Abstract
Purpose.
Mutations at some retinitis pigmentosa (RP) loci are associated with variable penetrance and expressivity, exacerbating diagnostic challenges. The purpose of this study was to dissect the genetic underpinnings of nonsyndromic RP with variable age of onset in a large Mexican family.
Methods.
We ascertained members of a large, multigenerational pedigree using a complete ophthalmic examination. We performed whole exome sequencing on two affected first cousins, an obligate carrier, and a married-in spouse. Confirmatory sequencing of candidate variants was performed in the entire pedigree, as well as genotyping and mRNA studies to investigate expression changes in the causal locus.
Results.
We identified a 14–base pair (bp) deletion in PRPF31, a gene implicated previously in autosomal dominant (ad) RP. The mutation segregated with the phenotype of all 10 affected females, but also was present in six asymptomatics (two females and four males). Studies in patient cells showed that the penetrance/expressivity of the PRPF31 deletion allele was concordant with the expression levels of wild-type message. However, neither the known PRPF31 modulators nor cis-eQTLs within 1 Mb of the locus could account for the variable expression of message or the clinical phenotype.
Conclusions.
We have identified a novel 14-bp deletion in PRPF31 as the genetic driver of adRP in a large Mexican family that exhibits nonpenetrance and variable expressivity, known properties of this locus. However, our studies intimate the presence of additional loci that can modify PRPF31 expression.
Keywords: retinitis pigmentosa, genetic diseases, autosomal dominant, PRPF31, exome sequencing
Mutations in PRPF31 are known to cause autosomal dominant retinitis pigmentosa frequently hallmarked by incomplete penetrance. Genetic and functional studies intimate that hitherto unknown modulators may be contributing to this phenomenon.
Introduction
Photoreceptor dysfunction is the most common cause of visual impairment and affects ∼1 in 3000 people.1 This group of disorders is underscored by extensive genetic heterogeneity, with approximately 190 primary disease loci reported (available in the public domain at http://www.sph.uth.tmc.edu/retnet/). Retinitis pigmentosa (RP) can be subcategorized further by the functional basis of disease. Although genetic lesions resulting in ciliary transport defects represent the most frequent cause of RP (∼25%), genes implicated in phototransduction, lipid metabolism, and pre-mRNA splicing comprise additional examples known to have a role in maintaining the architecture of the photoreceptor, and when rendered dysfunctional converge on the same cellular apoptotic fate.2
Within the RP spectrum, mutations in 55 different genes have been shown to contribute causal mutations in patients with autosomal dominant RP (adRP), with a significant fraction (∼30%) remaining unknown.3 Additional confounders include the observation that some RP genes, such as rhodopsin4,5 and RPE65,6–8 potentially can give rise to dominant and recessive RP; in other instances, digenic phenomena have been reported involving transmission of multiple dominant mutations.9 Finally, modifiers of penetrance and expressivity have been reported in humans and in mouse RP models for dominant and recessive RP.10,11
The extensive genetic heterogeneity of RP and its confounding genetic architecture have accelerated the transition of RP diagnostics to next-gen sequencing platforms that have ranged from targeted sequencing of known loci to whole exome sequencing,12 in the hope that a broader evaluation of at least the coding fraction of the human genome can improve diagnostic yield, and inform the mechanisms of causality and modification. Importantly, identification of molecular lesions has motivated retrospective and additional phenotypic evaluation of patients12; thus, offering the potential of better clinical management of overt and subclinical disorders.
Here, we have studied a large multigeneration pedigree of Mexican descent with dominant RP. The study was motivated in part by the observation that the disorder was expressed overtly exclusively in females (n = 10 patients), but, assuming a monogenic causality model, had at least four male obligate carriers. Subsequent to whole exome sequencing of two patients, one obligate carrier, and one married-in spouse, we identified a novel deletion in PRPF31, a known dominant RP locus that was present in all affected individuals and all four obligate carriers. In contrast to previous reports, however, we observed only a modest increase in the expression of the wild-type (WT) PRPF31 allele and no appreciable differences in the expression levels of the previously reported cis-modulator, CNOT3, suggesting that mechanisms other than regulation of expression of PRPF31 can modify the penetrance of haploinsufficiency-driven phenotypes.
Methods
Patient Samples
Subsequent to informed consent, we obtained peripheral blood samples from 31 members of an adRP pedigree. We extracted DNA from whole blood according to standard procedures using the Gentra Puregene Blood kit (Qiagen, Venlo, The Netherlands). Enrollment of research participants followed the tenets of the Declaration of Helsinki, and was approved by the Duke University Internal Review Board (IRB).
Retinal Phenotyping (Imaging and Electroretinograms)
Complete ophthalmic examination of each family member was performed initially as part of standard of care. Phenotyping included retinal imaging, autofluorescence, and electroretinograms. In general, and subsequent to an initial consultation, an annual ophthalmic exam was conducted for each member of the family. We observed three phenotypic groups: symptomatic-early onset, symptomatic-late onset, and asymptomatic (see Table). At the initial consultation, none of asymptomatics had functional complaints, but during follow-up ophthalmic exams two individuals displayed subclinical phenotypes.
