Abstract
The biogenic amine serotonin (5-HT, 5-hydroxytryptamine) exerts powerful, modulatory control over multiple physiological functions in the brain and periphery, ranging from mood and appetite to vasoconstriction and gastrointestinal motility. In order to gain insight into shared and distinct molecular and phenotypic networks linked to variations in 5-HT homeostasis, we capitalized on the stable genetic variation present in recombinant inbred (RI) mouse strains. This family of strains, all derived from crosses between C57BL/6J and DBA/2J (BXD) parents, represent a unique, community resource with ∼40 years of assembled phenotype data that can be exploited to explore and test causal relationships in silico. We determined levels of 5-HT and 5-hydroxyindoleacetic acid (5-HIAA) from whole blood, midbrain, and thalamus/hypothalamus (diencephalon) of 38 BXD lines and both sexes. All 5-HT measures proved highly heritable in each region, although both gender and region significantly impacted between-strain correlations. Our studies identified both expected and novel biochemical, anatomical, and behavioral phenotypes linked to 5-HT traits, as well as distinct quantitative trait loci (QTL). Analyses of these loci nominate a group of genes likely to contribute to gender- and region-specific capacities for 5-HT signaling. Analysis of midbrain mRNA variations across strains revealed overlapping gene expression networks linked to 5-HT synthesis and metabolism. Altogether, our studies provide a rich profile of genomic, molecular and phenotypic networks that can be queried for novel relationships contributing risk for disorders linked to perturbed 5-HT signaling.
Introduction
Serotonin (5-hydroxytryptamine, 5-HT) provides for powerful modulation of physiology and behavior throughout the body and across the lifespan. Approximately 90% of 5-HT stores in the human body derive from gut enterochromaffin cell 5-HT synthesis and release (Gershon 2013), with a large stable pool found in platelets (Rand and Reid 1951) that acquire the indoleamine during their passage through the enteric circulation. Whereas blood and brain pools of 5-HT in the adult are separated by the blood-brain barrier (Mann et al. 1992), Bonnin and colleagues recently reminded us that another peripheral site, the placenta, can produce and release 5-HT that can travel to the CNS and modulate axon guidance (Bonnin et al. 2011).
CNS regions that regulate mood, emotion, feeding and reward are prominent sites of 5-HT synthesis and release (Steinbusch 1981). As such, a number of disorders have been reported to display alterations in CNS 5-HT homeostasis, including anxiety, depression, obsessive-compulsive disorder (OCD) and addiction (Barondes 1994). Although a subject of significant debate (Risch et al. 2009), biochemical and genetic evidence continues to drive consideration that risk for depression in some individuals may be linked to a limited capacity for normal brain 5-HT signaling. For example, elevated brain 5-HT turnover has been repeatedly observed in unmedicated patients with depression (Gjerris et al. 1987; Sullivan et al. 2006), with levels returning to normal levels upon treatments with SSRIs (Barton et al. 2008). Removal of tryptophan, the essential precursor for 5-HT synthesis, induces relapse in subjects that previously remitted with SSRI therapy (Delgado et al. 1990; Barr et al. 1994). With respect to blood 5-HT, hyperserotonemia (elevated whole blood 5-HT) has been observed in a sizeable fraction (25-30%) of autistic children for over 50 years (Schain and Freedman 1961; Cook and Leventhal 1996; Cross et al. 2008). Genetic variation in the 5-HT transporter (SERT) promoter (5HTTLPR) has been associated with neuroticism (Lesch et al. 1996), the impact of early life stress on mood disorder risk (Caspi et al. 2003; Karg et al. 2011) and in SSRI response (Pollock et al. 2000; Arias et al. 2003; Serretti et al. 2007). With respect to 5-HT metabolism, deletion in humans of a region encompassing one or both of the 5-HT metabolizing genes, MAOA and MAOB, has been found in subjects who display a spectrum of developmental and behavioral disorders, including mental retardation, autism-like features and psychosis (Lenders et al. 1996; Whibley et al. 2010; Saito et al. 2013). Finally, functional variation in MAOA has been reported to represent a point of convergence for gene x environment interactions through the moderation of the long-lasting effects of childhood maltreatment (Caspi et al. 2002).
