Abstract
The occurrence of clonal perturbations and leukemia in patients transplanted with gamma-retroviral (RV) vector–transduced autologous hematopoietic stem and progenitor cells (HSPCs) has stimulated extensive investigation, demonstrating that proviral insertions may perturb adjacent proto-oncogene expression. Although enhancer-deleted lentiviruses are less likely to result in insertional oncogenesis, there is evidence that they may perturb transcript splicing, and one patient with a benign clonal expansion of lentivirally transduced HPSC has been reported. The rhesus macaque model provides an opportunity for informative long-term analysis to ask whether transduction impacts on long-term HSPC properties. We used two techniques to examine whether lentivirally transduced HSPCs from eight rhesus macaques transplanted 1–13.5 years previously are perturbed at a population level, comparing telomere length as a measure of replicative history and gene expression profile of vector positive versus vector negative cells. There were no differences in telomere lengths between sorted GFP+ and GFP− blood cells, suggesting that lentiviral (LV) transduction did not globally disrupt replicative patterns. Bone marrow GFP+ and GFP− CD34+ cells showed no differences in gene expression using unsupervised and principal component analysis. These studies did not uncover any global long-term perturbation of proliferation, differentiation, or other important functional parameters of transduced HSPCs in the rhesus macaque model.
Introduction
The two decades since the first patient was transplanted with genetically modified hematopoietic stem and progenitor cells (HSPCs) have progressed from initial expectations of success, through disappointment over insufficient gene transfer efficiency, to elation when improvements in vectors and transduction conditions resulted in clear clinical evidence for amelioration of serious genetic disorders, followed by major concerns with the realization that insertional activation of proto-oncogenes resulted in clonal expansion and leukemia in some patients.1,2,3 Intensive investigation of the integration patterns for standard gamma-retroviral (RV) vectors and the degree of risk related to proviral insertional mutagenesis when targeting HSPCs has resulted in a reassessment of the risk profile for these vectors, more regulatory constraints, and a search for safer vectors to use for further clinical gene therapy applications targeting HSPCs.4,5,6,7,8
Lentiviral (LV) vectors derived from the human immunodeficiency virus (HIV) or the simian immunodeficiency virus have been explored as alternatives. LV vectors have an integration pattern distinct from that of RV vectors, with a lower propensity to integrate near transcription start sites, and are thus predicted to have a lower risk of insertional gene activation.6,9,10,11 LV vectors also have the strong viral enhancer in the long-terminal repeat (LTR) deleted to avoid recombination with endogenous HIV, and to decrease the risk of enhancer-mediated adjacent proto-oncogene activation. LV vectors were significantly less likely than RV vectors to transform HSPCs, as assessed by in vitro immortalization of murine bone marrow or leukemia induction in tumor-prone mice.12,13 In rhesus macaques and other nonhuman primates, LV vector transduction of HSPCs resulted in long-term polyclonal vector–derived hematopoiesis, without dominance of clones containing insertions near proto-oncogenes, in contrast to results with RV vectors.6,10,14,15 However, there remains a finite risk of insertional mutagenesis associated with any integrating vector, given the thousands of genes being disrupted or potentially impacted by vector insertions, as revealed by murine and cell immortalization assays.12,13 Some overrepresentation of LV insertion sites near proto-oncogenes occurs in transduced HSPCs is expected, because proviral insertions in open areas of chromatin, within or near genes highly expressed in HSPCs, including proto-oncogenes, are favored.9,14
Despite the years of experimentation and optimization of RV vectors in the laboratory, and >10 years of clinical experience, it was a surprise when some patients in X-SCID gene therapy trial developed leukemia following RV gene therapy. It is important therefore to maximally and creatively use all available experimental models and early clinical trial data to ask whether LV gene therapy targeting HSPCs is associated with any significant oncogenic risk. To date, only a few patients have received HSPCs transduced with LV vectors, with limited follow-up. Two patients with adrenoleukodystrophy have received CD34+ cells transduced with LV vectors, and have been reported to maintain highly polyclonal LV-containing hematopoiesis for up to 2.5 years.16 A single patient treated with LV-transduced CD34+ cells for thalassemia has developed a highly dominant clone with aberrant splicing and overexpression of a gene linked to myeloproliferation in a murine model,17,18 and a recent in vitro study suggested that integrated LV vectors do have a propensity to disrupt transcript splicing.19
An alternative approach to investigating the impact of LV transduction on HSPC behavior is to compare fundamental properties of cell proliferation and gene expression in engrafted transduced versus nontransduced HSPCs long term. We have focused on the rhesus macaque as the best available model for predicting HSPC behavior in humans.20,21 Telomere shortening in HSPCs reflects the number of prior cell divisions, and can be used as a “replicative clock,”22,23 uncovering differences in replicative history between LV-transduced and nontransduced cells, in rhesus macaques transplanted years previously. Global gene expression profiling of rhesus CD34+ cells engrafted in vivo long term following transplantation, comparing LV-transduced and nontransduced cells, may uncover HSPC perturbations resulting from LV transduction. These two complementary approaches can provide insights into the impact of LV transduction on HSPCs, and inform risk/benefit assessments of future human clinical gene therapy trials.