Table.
Clinical Phenotypes of PRPF31 p.Arg289Profs*30 Mutation Carriers
|
Individual |
Pedigree Branch |
Finding |
Age of Onset |
Visual Acuity |
Age at Examination |
| Symptomatic, early onset | |||||
| IV-75-27 | 1 | Dark adaptation difficulty and visual field constriction | 6–10 y | 20/30 | 38 y |
| MA-So-78 | 1 | Photophobia, dark adaptation difficulty and visual field constriction | 10 y | 20/20 | 34 y |
| 29-AP-So | 1 | Visual field constriction | 2 y | 20/10 | 7 y |
| III-49-19 | 2 | Dark adaptation difficulty, visual field constriction | 6 y | CF 1.5 m | 63 y |
| III-54-19 | 2 | Light preference and visual field constriction | 10 y | 20/40 | 59 y |
| Symptomatic, late onset | |||||
| IV-95-15.12 | 1 | Dark adaptation difficulty | 16 y | 20/20 | 18 y |
| IV-81-25.13 | 2 | Dark adaptation difficulty | 16 y | 20/20 | 32 y |
| IV-79-6 | 2 | Dark adaptation difficulty | 16 y | 20/20 | 28 y |
| IV-75-8 | 2 | Visual field constriction | 15 y | 20/30 | 36 y |
| III-90-22 | 2 | Dark adaptation difficulty | 15 y | 20/20 | 23 y |
| Asymptomatic | |||||
| S48 | 1 | None | N/A | 20/20 | 64 y |
| III-52-19, 7/19 | 1 | None | N/A | 20/20 | 60 y |
| 58-So-09 | 1 | None, only ERG dysfunction | 52 y | 20/20 | 54 y |
| IV-86-13 | 1 | None | N/A | 20/20 | 16 y |
| III-52-19, 3/19 | 2 | Dark adaptation difficulty | Unknown | 20/20 | 60 y |
| IV-73-18 | 2 | None | N/A | 20/20 | 39 y |
Whole Exome Sequencing
Genomic DNA of four individuals was sheared to 150 base pairs (bp) using a Covaris S220 (Covaris, Inc., Woburn, MA, USA). Fragment sizes were verified with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), ligated to Illumina TrueSeq (Ilumina, Inc., San Diego, CA, USA) library prep adapters and captured using the NimbleGen EzExome V2 library (Roche NimbleGen, Madison, WI, USA) according to manufacturer's instructions. Paired-end 100 base reads were generated on an Illumina HiSeq2000 (Ilumina, Inc.).
Bioinformatic Analysis
Contaminating adapter sequence was removed from reads using CASAVA (BclToFastq; Illumina, Inc.), and aligned to the hg19 reference human genome using Burrows-Wheeler Aligner (BWA).13 Duplicates were marked with SAMtools14 and realigned around indels using the Genome Analysis Toolkit (GATK, RealignerTargetCreator/Indel Realigner; available in the public domain at http://www.broadinstitute.org/gatk).15 Variants were called with the GATK Unified Genotyper and annotated with ANNOVAR.16 We calculated exome coverage with Picard (CalculateHsMetrics; Supplementary Table S1).17 We then utilized Enlis Genome Research software (Enlis, LLC, Berkeley, CA, USA) to filter exome data within the four individuals sequenced, and against publicly available variant data (dbSNP137 and NHLBI GO Exome Sequencing Project [ESP], available in the public domain at http://evs.gs.washington.edu/EVS/; accessed June 2013; Supplementary Table S2).
Sanger Sequencing
We amplified the coding sequence and intron/exon boundaries of PRPF31 exon 9, and 26 predicted eQTLs by standard PCR amplification and sequenced amplicons with BigDye Terminator v3.1 chemistry on an ABI 3730 automated capillary sequencer (Applied Biosystems, Foster City, CA, USA). Sequences were aligned with Lasergene (DNASTAR, Madison, WI, USA), and variants were confirmed by visual assessment of chromatograms. Primer sequences are available upon request.
Lymphocyte Cell Culture
B-cell lymphocytes were separated from whole blood by gradient centrifugation with Ficoll and were immortalized by Epstein-Barr virus (EBV) infection according to standard procedures at the University of North Carolina Lineberger Comprehensive Cancer Center Tissue Culture Facility. Cells were cultured in RPMI-1640 with 10% fetal bovine serum.