Although the basic elements controlling 5-HT synthesis, storage, response and inactivation have been known for more many decades, novel features of the molecular and cellular landscape controlling or responding to 5-HT homeostasis continue to emerge. For example, only within the past decade did Walther and Bader (Walther et al. 2003) establish that distinct isoforms of tryptophan hydroxylase (TPH) differentially dictate 5-HT synthesis in the periphery (TPH1) and CNS (TPH2). Serotonylation, a process whereby 5-HT is covalently attached to small G-proteins via the actions of transglutaminase II, has also only recently been recognized as a mechanism by which intracellular 5-HT can impact secretion in platelets (Walther et al. 2003) and pancreas (Paulmann et al. 2009). To gain a better understanding of functionally relevant networks linked to 5-HT homeostasis and signaling, we exploited Recombinant Inbred (RI) mice, a community resource of clonal lines derived from C57BL/6J × DBA/2J parents (BXD lines) (Taylor et al. 1977; Taylor et al. 1999; Peirce et al. 2004). Thousands of anatomical, biochemical, physiological and behavioral phenotypes as well as single nucleotide polymorphisms (SNPs) have been collected on these lines and are publically accessible via a curated database (http://www.genenetwork.org). The present study represents the first systematic analysis of heritability, trait correlations and quantitative traits loci (QTL) based on BXD brain and blood 5-HT traits, efforts that reveal multiple, and in some cases gender-specific, associations. We identify a suite of phenotypes likely to influence, or be influenced by, 5-HT signaling capacity. Finally, we elucidate transcriptional networks linked to variation in 5-HT traits in the BXD lines and nominate candidate genes as determinants of 5-HT homeostasis and signaling.
Materials and Methods
Animals
BXD RI mice were obtained from the Oak Ridge National Laboratories (ORNL) and the Jackson Laboratory (JAX). A total of 126 BXD recombinant inbred mice from 38 strains were used for neurochemical analyses. The average age of mice used was 74 days (range from 51 to 89 days), and comprised a total of 67 male and 59 female mice, respectively. Females were used irrespective of estrous cycle though they were never exposed to males or male bedding after weaning, typically required to initiate synchronized estrous cycling. A total of 129 male mice from 37 strains, selected independently of the 38 strains chosen for 5-HT analyses based on availability, were used for to obtain transcriptome profiles. All lines utilized are provided in Suppl File 1. Animals were grouped house and maintained on a 12:12 light/dark cycle with food and water available ad libitum.
Blood and Brain Tissue Neurochemical Measurements
All tissue procurements were performed under approved IACUC protocols of the Oak Ridge National Laboratory or Vanderbilt University. Brains were procured following rapid decapitation in the absence of anesthesia to limit artifactual alterations in 5-HT, 5-HIAA and mRNA levels. Trunk blood was obtained from the same animals used for brain harvests. Regions dissected from brain were determined based on landmarks provided by the Paxinos and Watson Atlas of the Mouse Brain (Watson and Paxinos 2010). Midbrain was collected to provide measures of 5-HT and metabolite levels in the region of the dorsal and median raphe nuclei and their local projects and was defined as brain tissue lying between the anterior and posterior margins of the inferior and superior colliculi, respectively (AP approximately -3.25 to -5.02 mm relative to Bregma). The diencephalon, constituted primarily by the thalamus and hypothalamus, was isolated to provide a measure of 5-HT and metabolite levels in a significant projection area of raphe neurons, and was defined as tissue lying between the anterior margin of the third ventricle and the anterior margin of the midbrain segment noted above, minus the basal ganglia (AP approximately -0.80 to -3.25 mm relative to Bregma). For assessment of 5-HT and 5-HIAA levels, sections were homogenized, using an Omni Tissue Homogenizer, in 100-750 μl of 0.1M trichloroacetic acid (TCA), 10 mM sodium acetate, 0.1 mM EDTA, 5 ng/ml isoproterenol (as internal standard) and 10.5 % methanol (pH 3.8). Whole blood from BXD mice was collected in 1.5 mL venoject tubes containing dry EDTA. Equal amounts of sample were mixed with extraction solvent (0.8 M perchloric acid, 0.1 M ascorbic acid and 10 mM EDTA), vortexed for 15 seconds and spun in a microfuge at 10,000 ×g for 10 minutes with supernatants collected. 5-HT and 5-HIAA were determined by HPLC through the Neurochemistry Core of the Vanderbilt Brain Institute, utilizing an Antec Decade II (oxidation: 0.5) electrochemical detector operated at 33° C. Twenty μl samples of the supernatant were injected using a Water 717+ autosampler onto a Phenomenex Nucleosil (5u, 100A) C18 HPLC column (150 × 4.60 mm). Samples were eluted with a mobile phase consisting of 89.5% 0.1 M TCA, 10 mM sodium acetate, 0.1 mM EDTA and 10.5 % methanol (pH 3.8). Solvent was delivered at 0.6 ml/min using a Waters 515 HPLC pump. HPLC control and data acquisition were managed by Millennium 32 software.