Results
Subject selection
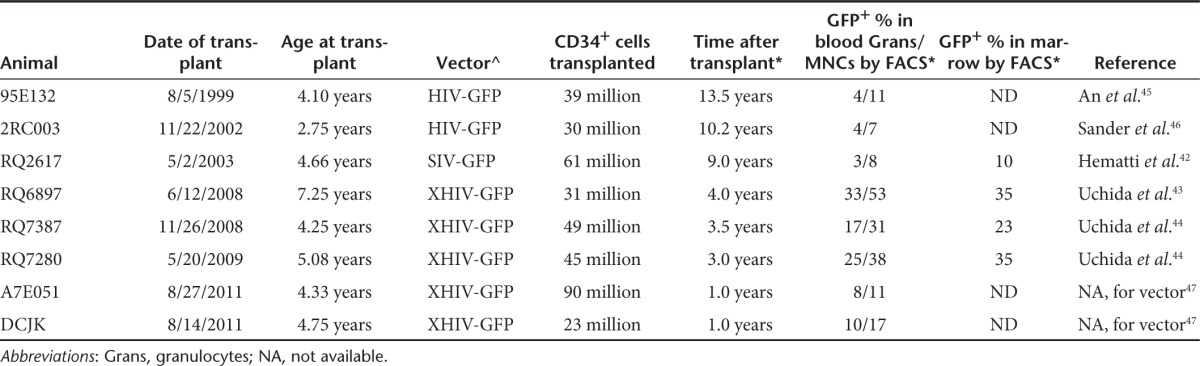
A total of eight rhesus macaques were studied. All were transplanted with LV-transduced autologous CD34+ cells a median of 3.75 years (range 1–13.5) before the collection of samples for the current study. Table 1 summarizes the characteristics of the animals, including the specific vectors used, cell doses transplanted, age at time of transplant, length of follow-up before sample collection for the current study, and the level of marking at the time of sample collection. All vectors were third generation and consisted of modified HIV or simian immunodeficiency virus backbones, an internal viral promoter, and a green fluorescent protein (GFP) transgene unlikely to impact on cellular behavior. The transduction conditions, vector characteristics, and follow-up on six of the eight animals have already been published, as listed in Table 1. Several of the animals have been followed in detail regarding the pattern of vector integration sites, and all remain highly polyclonal, without evidence for any development of progressively more dominant clones suggesting insertional mutagenesis.6,14,24
Table 1. Rhesus macaque transplantation and transduction characteristics.
Telomere length of GFP+ and GFP− peripheral blood cells
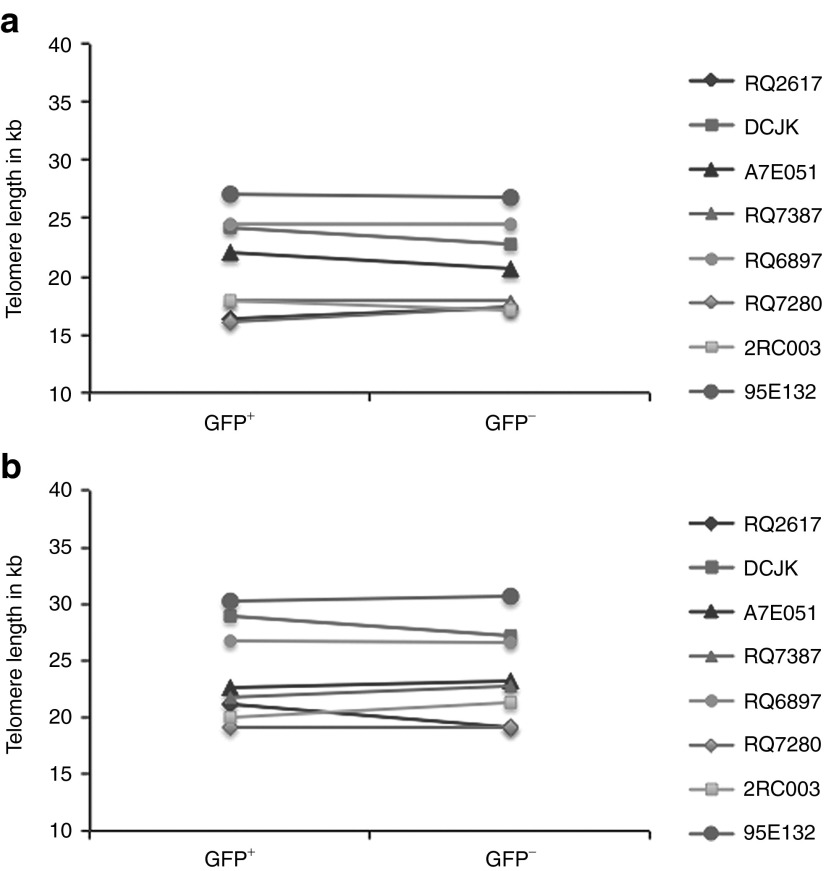
The mean telomere length was measured in sorted GFP+ and GFP− peripheral blood (PB) granulocytes and mononuclear cells (primarily lymphocytes) at the longest possible time point following transplantation of autologous lentivirally transduced CD34+ cells, following ablative total body irradiation. There were no significant differences in telomere length in the GFP+, vector-containing granulocytes or lymphocytes compared with paired GFP− samples from the same time point and cell lineage (Figure 1).