Gene Expression Studies (Quantitative RT-PCR [qRT-PCR])
We extracted total RNA from lymphocyte cell pellets using Trizol (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions. Total RNA (1 μg) was DNAse treated and reverse transcribed using oligo-dT priming according to the QuantiTect Reverse Transcription kit instructions (Qiagen). The qRT-PCR reactions were performed in triplicate using SYBR Green PCR Master Mix (Life Technologies, Carlsbad, CA) per manufacturer protocol; primers were designed using Primer3 with a target amplicon size 80 to 130 bp. Absolute quantification was performed on an ABI 7900H (Applied Biosystems), and Ct values were extracted using SDS 2.3 (Applied Biosystems) for β-Actin normalization. Triplicate assays were performed three separate times.
Cis-eQTL Analyses
The cis-eQTLs for PRPF31 were derived from the Avon Longitudinal Study of Parents and Children (ALSPAC)18 cohort. Gene expression measurements were performed in lymphoblastoid cell lines of a cohort of 864 children (ALSPAC) with the Illumina HT12 array v3. Values were quantile normalized, and outliers were removed based on principal component analysis of gene expression and genotyping data. Genotypes were based on an Illumina human Hap 550-quad and imputed to HapMap 2. The eQTL analysis was performed using Spearman rank correlation between any SNP in a 1 Mb window around the transcription start site of the gene and the expression values of each probe. Permutations were performed to assign a critical threshold for each gene ensuring an FDR of 5% (Bryois et al., unpublished data).
Results
We consulted a large nonsyndromic RP pedigree originating from the Yucatan peninsula; retinal degeneration was evident, either described by family members, ascertained through prior medical records, or through our direct clinical assessment, across five generations (Fig. 1). Of the three branches of the pedigree, two branches, consisting of four generations each, had multiple affected individuals (n = 4, Branch 1; n = 6, Branch 2). Among the 10 symptomatic individuals from whom we had detailed medical histories and fundus images with clear RP and hyperfluorescent regions according to auto-fluorescence imaging (Fig. 2, Supplementary Fig. S1), we stratified them as early onset, sharing onset of symptoms in the first decade of life, and later onset, with notable symptoms arising in the second decade of life (see Table). For all affected individuals with RP, early signs manifested as functional dark adaptation difficulties and led to constriction of visual fields. At the most recent clinical examination (2013), all 10 affected individuals still conserved fairly high visual acuity (ranging from 20/10–20/40; see Table), except for one who had undergone nonstandard treatments.
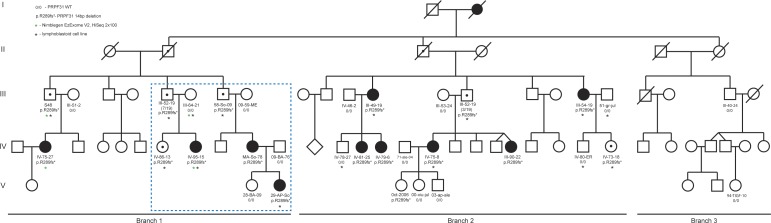
Figure 1.
Schematic representation of the Mexican adRP pedigree. We ascertained samples from three generations of a family in which two of three branches had individuals affected with RP. Samples that were whole exome sequenced are noted with a green star; samples used for lymphocyte cell culture isolation and gene expression studies are denoted with a black star. Dashed blue box indicates the section of the pedigree shown in Figure 2.
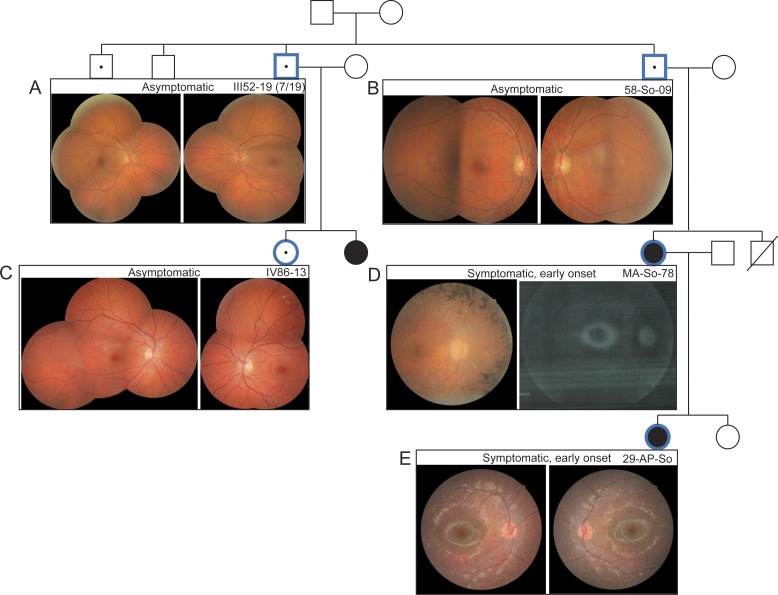
Figure 2.