RNA Isolation and Microarray Analysis
Midbrain RNA was extracted using an RNeasy mini kit (Qiagen) according to manufacture's instructions. Samples were fragmented and hybridized using Agilent hybridization kit (#5188-5242). Arrays were hybridized for 17 hours at 65 C. Microarray analysis was performed on an Agilent-028005 SurePrint G3 Mouse GE 8×60K platform in the VANTAGE (Vanderbilt Technologies for Advanced Genomics) core laboratory. To make gene expression data comparable across samples, the processed signal values (gProcessedSignal) were subjected to a quantile normalization. Next, the data were log transformed and then gene-wise normalized using Z-scores. Data were rescaled to a mean of 8 units with a standard deviation of 2 units (the 2Z + 8 normalization used in GeneNetwork).
A subset of ∼50 probes with bimodal Mendelian patterns of expression were used as internal (sample-specific) genetic markers. These Mendelian probes are almost always associated with QTLs with LOD scores above 20 that map precisely to location of the cognate gene. (Rare bimodal probes with trans expression QTLs are usually associated with annotation errors or genome assembly errors and do not materially affect this QC step.) Our transcriptome studies were blinded to strain ID and then assigned based on the internal cis eQTL markers noted above. The final verified data has been deposited in GEO and in GN and reflects this systematic genotype QC process. Transcriptome-derived gene networks were generated using Cytoscape 2.8.3 (Cytoscape Consortium). Input data files (node and network) were generated from WebQTL correlation analysis module.
Statistical Analyses
Biochemical data were analyzed using standard analysis of variance (ANOVA) and correlation analysis (Stata 11, College Station, TX) with P < 0.05 taken as significant. As RI strains lack heterozygous samples and thus overestimate additive genetic variance, adjusted heritability (h2) of the traits were estimated using Hegmann and Possidente's method (Hegmann and Possidente 1981). Significance of heritability was calculated by the F-test comparing within-strain vs. between-strain variances. Quantitative trait locus mapping was performed using the WebQTL module of GeneNetwork (http://www.genenetwork.org). Quantitative trait loci (QTL) were calculated from the likelihood ratio statistic (LRS) and logarithm of odds (LOD) scores. Statistical significance was determined on data sets subjected to 2,000 permutations. Significant QTLs were defined as the LRS value that represents a genome-wide P value of 0.05. Suggestive QTLs were defined as having an LRS value a genome-wide P value of 0.63 since this value yields, on average, one false positive per genome scan (Lander and Kruglyak 1995). Thus, roughly one-third of scans at this threshold will yield no false positive, one-third will yield one false positive, and one-third will yield two or more false positives. Although a permissive threshold, such estimates call attention to loci that may derive from underpowered analyses and can be pursued with additional lines/approaches. Confidence intervals were determined as the location at which the LOD value is reduced by 1.5. A 1.5 LOD cutoff has been estimated to reflect a 95% confidence that genes driving the QTL lay within this range (Visscher et al. 1996; Dupuis and Siegmund 1999). Phenotypic correlation analyses were performed by comparing measured 5-HT traits with archived BXD phenotypes in the GeneNetwork database, with correlation coefficients and P values calculated using Spearman's rank tests (uncorrected P < 0.05 was considered significant).
To assess whether midbrain gene products identified as correlates of 5-HT homeostasis are likely to reflect cell-autonomous associations with neurochemical measures, we asked whether these genes are 1) exhibit colocalization in the adult mouse brain with Tph2 or Slc6a4 gene expression (http://mouse.brain-map.org) using the Anatomic Gene Expression Atlas and the NeuroBlast tool (Hawrylycz et al. 2011), or 2) expressed selectively in day 12.5 embryonic 5-HT neurons (flow-sorted, ePET-1:YFP positive), relative to non-ePET-1:YFP expressing cells in the midbrain (Wylie et al. 2010).
Online Data Access
All neurochemical measurements and midbrain gene expression data for the BXD strains reported in this study have been deposited into the GeneNetwork database at http://www.genenetwork.org. Neurochemical traits are stored under the category of “Central nervous system: Neurochemistry”. Our midbrain gene expression data set “VU BXD Midbrain Agilent SurePrint G3 Mouse GE (May12) Quantile” is archived under “Midbrain mRNA” with GN accession number GN381.