Figure 1.
Comparison of telomere lengths in transduced and untransduced blood cells. Quantitative PCR for mean telomere length was performed on genomic DNA from peripheral blood (a) GFP+ or GFP− granulocytes and (b) GFP+ or GFP− lymphocytes from eight rhesus macaques transplanted with lentivirally transduced autologous CD34+ cells 1–13.5 years before. The panels show the telomere length in kilobases (kb) on the y axis for GFP+ and GFP− (a) granulocytes or (b) lymphocytes from each animal. The time of sample collection after transplantation for each animal is given in Table 1. Each sample was run in triplicate, and the mean values are shown as individual points on the plots. There was no significant change in telomere length, calculated as the mean of the triplicate averages for changes in telomere length, between the GFP+ and GFP− cells in either the granulocyte (P = 0.1336) or lymphocyte (P = 0.5183) populations, via a paired Student's t-test.
Although we do not have an equivalent set of sorted transduced versus untransduced samples to analyze from rhesus macaques transplanted with RV-transduced HSPCs, measurement of the telomere lengths in available samples from one animal that progressed to an overt clonal RV-vector–related myeloid tumor is potentially instructive. Rhesus macaque 95E113 had a telomere length of 22.2 kb in total granulocytes 3 years following transplantation with RV-transduced autologous CD34+ cells, at a time that ~3–5% of granulocytes were vector positive. Two years later, the animal developed a vector-related myeloid sarcoma in the abdomen characterized by insertional activation of the antiapoptotic gene BCL2A1, and also marked clonal dominance of granulocytes from the same clone in the PB.25 At that time point, the telomere length in both the blood granulocytes and the myeloid sarcoma tumor had dropped precipitously, to 7.9 and 9.0 kb respectively.
Gene expression profile of GFP+ and GFP− bone marrow CD34+ cells
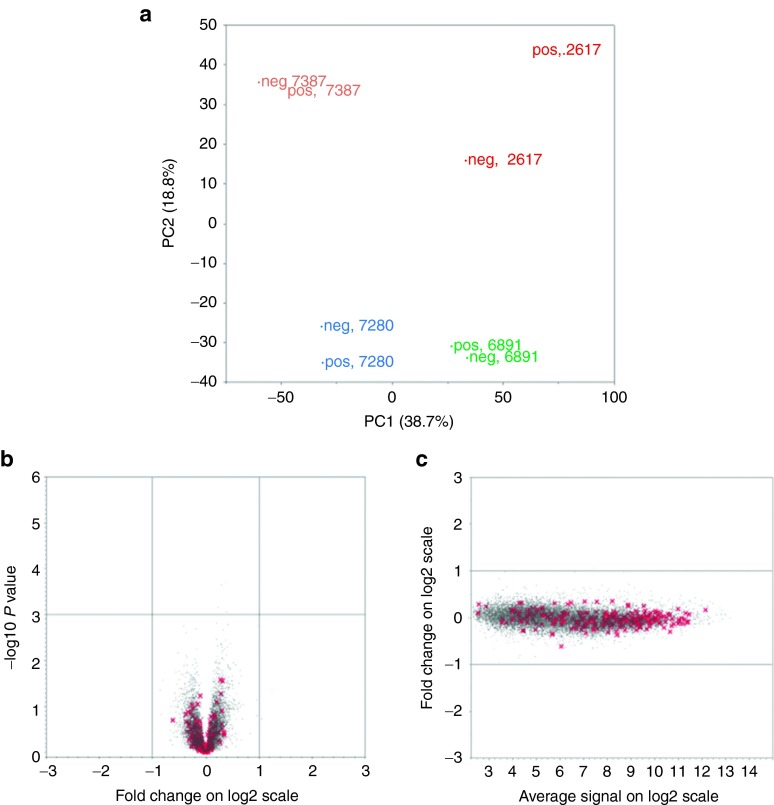
In four animals, we were able to obtain sufficient bone marrow to allow sorting of GFP+ and GFP− CD34+ cells (Table 1), collected a mean of 3.75 years after transplantation. Total RNA was extracted from each cell population, and used for gene expression profiling via rhesus Affymetrix arrays. Principle component analysis was used to group samples, and as shown in Figure 2a, no grouping of GFP+ versus GFP− samples occurred. Instead, the variation was much greater between both GFP+ and GFP− samples from different animals than between the GFP+ and GFP− samples from the same animal. A volcano plot (Figure 2b) does not show any genes with significant differences in expression between GFP+ and GFP− samples, including published Sanger cancer genes,26 as marked on the plot. An MvA plot (Figure 2c) shows that the Sanger cancer genes have quite high expression in CD34+ cells, but none are significantly differentially expressed between GFP+ and GFP− samples. The slightly greater distance between the GFP+/GFP− samples for animal RQ2617 in the principle component analysis was due to minor technical variations rather than true biologic differences. No Sanger Cancer genes were greater than twofold altered in this animal, and none of the 138 genes that were differentially expressed in GFP+ versus GFP− CD34+ cells in this animal were differentially expressed in the other three animals.