Phenotypic variability within Branch 1 of the Mexican adRP pedigree. (A–C) Fundus images of eyes are shown for three asymptomatic PRPF31 mutation carriers in Branch 1. (D, E) Fundus images for two early-onset individuals who harbor the PRPF31 mutation display retinitis pigmentosa and hyperfluorescent regions upon autofluorescence imaging (D). Age at retinal imaging: III52-19 (7/19), 60 years; 58-So-09, 54 years; IV86-13, 16 years; MA-So-78, 34 years; 29-AP-So, 7 years.
Based on observations of disease transmission, we hypothesized an autosomal dominant mode of inheritance with incomplete penetrance. First, we noted transmission of RP status from affected parents to offspring (in each case affected mother to affected daughter, n = 3). Second, we observed multiple instances of transmission from unaffected parents to affected offspring (asymptomatic father to affected daughter, n = 5), lending support to the possibility of incomplete penetrance. Notably, clinical re-examination of the four asymptomatic obligate carriers revealed subtle, subclinical phenotypes in two of the four as measured either by electroretinography or dark adaptation studies (see Table; Supplementary Fig. S2). Additionally, we noted an apparent bias in the sex of affected individuals exclusively toward females and wondered if the genetic basis of disease was driven by a sex-specific mechanism. This was not because of a chance skewing of the representation of the two sexes in our family. Counting the total number of relevant males and females in generations III, IV, and V (either with RP or with an affected parent or offspring (excluding married-in spouses), we observed an approximate 1:1 ratio (14 females [56%], 11 males [44%]). By contrast, we found significant sex bias associated with disease (10 symptomatic females [71%], zero symptomatic males [0%]).
To understand the inheritance paradigm in this family, and to identify their genetic lesion(s) underscoring RP, we employed whole exome sequencing (WES) of four individuals from Branch 1. We selected one asymptomatic male (S48), an obligate carrier by virtue of his affected offspring; his affected daughter and her affected cousin (IV-75-27, early onset; IV95-15, late onset, respectively), and a married in spouse (III-64-21; Figs. 1, 2). We conducted whole exome capture and generated 58 to 149 million mapped Illumina sequencing reads per individual, resulting in mean exome coverage of 74× to 97× (Supplementary Table S1). Using a tiered filtering strategy (Supplementary Table S2), we first considered all functional variants with ≥10× coverage that were shared between affected cousins (n = 6015). Next, we retained all variants that were shared between affecteds and the obligate carrier, but not the married-in spouse (n = 947). We narrowed the list further by comparing all 947 variants to the ESP6500 and dbSNP137 datasets, and filtering out all variants with an alternate allele frequency of >1%, which eliminated all but 26 alleles. We then conducted bidirectional dideoxy terminator sequencing of all 26 variants in all 31 family members from whom we had DNA available. The only allele that was shared between all 10 affected individuals in the pedigree was a single heterozygous 14 bp deletion in exon 9 of the U4/U6 small nuclear ribonucleoprotein Prp31 (PRPF31/RP11) encoding a frameshift, p.Arg289Profs*30, and predicted to truncate the C-terminal Prp31_C domain. This mutation also was present in each of the four asymptomatic obligate carriers, the unaffected sibling of IV-95-15, and the unaffected daughter of III-54-19 (6/16 asymptomatic mutation carriers; 37.5%). To our knowledge, this deletion has not been reported previously in individuals with adRP, was absent from 13,000 chromosomes in the Exome Variant Server, and was absent from an additional 192 ethnically matched chromosomes.
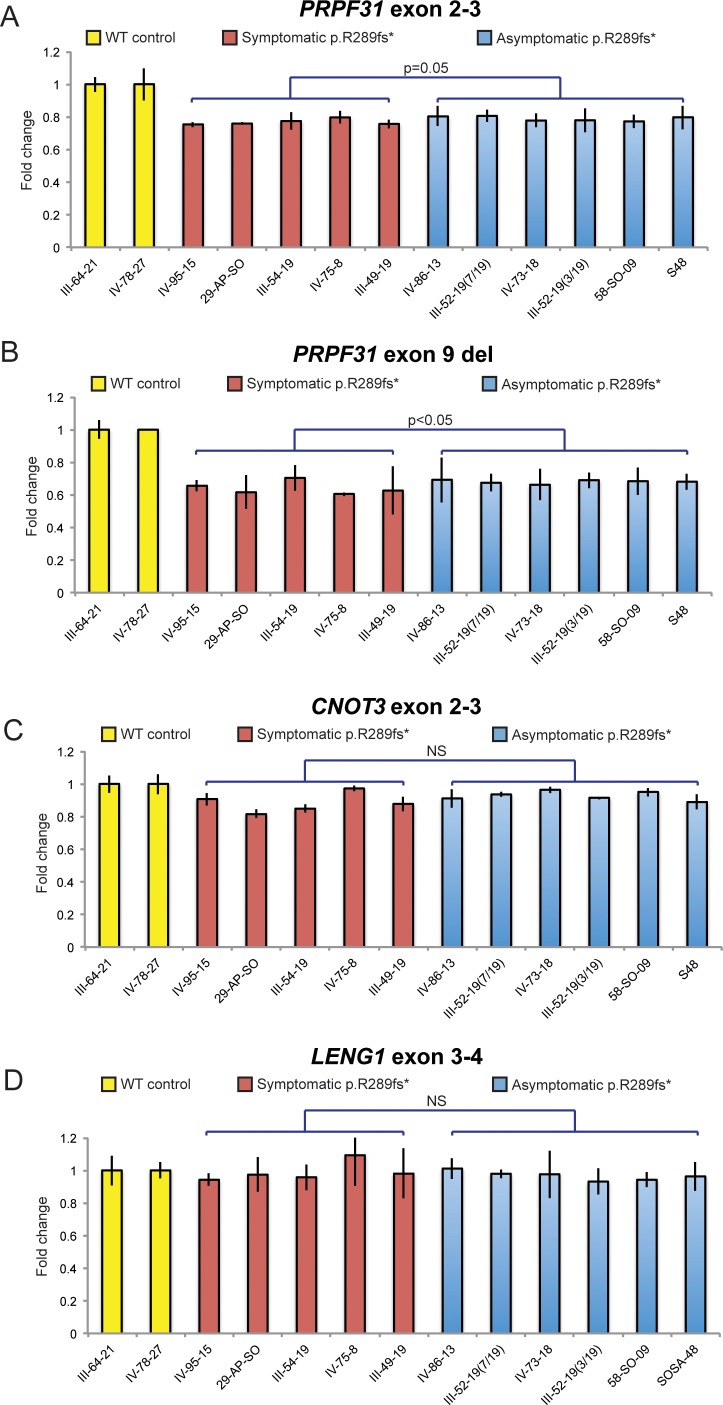