Results
BXD strain variation and heritability of brain and blood 5-HT measures
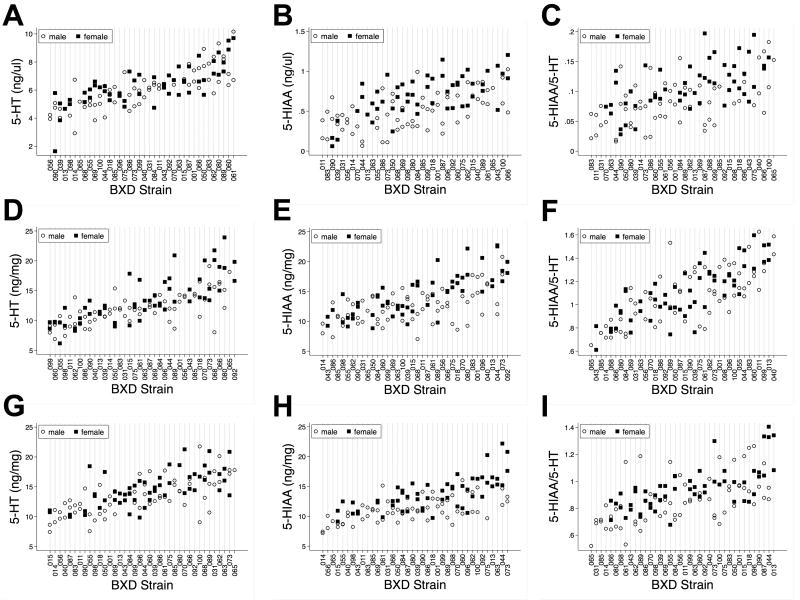
Using HPLC analysis of rapidly dissected, flash-frozen blood, midbrain and diencephalon, we determined levels of 5-HT and the 5-HT metabolite 5-HIAA from 38 BXD strains. Levels of both 5-HT and 5-HIAA, as well as the computed 5-HT turnover (5-HIAA/5-HT ratio) showed significant variation across strains for all tissues and for both genders (Figure 1), varying ∼2-3 fold. When evaluating the heritability of these measures, we found significantly greater (P < 0.05) between- versus intra-strain variation for some but not all of these measures (Suppl Tables 1 and 2), with adjusted heritability (h2) estimates of those measures reaching significance to range between 50-72%. We found 5-HT levels to be significantly heritable in all three regions, for both genders, whereas 5-HIAA levels were significantly heritable in all three tissues for females, but only significantly heritable in midbrain for males. Computed 5-HT turnover estimates for females showed significant heritability in both brain regions, but not in blood, whereas for males, only midbrain values demonstrated significant heritability.
Figure 1.
Levels of 5-HT, 5-HIAA and computed 5-HIAA/5-HT ratios across BXD RI lines. A) Blood 5-HT; B) Blood 5-HIAA; C) Blood 5-HIAA/5-HT; D) Midbrain 5-HT; E) Midbrain 5-HIAA; F) Midbrain 5-HIAA/5-HT; G) Diencephalon 5-HT; H) Diencephalon 5-HIAA and I) Diencephalon 5-HIAA/5-HT. Male animals are represented by open circles and female animals are represented by solid black squares.
Gender-dependent relationships between 5-HT traits
Since significant heritability of multiple 5-HT traits was evident when analyzing male and female animals separately, we asked whether these measures correlate between genders, and therefore whether they may be modulated by common genetic influences. In assessing lines where both male and female samples were available, we found that male and female 5-HT levels within midbrain or blood, but not diencephalon, to be significantly correlated (Spearman Rank Test: midbrain (N = 24), r = 0.54, P = 0.006; blood (N = 25), r = 0.62, P < 0.001; diencephalon (N = 25), r = 0.06, P = 0.77). In contrast, 5-HIAA levels were not significantly correlated across any region (midbrain, r = -0.04, P = 0.87; diencephalon, r = 0.05, P = 0.81; blood, r = 0.14, P = 0.50). Similarly, we found no correlation between genders for 5-HT turnover (midbrain, r = 0.33, P = 0.11; diencephalon, r = 0.09, P = 0.67; blood, r = -0.07, P = 0.74).