Figure 2.
Comparison of gene expression profiles between transduced and untransduced bone marrow CD34+ cells. (a) Principal component analysis of global gene expression patterns in GFP+ (pos) and GFP− (neg) CD34+ cells sorted from the bone marrow of four rhesus macaques transplanted with lentivirally transduced autologous CD34+ cells 3–9 years before. The principal component analysis shows that the variation between the four animals was much greater than the variation between GFP+ and GFP− CD34+ cells overall. The first component (PC1) on the x axis and the second component (PC2) on the y axis explain a cumulative 57.5% of the variance between the four animals. (b) A Volcano plot of log2 fold changes versus one-way analysis of variance P value (-log10 scale) comparing the expression of each individual gene (red x marks Sanger cancer genes, gray dots mark all other genes) in the GFP+ and GFP− cells from all four animals. No genes were up- or downregulated more than twofold (log2 = 1) between the cell populations, as demonstrated by lack of any dots outside the -1 and 1 thresholds on the x axis, and there was no significant difference in level of expression for any genes comparing GFP+ and GFP− cells. (c) MvA plot of the average log2 intensities of expression for individual genes on the x axis versus the log2 fold change in GFP(+) versus GFP(−) cells on the y axis. Each dot represents one transcript on the plot, red x's for Sanger cancer genes and gray dots for other genes. The majority of Sanger cancer genes show high expression level in both GFP(+) and GFP(−) two conditions.
Discussion
In this study, we have used telomere length primarily as a “mitotic clock” to ask whether the in vivo behavior of primitive nonhuman primate HSPCs are altered by transduction with LV vectors.21,27 A direct relationship between hematopoietic stem cell cycling and telomere shortening has been demonstrated via serial transplantation in mice.28 Laboratory mice start with much longer telomeres as compared with other mammals, and humans have somewhat shorter telomeres than macaques; however, the degree of shortening with each somatic cell division is similar in all species. In rhesus macaques followed for as long as 13 years following an ablative autologous PB HSPC transplant with LV vector–transduced CD34+ cells, there were no significant or consistent differences in telomere lengths between circulating granulocytes and lymphocytes containing vector, and thus derived from LV-transduced CD34+ cells, and hematopoietic cells without vector, likely derived from CD34+ cells in the graft that remained nontransduced, given the high dose total body irradiation used for pretransplant conditioning. We have previously documented that LV vectors are not silenced in macaque HPSCs or their progeny long term, validating use of GFP+ versus GFP− cell sorting as a methodology to separate transduced versus nontransduced cells.14 A number of studies have confirmed that telomere length in all blood lineages are closely correlated, and that mature PB telomere length corresponds closely with CD34+ HSPC telomere length.29,30 Our results suggest that LV-transduced macaque HSPCs engraft and cycle equivalently to nontransduced HSPCs.