The PRPF31 gene encodes a ubiquitous splicing factor.19 and mutations at this locus account for approximately 6% of adRP pedigrees.20 Previous reports of incomplete penetrance with up to approximately 35% of carriers reported as asymptomatic are consistent with our observations of this adRP family21–24 (Supplementary Table S3). The prevailing hypothesis to explain this phenomenon is that asymptomatic individuals are able to compensate for PRPF31 haploinsufficiency by exceeding an expression threshold with their wild-type (WT) allele.23–25 We tested this notion in our adRP pedigree by establishing lymphocyte cell lines (LCLs) from 13 individuals (WT, n = 2; asymptomatic carriers, n = 6; symptomatics, n = 5), a cell type in which PRPF31 expression has been shown to be a surrogate model for the retina.21,25–28 We performed qRT-PCR amplifying two regions of PRPF31 from LCL cDNA (Figs. 3A, 3B): one region spanning the 14 bp deletion in exon 9 (with a forward primer annealing to the 14 bp deletion, thereby detecting only the WT transcript), and a second amplicon spanning the exon 2-3 junction (detecting WT and mutant alleles).
Figure 3.
Expression studies in asymptomatic versus symptomatic carriers of p.Arg289Profs*30 demonstrated modestly augmented PRPF31 in asymptomatics, but similar expression of CNOT3 and LENG1. (A, B) PRPF31 expression is decreased in mutation carriers versus WT control individuals when alleles are monitored (exon 2–3 primers; [A]) or when only the WT allele is monitored (exon 9 del; [B]). (C, D) Mean differences in either CNOT3 or LENG1 expression were not significantly (NS) different between symptomatics and asymptomatics. Expression for each of the four primer pairs was normalized to β-ACTIN. Assays were performed three times, with similar results.
Consistent with a haploinsufficiency model of causality, PRPF31 expression in all carriers of the heterozygous deletion was reduced on average by 40% compared to the WT cell lines, as detected by the exon 9-specific primers (Fig. 3B). Additionally, qPCR from the exon 2-3 amplicon suggested rapid degradation of mutant PRPF31 transcript, likely through nonsense-mediated decay. However, the exon 2-3 qRT primers also reported a borderline significant, reduction of overall PRPF31 message in symptomatic versus asymptomatic individuals (P = 0.05; relative to WT and normalized to β-ACTIN (Fig. 3A), with no appreciable differences on Western blotting (data not shown). We interpreted this observation as a potential contribution of increased expression of the wild-type PRPF31 allele from asymptomatic individuals; however, the magnitude of the effect was markedly lower from the 20% to 50% differences in expression levels of the WT allele reported in prior analyses of this locus.27,28
Nonetheless, to probe the question further, we searched for genetic evidence of modification. One study proposed two different trans-acting expression quantitative trait locus eQTLs through the expression analysis of LCLs from multiple individuals of northern European descent; an 8.2 Mb region on 14q21-23, and a second locus proximal to PRPF31 at 19q13.4.25 More recently, CNOT3 was proposed as a modifier of PRPF31 as supported by an inverse correlation between CNOT3 transcript levels and WT PRPF31 expression in asymptomatic mutation carriers in multiple RP families, and also chromatin immunoprecipitation studies demonstrating direct binding of CNOT3 to the PRPF31 promoter sequence.24 The variable CNOT3 expression was associated subsequently with a common intronic SNP (rs4806718) in two unrelated pedigrees of differing ethnicity.24 To test whether this mechanism also might contribute a more subtle, but still relevant mechanism to disease pathogenesis in our family, we first genotyped rs4806718 in all 16 individuals harboring the p.Arg289Profs*30 change. We observed no significant enrichment of the minor allele (T) in symptomatics versus asymptomatics (6/20 and 5/12 chromosomes, respectively). These findings were corroborated by similar CNOT3 transcript levels in LCLs from each respective group of PRPF31 mutation carriers (Fig. 3C).