Region-dependent relationships between 5-HT traits
Next, we sought to assess whether 5-HT, 5-HIAA and 5-HT turnover are correlated with each other, and if so, whether these relationships vary by region and by gender. In all three regions, regardless of gender, 5-HT levels significantly correlated with 5-HIAA levels, consistent with their relationship in a common metabolic pathway (Tables 1 and 2). Within the two brain regions, 5-HT levels also correlated significantly with 5-HT turnover, whereas in blood this relationship was absent, possibly a reflection of the spatial separation between the major sites of 5-HT synthesis (gut enterochromaffin cells) and blood storage/release (platelets).
Table 1.
Correlations of male serotonergic traits between blood, midbrain and diencephalon. Values are pair-wise correlation coefficients between strain means.
| Males (34 Strains) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain Mean Correlations | Midbrain | Diencephalon | Blood | |||||||
| 5HT | 5HIAA | 5HIAA/5HT | 5HT | 5HIAA | 5HIAA/5HT | 5HT | 5HIAA | 5HIAA/5HT | ||
| Midbrain | 5HT | - | ||||||||
| 5HIAA | 0.41* | - | ||||||||
| 5HIAA/5HT | -0.60* | 0.47* | - | |||||||
| Diencephalon | 5HT | 0.37* | -0.01 | -0.33 | - | |||||
| 5HIAA | -0.02 | 0.24 | 0.20 | 0.59* | - | |||||
| 5HIAA/5HT | -0.40* | 0.30 | 0.59* | -0.61* | 0.25 | - | ||||
| Blood | 5HT | 0.11 | 0.20 | 0.17 | 0.07 | 0.20 | 0.17 | - | ||
| 5HIAA | 0.13 | -0.02 | -0.08 | 0.16 | 0.07 | -0.01 | 0.35* | - | ||
| 5HIAA/5HT | -0.001 | -0.15 | -0.16 | 0.11 | -0.03 | -0.10 | -0.17 | 0.85* | - | |
p < 0.05.
Table 2.
Correlations of female serotonergic traits between blood, midbrain and diencephalon. Values are pair-wise correlation coefficients between strain means.
| Females (29 Strains) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain Mean Correlations | Midbrain | Diencephalon | Blood | |||||||
| 5HT | 5HIAA | 5HIAA/5HT | 5HT | 5HIAA | 5HIAA/5HT | 5HT | 5HIAA | 5HIAA/5HT | ||
| Midbrain | 5HT | - | ||||||||
| 5HIAA | 0.68* | - | ||||||||
| 5HIAA/5HT | -0.53* | 0.24 | - | |||||||
| Diencephalon | 5HT | 0.24 | 0.09 | -0.14 | - | |||||
| 5HIAA | 0.12 | 0.41* | 0.28 | 0.58* | - | |||||
| 5HIAA/5HT | -0.09 | 0.37* | 0.45* | -0.39* | 0.52* | - | ||||
| Blood | 5HT | 0.10 | 0.16 | 0.13 | 0.21 | 0.10 | 0.03 | - | ||
| 5HIAA | 0.08 | 0.21 | 0.23 | -0.06 | 0.05 | 0.15 | 0.63* | - | ||
| 5HIAA/5HT | 0.02 | 0.09 | 0.14 | -0.31 | -0.06 | 0.12 | 0.11 | 0.83* | - | |
p < 0.05
Inter-regional correlations of 5-HT traits
Midbrain raphe 5-HT neurons elaborate axonal projections that both remain within the midbrain and/or project to higher forebrain areas, including multiple nuclei of the diencephalon. In the latter area, 5-HT output is regulated by multiple presynaptic inputs that differ from those that regulate the excitability and 5-HT synthesis/metabolism capacity of midbrain 5-HT cell bodies (Pineyro et al. 1995; Barnes and Sharp 1999; Stamford et al. 2000). Nonetheless, 5-HT levels in male, but not female midbrain samples, were significantly correlated with diencephalon 5-HT levels (Tables 1 and 2). In contrast, female, but not male midbrain 5-HIAA levels are correlated between the two tissues. These gender distinctions in regional relationships were lost when examining 5-HT turnover, where for both males and females we found midbrain and diencephalon values to be significantly correlated. In contrast to the significant correlations that exist for 5-HT traits when comparing brain regions, no significant correlations, for either gender or using gender-pooled samples, were evident comparing blood and midbrain or blood and diencephalon (Tables 1 and 2).
Correlations with archived BXD phenotypes
Next we sought to determine how 5-HT traits associated with archived phenotypic measures deposited in the GeneNetwork BXD database, with a cutoff value set a P value < 0.05 (Spearman Rank Test) (Suppl File 2). We chose to focus on the analyses of midbrain and blood 5-HT levels using combined genders, as 5-HT levels were significantly correlated between males and females in both regions, thereby providing more statistical power (Suppl File 3 and 4).