Rhesus macaque telomeres shorten ~100–120 bps per cell division, as assessed in primary cells cultured in vitro.31,32,33,34 More relevant are estimates from population studies of telomere lengths in normal hematopoietic cells from monkeys across age cohorts. Rhesus and baboon adult hematopoietic stem cells have been estimated to cycle every 23–52 weeks, with a loss of 100–150 bp at the telomeres per year in adult animals.21,33 Our qPCR assay reliably detects differences of 500–1,000 bp or greater in average telomere length within a population. Thus, in steady state, a global increasing in cycling of LV-transduced HSPCs of threefold would be detectable only after 3–4 years. However, a number of studies have demonstrated that very rapid telomere shortening occurs in the first year following autologous or allogeneic hematopoietic stem cell transplantation, due to rapid early sustained proliferation of the HSPCs to fill an empty compartment and regenerate sufficient HSPCs able to support robust hematopoiesis long term.35,36 Hematopoietic cells from human recipients of allogeneic HPSCs have telomeres that are 0.4–1.0 kb shorter than telomeres in donor hematopoietic cells before transplant, equivalent to an extra 10–15 years worth of telomere shortening. Assuming a similar magnitude HSPC expansion in rhesus recipients following ablative autologous transplantation, corresponding to 10 years of steady-state telomere shortening, even a twofold global increase in cycling of LV-transduced cells would be detectable even as early as 1 year after transplant. It would be of interest to examine telomere length in the patients transplanted with LV-transduced HSPCs to date, although neither the thalassemia nor the adrenoleukodystrophy trial used LV containing a cell surface or other marker allowing separation of vector-containing versus untransduced cells. The patient with thalassemia who recovered hematopoiesis and engrafted very slowly following busulfan conditioning would be predicted to have acceleration of telomere shortening, particularly in the single dominant clone, in contrast to the highly polyclonal pattern seen in our macaques and in the adrenoleukodystrophy clinical trial.16,17
Our prior extensive experience using gamma-RV vectors in the rhesus macaque autologous transplantation model did not incorporate a cell surface or fluorescent selectable marker allowing separation of transduced versus nontransduced cells, to ask whether with this more genotoxic vector system a difference in proliferative history manifested by telomere shortening could be detected. However, the limited data we do have on animal 93E113, which progressed to clonal dominance and full malignant transformation of a subclone, demonstrates remarkable telomere shortening (equivalent to 30 years of steady-state hematopoiesis) in the clonal granulocytes and the tumor itself compared with 2 years previously, looking at overall hematopoiesis when the marking level overall was only 5%. This result suggests that accelerated proliferation and telomere shortening is at least a marker of, and potentially contributes to, malignant transformation, as we have previously reported in patients with bone marrow failure and excessive proliferative demand who progress to myeloid malignancies.37 It is also possible that cells with accelerated proliferation could shorten telomeres to the extent that the clone exhausts and is lost, but this possibility seems unlikely from our data to date.
Gene expression profiling of bone marrow CD34+ cells from a subset of animals revealed no significant differences in gene expression of any single gene or class of genes between the entire populations of LV-transduced and nontransduced cells. These results are reassuring and suggest that LV vector integration does not routinely result in upregulation of proto-oncogenes or other genes with impact on growth and development in the population of transduced cells as a whole. These results are not compatible with a hypothesis that only transduced cells with integration events impacting on cell proliferation or survival engraft efficiently. There was no upregulation of the genes involved in telomere elongation, indicating the validity of conclusions from comparisons of telomere lengths in transduced and untransduced cells. Taken together, these findings suggest that LV vectors transduce CD34+ cells without changing their phenotype, and that these vectors do not target only a restricted CD34+ subpopulation at a particular stage of differentiation. Lack of 100% efficiency of transduction may reflect transient properties of the target cells during culture and transduction, for instance cell cycle status or spatial constraints. Only one prior study has analyzed gene expression in transduced versus untransduced cells engrafted long term. Patients received T cells transduced with a gamma-RV vector expressing a suicide gene and a cell surface marker, and had transduced and untransduced T cells sorted from the blood 4–6 months following infusion. Approximately 2–3% of genes was significantly up- or downregulated. None of these genes were ones that had been previously identified as close to a vector insertion site in the infused T cells, but it is also plausible that not all vector sites had been identified.38 It is reassuring that in contrast we did not detect significantly dysregulated genes, at least at a population level, comparing transduced and untransduced CD34+ cells engrafted in vivo long term.
In conclusion, our study used a nonhuman primate model with high relevance to human clinical gene therapy applications to provide evidence that LV vector transduction of HSPCs is not associated with perturbations of cell cycle/proliferation or gene expression, at least on a global level. These approaches are not designed or able to detect minor subclones that might arise due to insertional mutagenesis, because telomere length and gene expression are being assessed at a mean population level in transduced versus untransduced cells, for instance a tenfold increase in cell cycling or expression of a specific gene or pathway in 10% of the cells would not impact on the mean values. We are instead searching for any final common pathways that are shared by HSPCs able to be transduced or resulting from transduction. The final common EVI1 pathways noted in the gamma-RV clinical trials and experimental transformation studies would have been interesting to examine using these approaches.39 This information is complementary to insertion site profiling of transduced hematopoietic cells in nonhuman primate, canine, and limited human clinical trials suggesting that LV transduction of HSPCs is associated with a lower risk of genotoxicity and perturbation of hematopoietic stem and progenitor cell homeostasis than gamma-RV vectors.
Materials and Methods
Rhesus CD34+ cell collection, transduction and transplantation. Rhesus macaques (Macaca mulatta) were housed and handled in accordance with guidelines set by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council, and all animal protocols were approved by the Animal Care and Use Committee of the National Heart, Lung, and Blood Institute. The methodology used for rhesus macaque CD34+ cell collection, transduction, conditioning with total body irradiation, transplantation, and posttransplantation care has been published.40 Details including the vector used for transduction, transduction conditions, and initial gene marking results following transplantation have been published for animals 93E113,25,41 RQ2617,42 RQ6897,43 RQ7387 and RQ7280,44 95E132,45 and 2RC003.46 Details for animals DCJK and A7E051 have not previously been published. These two animals were transplanted with autologous CD34+ cells collected from the PB by apheresis following mobilization with recombinant human G-CSF and recombinant human stem cell factor (Amgen, Thousand Oaks, CA) as described.40 Following transduction with LeGO LV vectors expressing GFP or Tomato fluorescent proteins47 under normoxic or low oxygen (2%) conditions, the autologous cells were reinfused into the animals following 1,000 rads total body irradiation. Relevant information for each animal is summarized in Table 1.