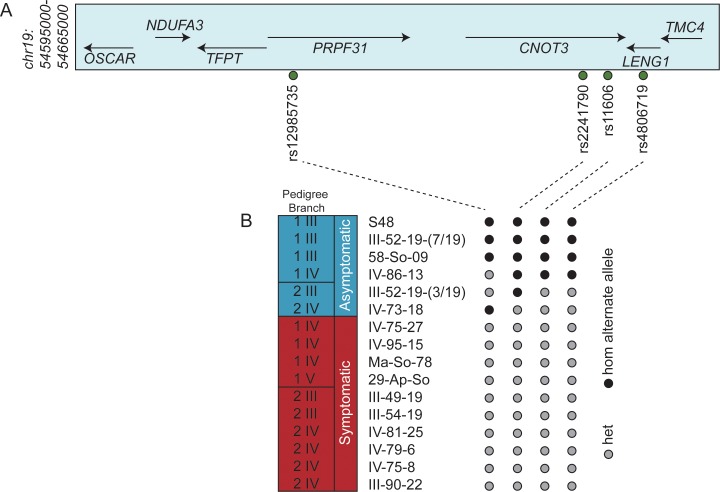
Finally, we explored the possibility of other eQTLs that might be relevant to the penetrance of the PRPF31 deletion. We extracted eQTL data from ALSPAC cohort LCLs (n = 864 individuals; Bryois et al., unpublished data) with FDR of 5% as described.29 The 25 eQTLs with rho P values < 10E-05 were within 1 Mb of the PRPF31 transcriptional start site (TSS; Supplementary Table S4), and two additional eQTLs mapped to different chromosomes (rs7629060 on chromosome 3 and rs9599183 on chromosome 13). We genotyped all 27 SNPs in the 16 individuals bearing the heterozygous p.Arg289Profs*30 mutation. We did not identify any SNPs that were shared among all six asymptomatic individuals. By contrast, we observed a consistent set of genotypes at four cis-eQTL SNPs that were shared uniquely by all 10 affected individuals (Fig. 4). For those markers, haplotype phasing indicated that the minor SNP allele was on the same chromosome as the p.Arg289Profs*30 deletion. However, none of these SNPs, alone or in tandem, correlated with the expression of PRPF31, CNOT3, or LENG1, three of the prime modification candidates in the region (Figs. 3A–D). Together with our expression data from the symptomatic individuals, these data suggested that a different cis event(s), possibly tagged by these SNPs, might be relevant to the expressivity of PRPF31 deletion, possibly in an expression-independent mechanism.
Figure 4.
The cis-eQTL genotypes for rs12985735, rs2241790, rs11606, and rs4806719 in PRPF31 p.Arg289Profs*30 mutation carriers. (A) Schematic of the PRPF31 locus on chr19; hg19 coordinates are shown corresponding to 1 Mb from the TSS of PRPF31. (B) Genotypes at the four SNPs are shown with circles; black, homozygous minor allele; gray, heterozygous. We did not identify a consistent haplotype on the WT allele that was shared among the six asymptomatic individuals within 1 Mb of the PRPF31 TSS.
Discussion
Here we have described a multigeneration pedigree with ad RP and an unusual pattern of inheritance, wherein there is variable age of onset, apparent female-specific bias and nonpenetrance. Our genetic studies have identified an inherited heterozygous deletion in PRPF31 as the primary driver of RP in this pedigree, with nonpenetrance and variable expressivity documented previously for this locus.21–24,30–34 We cannot differentiate between variable expressivity and nonpenetrance as a function of time. Two of the initially considered nonpenetrant males were found, subsequent to our discovery, to have subtle, suggestive features of RP with no clinical involvement that should be tested further using electroretinography to eliminate subjectiveness. Nonetheless, despite the imperfect longitudinal phenotype data that typify most genetic and genomic studies on historical cohorts, we observed a clear dichotomization of the phenotype, which in this family manifests with significant sex bias (Supplementary Table S3).