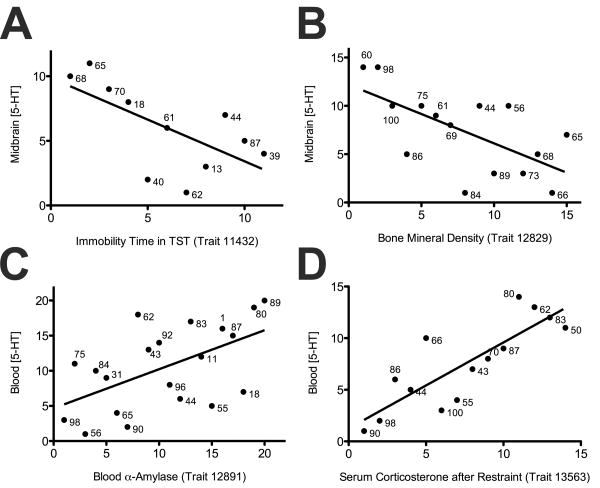
For pooled midbrain 5-HT levels, we observed significant, negative correlations between 5-HT levels and the duration of immobility in tail suspension test from two independent studies (trait 11432: r = -0.65, P = 0.03; trait 12554: r = -0.83, P = 0.04, Figure 2A), a measure known to be responsive to acute SSRI administration (Cryan et al. 2005). We also found a number of iron phenotypes to exhibit significant correlations, including liver iron levels (r = 0.86, P = 0.01) and blood iron binding capacity (r = 0.68, P = 0.04) (Suppl File 3). Handling-induced convulsion (HIC) exhibited a significant, negative correlation (r = -0.49, P = 0.03) and several traits measuring bone mineral densities (BMDs) showed negative correlations with midbrain 5-HT levels (Figure 2B).
Figure 2.
Correlations between midbrain or blood 5-HT levels and selected archived BXD phenotypes. (A) midbrain 5-HT levels and immobility time in tail suspension test (trait 11432, r = -0.65, P = 0.03); (B) midbrain 5-HT levels and bone mineral density (trait 12829, r = -0.63, P = 0.01); (C) blood 5-HT levels and blood α-amylase levels (trait 12891, r = 0.55, P = 0.01) and (D) blood 5-HT levels and serum corticosterone levels after restraint (trait 13563, r = 0.83, P < 0.0001). All P values are calculated using Spearman's rank test.
Interestingly, as with midbrain 5-HT, we found correlations for pooled blood 5-HT levels with several iron-associated traits including plasma iron levels (r = 0.82, P = 0.02) and liver iron levels (r = 0.96, P < 0.0001) (Suppl File 4). Multiple blood measures also exhibited unique correlations with blood 5-HT traits, including TRBV4+ T cell (r = 0.89, P = 0.004), alpha-amylase (r = 0.55, P < 0.01, Figure 2C), triglyceride (r = 0.43, P = 0.03) and hematocrit (r = 0.48, P = 0.04) levels. Serum corticosterone levels after restraint stress, a paradigm used to examine roles of 5-HT in despair behavior (Ayada et al. 2002), also showed significant positive correlation with blood 5-HT levels (r = 0.83, P < 0.0001, Figure 2D).
Lastly, we inspected phenotypes that correlate with both brain and blood serotonergic traits. Deoxycorticosterone (DOC) levels in cerebral cortex exhibited significant negative correlations with both midbrain and blood 5-HT levels (r = -0.54, P = 0.009 for midbrain; r = -0.42, P = 0.05 for blood). When inspecting individual traits separated by genders, we found that intraocular pressure (IOP) correlated positively with both male midbrain and male blood 5-HT levels (midbrain: r = 0.46, P = 0.01; blood: r = 0.45, P = 0.01). Morphine response traits (locomotion and vertical activity) displayed significant positive correlations with female diencephalon 5-HIAA levels, but significant negative correlations with female blood 5-HIAA levels (diencephalon: r = 0.38 to 0.42, P = 0.02 to 0.04; blood: r = -0.38 to -0.40, P = 0.03 to 0.04). Finally, we found several traits correlating with both male diencephalon and male blood 5-HT turnover ratios, including liver iron levels (diencephalon: r = -0.86, P = 0.004, blood: r = -0.74, P = 0.03), brain weight (diencephalon: r = 0.81, P = 0.01, blood: r = 0.76, P = 0.03), hippocampus weight (diencephalon: r = 0.71, P = 0.05, blood: r = 0.86, P = 0.004), as well as adult neurogenesis (diencephalon: r = -0.79, P = 0.02, blood: r = -0.76, P = 0.03).