Cell collection and processing. For telomere length analysis, 40 ml of rhesus macaque PB was collected in acid citrate dextrose tubes. The mononuclear cells and granulocytes were separated via density gradient centrifugation over lymphocyte separation medium (MP Biomedicals, Solon, OH). The cells were then sorted via forward and side scatter and GFP expression into four populations on a FACS AriaII instrument (BD Biosciences, San Jose, CA): GFP+ and GFP− mononuclear cells and GFP+ and GFP− granulocytes. DNA extraction on sorted populations was carried out using the Maxwell 16 LEV Blood DNA Kit (Promega, Madison, WI).
For gene expression analysis, 30 ml of bone marrow was aspirated from the posterior iliac crests of rhesus macaques into acid citrate dextrose tubes. The mononuclear cells were isolated by density gradient centrifugation over lymphocyte separation medium (MP Biomedicals). Enrichment for CD34+ cells was performed using the Miltenyi Biotec MACS cell separation system (Miltenyi Biotec, Auburn, CA) and the biotynylated 12.8 anti-CD34 antibody as described.40 The cells were then sorted for CD34+GFP+ and CD34+GFP− populations on a FACS AriaII instrument (BD Biosciences). RNA extraction on sorted populations was carried out using the Qiagen RNeasy Micro Kit (Qiagen, Valencia, CA).
Telomere length analysis. Mean telomere length was measured in PB neutrophils and mononuclear cells by a validated and CLIA-certified quantitative PCR (qPCR), as originally described.48,49 A modification of this method using the Qiagen Rotor-Gene Q real-time cycler was developed by our group and a detailed description can be accessed on the Qiagen website at http://b2b.qiagen.com/literature/qiagennews/weeklyarticle/11_04/telomere/default.aspx. Experimental samples of 1.6 ng genomic DNA were run in triplicate using a Rotor-Gene Q real-time instrument with the Rotor-Gene SYBR Green Kit (Qiagen). Primers for telomeric repeats (FP 5′-GGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′ and RP 5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3′) and human 60S acidic ribosomal protein P0 as single gene control (FP 5′-CCCATTCTATCATCAACGGGTACAA-3′ and RP 5′-CAGCAAGTGGGAAGGTGTAATCC-3′) were used. These sequences are identical in the rhesus genome. The telomere length for each sample is based on the telomere to single copy gene ratio (T/S ratio) calculated as deltaCt [Ct(telomere)/Ct(single gene)]. Telomere length for each sample was calculated from the T/S ratio for each sample normalized to the mean T/S ratio of a reference sample, which was used for the standard curve, using [2−(ΔCtx − ΔCtr) = 2−ΔΔCt] equation. The reference sample was used both as a reference sample, and between runs as a validation sample, to ensure reproducibility between individual runs of the assay.
Statistical analysis comparing mean telomere length in GFP+ versus GFP− samples was done using a paired Student's t-test.
Gene expression analysis. Biotin Labeled sense targets were prepared with total RNA using the Affymetrix Whole-Transcript Sense Target Labeling Protocol without rRNA reduction following the manufacturer's directions (Affymetrix, Santa Clara, CA). Briefly, 50 ng of total RNA was used for synthesizing double-stranded cDNA with random hexamers coupled with a T7 promoter sequence. The cDNA was then used as a template for in vitro transcription amplification with T7 RNA Polymerase, producing multiple copies of cRNAs. Sense Strand cDNA was then prepared from purified antisense cRNA using random hexamers. ST-cDNA of 10 µg was fragmented and biotin labeled with terminal deoxynucleotidyl transferase and hybridized to Affymetrix Rhesus Gene 1.0 ST microarrays (Affymetrix). Hybridization was performed at 45 °C overnight, followed by washing and staining using a FS450 fluidics station. Hybridization, washing, and laser scanning of microarrays on a 7G GCS3000 scanner were performed according to the manufacturer's protocol.
Gene-level intensity values for each of the chips were collected using Affymetrix Expression Console (EC) Software (Affymetrix). The RMA-sketch workflow was applied for raw data preprocessing, which included global background correction, quantile normalization and median polish summarization. Principal component analysis was performed for detecting the outlier across all eight chips. The comparison between GFP(+) and GFP(−) groups was assessed using one-way analysis of variance implemented in the MSCL Analyst's Toolbox (http://abs.cit.nih.gov/MSCLtoolbox/) and JMP statistical software package (SAS, http://www.jmp.com). Significant changes for individual genes required a greater than twofold increase or decrease in expression and a P value of <0.001.