In contrast to most genetic disorders, in which sex-bias favors disease pathogenesis in males,35 females with the PRPF31 deletion appear more likely to develop RP. We considered two possibilities for this observation. First, we have ruled out the chance skewing of the representation of the two sexes in our family; we observed an approximate 1:1 ratio of females versus males (14 females, 11 males). However, considering the simplest scenario of a single modulator gene in trans with PRPF31, an asymptomatic individual would necessarily carry at least one of such alleles and generate offspring where 50% to 100% of mutation carriers would be asymptomatic. Since three of the four asymptomatic mutation carriers within the family are brothers, the observed bias is likely not due to sex, but to the fact that three of the asymptomatic males are siblings and sons of an asymptomatic parent (Branch 1, Fig. 1). We considered it unlikely that the presence of this particular PRPF31 mutation is associated with lethality in males, therefore the augmented prevalence of symptomatic females can be attributed to an observational bias deriving from an almost triple prevalence of females carrying the mutation in comparison to males carrying the mutation (11 vs. 4) and to the fact that females with the mutation are less related than males with the mutation.
Genetic and expression analyses of the PRPF31 locus in this pedigree suggested that although there are modest changes in the expression levels of the WT allele that might contribute a protective effect, the magnitude of protection is significantly lower compared to published reports27,28 to the point where we do not observe differences at the level of protein abundance. We considered the possibility that this difference relates to the fact that our studies were conducted, by necessity, in LCLs. However, several prior studies have used similar material, largely discounting this hypothesis.27,28 Further, genotyping of all plausible eQTLs at the PRPF31 locus did not identify any SNPs with genotypes unique to asymptomatic mutation carriers in the neighboring genes CNOT3 and LENG1, nor did we observe marked differences in the expression levels of these two transcripts between symptomatic and asymptomatic deletion carriers. Haplotype analysis was consistent with that observation; we found a unique haplotype block on the allele bearing the deletion, but there was no consistent haplotype on the WT allele that was shared among the six asymptomatic individuals within 1 Mb of the PRPF31 transcriptional start site, suggesting that it is unlikely that a single cis modifier within that region can account for the differences in penetrance/expressivity.
The PRPF31 gene exemplifies the complexity of disease-causing loci and the challenges they can pose for patient management. Despite these limitations, even imperfect understanding of such genetic data has value. First, returning these results to our family is of innate emotional value. Second, and just as importantly, we can improve the predictive power of the genotype of this locus for the following generations, since they are likely to develop RP in their first or second decade of life. Finally, studies such as ours contribute to our appreciation of the natural history of the disorder; the aggregation of additional pedigrees, particularly families from ethnic groups that have been undersampled in RP studies to date, will be of value to dissecting the effect of cis and trans alleles on the phenotypic expressivity of PRPF31 haploinsufficiency.
Acknowledgments
The authors thank this adRP family for their active participation in this research, Xue Qin and Allison Ashley-Koch for helpful discussions pertaining to statistical analyses, and James Bensen and Camila Lopez at the University of North Carolina Lineberger Comprehensive Cancer Center Tissue Culture Facility for their assistance with the establishment of lymphocyte cell lines. NK is a distinguished Jean and George W. Brumley Professor.
Supported by United States National Institutes of Health Grant R01-EY021872 (EED).
Disclosure: A. Villanueva, None; J.R. Willer, None; J. Bryois, None; E.T. Dermitzakis, None; N. Katsanis, None; E.E. Davis, None
References
- 1. Buch H, Vinding T, La Cour M, Appleyard M, Jensen GB, Nielsen NV. Prevalence and causes of visual impairment and blindness among 9980 Scandinavian adults: the Copenhagen City Eye Study. Ophthalmology. 2004; 111: 53–61 [DOI] [PubMed] [Google Scholar]
- 2. Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010; 11: 273–284 [DOI] [PubMed] [Google Scholar]
- 3. Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013; 84: 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dryja TP, McGee TL, Hahn LB, et al. Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. N Engl J Med. 1990; 323: 1302–1307 [DOI] [PubMed] [Google Scholar]
- 5. Rosenfeld PJ, Cowley GS, McGee TL, Sandberg MA, Berson EL, Dryja TP. A null mutation in the rhodopsin gene causes rod photoreceptor dysfunction and autosomal recessive retinitis pigmentosa. Nat Genet. 1992; 1: 209–213 [DOI] [PubMed] [Google Scholar]
- 6. Bowne SJ, Humphries MM, Sullivan LS, et al. A dominant mutation in RPE65 identified by whole-exome sequencing causes retinitis pigmentosa with choroidal involvement. Eur J Hum Genet. 2011; 19: 1074–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marlhens F, Bareil C, Griffoin JM, et al. Mutations in RPE65 cause Leber's congenital amaurosis. Nat Genet. 1997; 17: 139–141 [DOI] [PubMed] [Google Scholar]