QTL Analysis of 5-HT Levels on Pooled Midbrain and Blood Samples
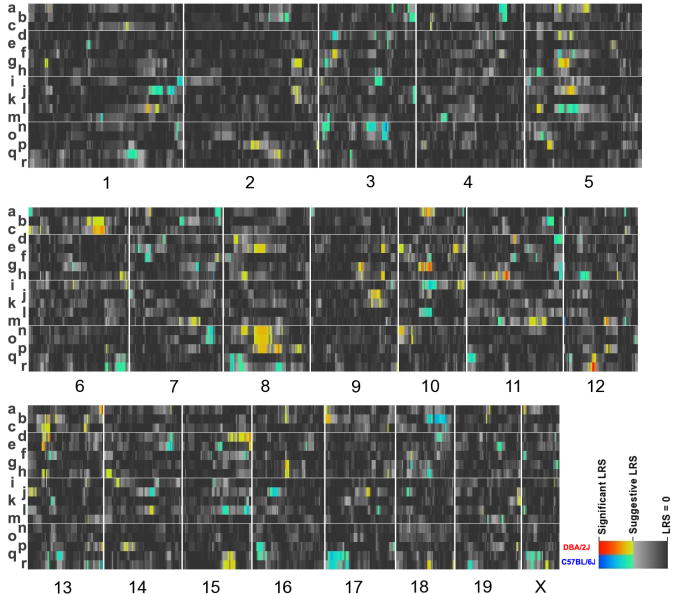
As both CNS and blood 5-HT measures exhibited significant heritability, we next sought to nominate genomic loci associated with variation in 5-HT, 5-HIAA and 5-HT turnover using SNP-based interval mapping (Lander and Botstein 1989). We analyzed each gender separately, and provide a heat map representation of QTLs for each trait in Figure 3, affording an overview of region, trait and gender-specific loci. Similar to the phenotype correlation analysis noted above, we also pursued QTL analysis of midbrain and blood 5-HT using combined genders.
Figure 3.
Heatmap of QTL intervals for serotonergic traits: a) female diencephalon 5-HT; b) female diencephalon 5-HIAA; c) female diencephalon 5-HIAA/5-HT; d) male diencephalon 5-HT; e) male diencephalon 5-HIAA; f) male diencephalon 5-HIAA/5-HT; g) female midbrain 5-HT; h) female midbrain 5-HIAA; i) female midbrain 5-HIAA/5-HT; j) male midbrain 5-HT; k) male midbrain 5-HIAA; l) male midbrain 5-HIAA/5-HT; m) female blood 5-HT; n) female blood 5-HIAA; o) female blood 5-HIAA/5-HT; p) male blood 5-HT; q) male blood 5-HIAA and r) male blood 5-HIAA/5-HT. Numbers on the bottom specify the mouse chromosome linked to the heat map of QTLs above. Genomic loci are colored according to the strength of correlation. Genomic regions where DBA/2J alleles associate with higher trait values are shown in red whereas genomic regions where C57BL/6J alleles associate with higher trait values are shown in blue.
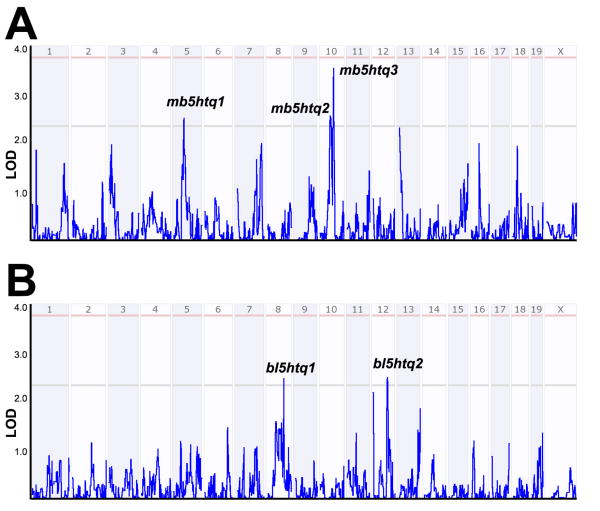
From the interval analysis of the pooled midbrain 5-HT traits we identified three suggestive loci (Figure 4A): mb5htq1 centered at 56 Mb on chromosome 5 (LOD = 2.52), mb5htq2 centered at 60 Mb on chromosome 10 (LOD = 2.56), and mb5htq3 centered at 73 Mb on chromosome 10 (LOD = 3.60). mb5htq1 was also identified as a potential QTL for diencephalon 5-HT (both genders) and 5-HT turnover (males). Both mb5htq2 and mb5htq3 also showed strong correlation with pooled midbrain 5-HT turnover ratios, with mb5htq3 reaching a genome-wide significance level (LOD = 3.75). In addition, mb5htq3 was also associated with several other brain 5-HT traits including female diencephalon 5-HT levels and male diencephalon 5-HT turnover ratios.
