin YS, Hwang K, Qin L, Krom R-J, Heath JR. A robotics platform for automated batch fabrication of high density, microfluidics-based DNA microarrays, with applications to single cell, multiplex assays of secreted proteins. Rev Sci Inst. 2011;5:094301. doi: 10.1063/1.3636077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Chen JJ, Mu L, Xue Q, Wu Y, Wu P-H, Li J, Vortmeyer AO, Miller-Jensen K, Wirtz D, Fan R. High-throughput secretomic analysis of single cells to assess functional cellular heterogeneity. Anal Chem. 2013;5:2548–2556. doi: 10.1021/ac400082e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan N, Julg B, Yamanaka YJ, Chen H, Ogunniyi AO, McAndrew E, Porter LC, Piechocka-Trocha A, Hill BJ, Douek DC, Pereyra F, Walker BD, Love JC. A high-throughput single-cell analysis of human CD8+ T cell functions reveals discordance for cytokine secretion and cytolysis. J Clin Invest. 2011;5:4322–4331. doi: 10.1172/JCI58653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;5:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah PH, Patel DT, De Luca PM, Lindsay RW, Flynn BJ, Hoff ST, Anderson P, Reed SG, Morris SL, Roederer M, Seder RA. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;5:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- Newell EW, Sigal N, Bendall Sean C, Nolan Garry P, Davis Mark M. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;5:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, Zheng Z, Nahvi A, de Vries CR, Rogers-Freezer LJ, Mavroukakis SA, Rosenberg SA. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;5:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;5:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- Han Q, Bagheri N, Bradshaw EM, Hafler DA, Lauffenburger DA, Love JC. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc Nat Acad Sci USA. 2012;5:1607–1612. doi: 10.1073/pnas.1117194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YS, Remacle F, Fan R, Hwang K, Wei W, Ahmad H, Levine RD, Heath JR. Protein signaling networks from single cell fluctuations and information theory profiling. Biophys J. 2011;5:2378–2386. doi: 10.1016/j.bpj.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue JS, Morrison DK. Complexity in the signaling network: insights from the use of targeted inhibitors in cancer therapy. Genes Dev. 2012;5:641–650. doi: 10.1101/gad.186965.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB. The scientific drunk and the lamppost: massive sequencing efforts in cancer discovery and treatment. Sci Signaling. 2013;5:1–3. doi: 10.1126/scisignal.2003684. [DOI] [PubMed] [Google Scholar]
- Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Canc. 2012;5:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud Ø, Gjertsen BT, Nolan GP. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;5:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Gibbs KD Jr, Gilbert PM, Sachs K, Zhao F, Blau HM, Weissman IL, Nolan GP, Majeti R. Single-cell phospho-specific flow cytometric analysis demonstrates biochemical and functional heterogeneity in human hematopoietic stem and progenitor compartments. Blood. 2011;5:4226–4233. doi: 10.1182/blood-2010-07-298232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;5:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- Wei W, Remacle F, Qin L, Shackelford D, Shin YS, Mischel PS, Levine RD, Heath JR. Hypoxia induces a phase transition within a kinase signaling network in cancer cells. Proc Natl Acad Sci USA. 2013;5:E1352–E1360. doi: 10.1073/pnas.1303060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko B, Yaffe MB, Kolch W. Computational approaches for analyzing information flow in biological networks. Sci Signal. 2012;5:1–14. doi: 10.1126/scisignal.2002961. [DOI] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;5:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek P, Ryan S, Ashall L, Sillitoe K, Harber CV, Spiller DG, Rand DA, White MRH. Population robustness arising from cellular heterogeneity. Proc Nat Acad Sci USA. 2010;5:11644–11649. doi: 10.1073/pnas.0913798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TL, Wulf G, Burga L, Cantley LC. Cell-to-cell variability in PI3K protein level regulates PI3K-AKT pathway activity in cell populations. Curr Biol. 2011;5:173–183. doi: 10.1016/j.cub.2010.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisan DR, Halter M, Hubbard JB, Plant AL. Predicting rates of cell state change caused by stochastic fluctuations using a data-driven landscape model. Proc Natl Acad Sci USA. 2012;5:19262–7. doi: 10.1073/pnas.1207544109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Can. 2008;5:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Chen CS. Cell-cell signaling by direct contact increases cell proliferation via a PI3K-dependent signal. FEBS Lett. 2002;5:238–242. doi: 10.1016/s0014-5793(02)02370-0. [DOI] [PubMed] [Google Scholar]
- Liu WF, Nelson CM, Pirone DM, Chen CS. E-cadherin engagement stimulates proliferation via RAC 1. J Cell Biol. 2006;5:431–441. doi: 10.1083/jcb.200510087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui EE, Bhatia SN. Micromechanical control of cell-cell interactions. Proc Nat Acad Sci USA. 2007;5:5722–5726. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puliafito A, Hufnagel L, Neveu P, Streichan S, Sigal A, Fygenson DK, Shraiman BI. Collective and single cell behavior in epithelial contact inhibition. Proc Nat Acad Sci USA. 2011;5:739–744. doi: 10.1073/pnas.1007809109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sear RP, Chung S-W, Markovich G, Gelbart WM, Heath JR. Spontaneous patterning of quantum dots at the air-water interface. Phys Rev E. 1999;5:R6255–R6258. doi: 10.1103/PhysRevE.59.R6255. [DOI] [PubMed] [Google Scholar]
- Tian R, Wang S, Elisma F, Li L, Zhou H, Wang L, Figeys D. Rare cell proteomic reactor applied to stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics study of human embryonic stem cell differentiation. Mol Cell Proteomics. 2011;5:M110.000679. doi: 10.1074/mcp.M110.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur D, Rejtar T, Wang D, Bones J, Cha S, Clodfelder-Miller B, Richardson E, Binns S, Dahiya S, Sgroi D, Karger BL. Microproteomic analysis of 10,000 laser captured microdissected breast tumor cells using short-range sodium dodecyl sulfate-polyacrylamide gel electrophoresis and porous layer open tubular liquid chromatography tandem mass spectrometry. J Chromatogr A. 2011;5:8168–8174. doi: 10.1016/j.chroma.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]