- 8. Morimura H, Fishman GA, Grover SA, Fulton AB, Berson EL, Dryja TP. Mutations in the RPE65 gene in patients with autosomal recessive retinitis pigmentosa or leber congenital amaurosis. Proc Nat Acad Sci U S A. 1998; 95: 3088–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim KP, Yip SP, Cheung SC, Leung KW, Lam ST, To CH. Novel PRPF31 and PRPH2 mutations and co-occurrence of PRPF31 and RHO mutations in Chinese patients with retinitis pigmentosa. Arch Ophthalmol. 2009; 127: 784–790 [DOI] [PubMed] [Google Scholar]
- 10. Khanna H, Davis EE, Murga-Zamalloa CA, et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 2009; 41: 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Louie CM, Caridi G, Lopes VS, et al. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat Genet. 2010; 42: 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang F, Wang H, Tuan HF, et al. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet. 2014; 133: 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38: e164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rio Frio T, Wade NM, Ransijn A, Berson EL, Beckmann JS, Rivolta C. Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J Clin Invest. 2008; 118: 1519–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boyd A, Golding J, Macleod J, et al. Cohort Profile: the ‘children of the 90s'--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013; 42: 111–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weidenhammer EM, Ruiz-Noriega M, Woolford JL Jr. Prp31p promotes the association of the U4/U6 x U5 tri-snRNP with prespliceosomes to form spliceosomes in Saccharomyces cerevisiae. Mol Cell Biol. 1997; 17: 3580–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Audo I, Bujakowska K, Mohand-Said S, et al. Prevalence and novelty of PRPF31 mutations in French autosomal dominant rod-cone dystrophy patients and a review of published reports. BMC Med Genet. 2010; 11: 145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu JY, Dai X, Sheng J, et al. Identification and functional characterization of a novel splicing mutation in RP gene PRPF31. Biochem Biophys Res Commun. 2008; 367: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pomares E, Riera M, Permanyer J, et al. Comprehensive SNP-chip for retinitis pigmentosa-Leber congenital amaurosis diagnosis: new mutations and detection of mutational founder effects. Eur J Hum Genet. 2010; 18: 118–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rose AM, Shah AZ, Venturini G, Rivolta C, Rose GE, Bhattacharya SS. Dominant PRPF31 mutations are hypostatic to a recessive cnot3 polymorphism in retinitis pigmentosa: a novel phenomenon of “linked trans-acting epistasis.” Ann Hum Genet. 2013; 78: 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Venturini G, Rose AM, Shah AZ, Bhattacharya SS, Rivolta C. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet. 2012; 8: e1003040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rio Frio T, Civic N, Ransijn A, Beckmann JS, Rivolta C. Two trans-acting eQTLs modulate the penetrance of PRPF31 mutations. Hum Mol Genet. 2008; 17: 3154–3165 [DOI] [PubMed] [Google Scholar]
- 26. Al-Maghtheh M, Vithana E, Tarttelin E, et al. Evidence for a major retinitis pigmentosa locus on 19q13.4 (RP11) and association with a unique bimodal expressivity phenotype. Am J Hum Genet. 1996; 59: 864–871 [PMC free article] [PubMed] [Google Scholar]
- 27. Rivolta C, McGee TL, Rio Frio T, Jensen RV, Berson EL, Dryja TP. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat. 2006; 27: 644–653 [DOI] [PubMed] [Google Scholar]
- 28. Vithana EN, Abu-Safieh L, Pelosini L, et al. Expression of PRPF31 mRNA in patients with autosomal dominant retinitis pigmentosa: a molecular clue for incomplete penetrance? Invest Ophthalmol Vis Sci. 2003; 44: 4204–4209 [DOI] [PubMed] [Google Scholar]
- 29. Stranger BE, Montgomery SB, Dimas AS, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012; 8: e1002639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kohn L, Bowne SJ, Sullivan L, et al. Breakpoint characterization of a novel approximately 59 kb genomic deletion on 19q13.42 in autosomal-dominant retinitis pigmentosa with incomplete penetrance. Eur J Hum Genet. 2009; 17: 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rio Frio T, McGee TL, Wade NM, et al. A single-base substitution within an intronic repetitive element causes dominant retinitis pigmentosa with reduced penetrance. Hum Mutat. 2009; 30: 1340–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saini S, Robinson PN, Singh JR, Vanita V. A novel 7 bp deletion in PRPF31 associated with autosomal dominant retinitis pigmentosa with incomplete penetrance in an Indian family. Exp Eye Res. 2012; 104: 82–88 [DOI] [PubMed] [Google Scholar]
- 33. Xia K, Zheng D, Pan Q, et al. A novel PRPF31 splice-site mutation in a Chinese family with autosomal dominant retinitis pigmentosa. Mol Vis. 2004; 10: 361–365 [PubMed] [Google Scholar]
- 34. Yang L, Yin X, Wu L, et al. Targeted exome capture and sequencing identifies novel PRPF31 mutations in autosomal dominant retinitis pigmentosa in Chinese families. BMJ Open. 2013; 3: e004030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008; 9: 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]