Acknowledgments
The authors thank Keyvan Kevanfar for flow cytometric sorting, and Mark Metzger, Alan Krouse, Barrington Thompson, and Sandra Price for excellent animal care. Funding for these studies was provided by the intramural research program of the NHLBI. The authors declared no conflict of interest.
References
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Ferguson C, Larochelle A, Dunbar CE. Hematopoietic stem cell gene therapy: dead or alive. Trends Biotechnol. 2005;23:589–597. doi: 10.1016/j.tibtech.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Corrigan-Curay J, Cohen-Haguenauer O, O'Reilly M, Ross SR, Fan H, Rosenberg N, et al. Challenges in vector and trial design using retroviral vectors for long-term gene correction in hematopoietic stem cell gene therapy. Mol Ther. 2012;20:1084–1094. doi: 10.1038/mt.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hematti P, Hong BK, Ferguson C, Adler R, Hanawa H, Sellers S, et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzwaelder K, Howe SJ, Schmidt M, Brugman MH, Deichmann A, Glimm H, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Beard BC, Dickerson D, Beebe K, Gooch C, Fletcher J, Okbinoglu T, et al. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol Ther. 2007;15:1356–1365. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- De Palma M, Montini E, Santoni de Sio FR, Benedicenti F, Gentile A, Medico E, et al. Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells. Blood. 2005;105:2307–2315. doi: 10.1182/blood-2004-03-0798. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther. 2009;17:1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Kim YS, Larochelle A, Renaud G, Wolfsberg TG, Adler R, et al. Sustained high-level polyclonal hematopoietic marking and transgene expression 4 years after autologous transplantation of rhesus macaques with SIV lentiviral vector-transduced CD34+ cells. Blood. 2009;113:5434–5443. doi: 10.1182/blood-2008-10-185199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calmels B, Ferguson C, Laukkanen MO, Adler R, Faulhaber M, Kim HJ, et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, et al. Transfusion independence and HMGA2 activation after gene therapy of human ß-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Mason PJ, Bessler M. 3'UTR-truncated Hmga2 cDNA causes MPN-like hematopoiesis by conferring a clonal growth advantage at the level of HSC in mice. Blood. 2011;117:5860–5869. doi: 10.1182/blood-2011-02-334425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana D, Sgualdino J, Rudilosso L, Merella S, Naldini L, Montini E. Whole transcriptome characterization of aberrant splicing events induced by lentiviral vector integrations. J Clin Invest. 2012;122:1667–1676. doi: 10.1172/JCI62189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue RE, Dunbar CE. Update on the use of nonhuman primate models for preclinical testing of gene therapy approaches targeting hematopoietic cells. Hum Gene Ther. 2001;12:607–617. doi: 10.1089/104303401300057289. [DOI] [PubMed] [Google Scholar]
- Shepherd BE, Kiem HP, Lansdorp PM, Dunbar CE, Aubert G, LaRochelle A, et al. Hematopoietic stem-cell behavior in nonhuman primates. Blood. 2007;110:1806–1813. doi: 10.1182/blood-2007-02-075382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM. Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci USA. 1994;91:9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp PM. Telomeres, stem cells, and hematology. Blood. 2008;111:1759–1766. doi: 10.1182/blood-2007-09-084913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim N, Presson AP, An DS, Mao SH, Bonifacino AC, et al. High-throughput, sensitive quantification of repopulating hematopoietic stem cell clones. J Virol. 2010;84:11771–11780. doi: 10.1128/JVI.01355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggewiss R, Pittaluga S, Adler RL, Guenaga FJ, Ferguson C, Pilz IH, et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood. 2006;107:3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd BE, Guttorp P, Lansdorp PM, Abkowitz JL. Estimating human hematopoietic stem cell kinetics using granulocyte telomere lengths. Exp Hematol. 2004;32:1040–1050. doi: 10.1016/j.exphem.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Allsopp RC, Cheshier S, Weissman IL. Telomere shortening accompanies increased cell cycle activity during serial transplantation of hematopoietic stem cells. J Exp Med. 2001;193:917–924. doi: 10.1084/jem.193.8.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada O, Oshimi K, Motoji T, Mizoguchi H. Telomeric DNA in normal and leukemic blood cells. J Clin Invest. 1995;95:1117–1123. doi: 10.1172/JCI117759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoff JA, De Waal E, Garg MB, Denham J, Scorgie FE, Enno A, et al. Telomere length in haemopoietic stem cells can be determined from that of mononuclear blood cells or whole blood. Leuk Lymphoma. 2002;43:2017–2020. doi: 10.1080/1042819021000015970. [DOI] [PubMed] [Google Scholar]
- Steinert S, White DM, Zou Y, Shay JW, Wright WE. Telomere biology and cellular aging in nonhuman primate cells. Exp Cell Res. 2002;272:146–152. doi: 10.1006/excr.2001.5409. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Suzuki J, Terao K, Ishida T. In vitro aging of macaque adherent cells: similar pattern of cellular aging between human and macaque. Mech Ageing Dev. 2003;124:237–244. doi: 10.1016/s0047-6374(02)00186-0. [DOI] [PubMed] [Google Scholar]
- Baerlocher GM, Mak J, Röth A, Rice KS, Lansdorp PM. Telomere shortening in leukocyte subpopulations from baboons. J Leukoc Biol. 2003;73:289–296. doi: 10.1189/jlb.0702361. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Notaro R, Cimmino A, Tabarini D, Rotoli B, Luzzatto L. In vivo telomere dynamics of human hematopoietic stem cells. Proc Natl Acad Sci USA. 1997;94:13782–13785. doi: 10.1073/pnas.94.25.13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn RF, Cross MA, Hatton C, Will AM, Lashford LS, Dexter TM, et al. Accelerated telomere shortening in young recipients of allogeneic bone-marrow transplants. Lancet. 1998;351:178–181. doi: 10.1016/S0140-6736(97)08256-1. [DOI] [PubMed] [Google Scholar]
- Calado RT, Cooper JN, Padilla-Nash HM, Sloand EM, Wu CO, Scheinberg P, et al. Short telomeres result in chromosomal instability in hematopoietic cells and precede malignant evolution in human aplastic anemia. Leukemia. 2012;26:700–707. doi: 10.1038/leu.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchia A, Bonini C, Magnani Z, Urbinati F, Sartori D, Muraro S, et al. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc Natl Acad Sci USA. 2006;103:1457–1462. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Donahue RE, Kuramoto K, Dunbar CE. Large animal models for stem and progenitor cell analysis. Curr Protoc Immunol. 2005;Chapter 22:Unit 22A.1. doi: 10.1002/0471142735.im22a01s69. [DOI] [PubMed] [Google Scholar]
- Kelly PF, Donahue RE, Vandergriff JA, Takatoku M, Bonifacino AC, Agricola BA, et al. Prolonged multilineage clonal hematopoiesis in a rhesus recipient of CD34 positive cells marked with a RD114 pseudotyped oncoretroviral vector. Blood Cells Mol Dis. 2003;30:132–143. doi: 10.1016/s1079-9796(03)00005-6. [DOI] [PubMed] [Google Scholar]
- Hanawa H, Hematti P, Keyvanfar K, Metzger ME, Krouse A, Donahue RE, et al. Efficient gene transfer into rhesus repopulating hematopoietic stem cells using a simian immunodeficiency virus-based lentiviral vector system. Blood. 2004;103:4062–4069. doi: 10.1182/blood-2004-01-0045. [DOI] [PubMed] [Google Scholar]
- Uchida N, Washington KN, Hayakawa J, Hsieh MM, Bonifacino AC, Krouse AE, et al. Development of a human immunodeficiency virus type 1-based lentiviral vector that allows efficient transduction of both human and rhesus blood cells. J Virol. 2009;83:9854–9862. doi: 10.1128/JVI.00357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Hargrove PW, Lap CJ, Evans ME, Phang O, Bonifacino AC, et al. High-efficiency transduction of rhesus hematopoietic repopulating cells by a modified HIV1-based lentiviral vector. Mol Ther. 2012;20:1882–1892. doi: 10.1038/mt.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An DS, Kung SK, Bonifacino A, Wersto RP, Metzger ME, Agricola BA, et al. Lentivirus vector-mediated hematopoietic stem cell gene transfer of common gamma-chain cytokine receptor in rhesus macaques. J Virol. 2001;75:3547–3555. doi: 10.1128/JVI.75.8.3547-3555.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander WE, Metzger ME, Morizono K, Bonifacino A, Penzak SR, Xie YM, et al. Noninvasive molecular imaging to detect transgene expression of lentiviral vector in nonhuman primates. J Nucl Med. 2006;47:1212–1219. [PubMed] [Google Scholar]
- Weber K, Mock U, Petrowitz B, Bartsch U, Fehse B. Lentiviral gene ontology (LeGO) vectors equipped with novel drug-selectable fluorescent proteins: new building blocks for cell marking and multi-gene analysis. Gene Ther. 2010;17:511–520. doi: 10.1038/gt.2009.149. [DOI] [PubMed] [Google Scholar]
- Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouilette SW, Moore JS, McMahon AD, Thompson JR, Ford I, Shepherd J, West of Scotland Coronary Prevention Study Group et al. Telomere length, risk of coronary heart disease, and statin treatment in the West of Scotland Primary Prevention Study: a nested case-control study. Lancet. 2007;369:107–114. doi: 10.1016/S0140-6736(07)60071-3. [DOI] [PubMed] [Google Scholar]