Figure 4.
Interval analysis of 5-HT levels in (A) midbrain and (B) blood. Numbers above red lines designate mouse chromosomes. Red lines designate LOD (Log of Odds) scores where gene variation achieves significance (see Methods) whereas gray lines designate LOD scores achieving suggestive threshold.
As expected from the lack of trait correlation found between blood and brain 5-HT traits, none of these loci were identified in the QTL analysis of blood 5-HT traits (Figure 3, see below). Interval analysis of pooled blood 5-HT levels revealed two suggestive loci: bl5htq1 on chromosome 8 (92 Mb, LOD = 2.50) and bl5htq2 on chromosome 12 (82 Mb, LOD = 2.44) (Figure 4B). When separated by gender, we found bl5htq1 was primarily driven by male samples, whereas bl5htq2 was mainly contributed by female samples. Genes that fall in the midbrain and blood QTLs using a 1.5 LOD based confidence interval can be found in Suppl Files 5 and 6, respectively. The reader is reminded that specific genes driving associations may lie outside the QTL borders, and thus additional experimental efforts are needed to validate candidates.
Association of 5-HT traits with the midbrain transcriptome
To inspect genes lying within the QTLs noted above where changes in expression also correlate with 5-HT traits, we pursued a transcriptome analysis. We chose to focus on the male midbrain for our transcriptome analysis, owing to the presence in this region of neurons that provide the bulk of forebrain 5-HT. Male samples were selected for practical reasons, though one could argue that female samples might possess higher variation owing to estrous cycling, possibly confounding analyses.
In Suppl Figure 1, we provide networks of genes whose expression in the male midbrain significantly correlated with 5-HT traits (the complete list can be found in Suppl File 7). Surprisingly, many of the genes that might be expected from prior studies to correlate in expression, such as the transcription factors Fev and Lmx1b, as well as Tph2 and Slc6a4, were not among the significantly correlated genes (uncorrected P < 0.05). We did identify Gch1 (GTP cyclohroxylase), an enzyme involved in TPH2 cofactor synthesis (positive correlation with 5-HT), Maoa (produces MAOA; positive correlation with 5-HT), and Slc18a2 (produces VMAT2; positive correlation with 5-HIAA). To provide insight into networks that support expression of these genes, and therefore could undergird the relationship each has with 5-HT homeostasis, we determined the identity of genes whose expression co-varies across BXD lines (Suppl Figure 2).
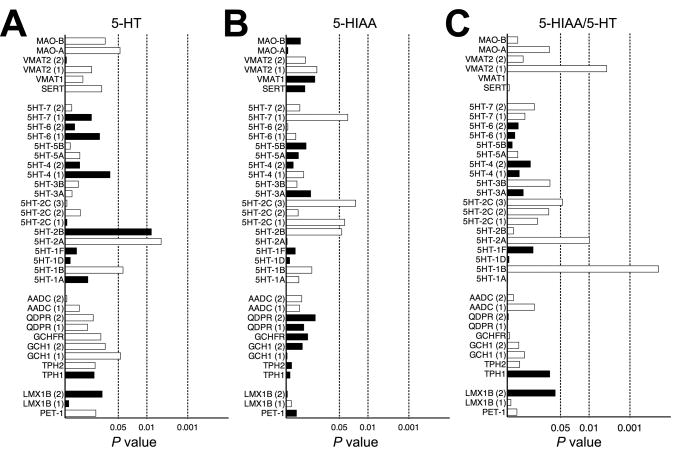
In our transcriptome analysis, we also identified multiple 5-HT receptors whose expression correlated with specific 5-HT traits. Thus, 5-HT1B and 5-HT2A correlated positively with midbrain 5-HT whereas 5-HT2B exhibited a negative correlation. 5HT1B, 5-HT2A and 5-HT7 receptors gene expression also positively correlated with 5-HT turnovers. Finally, mRNA expression of 5-HT2B, 5-HT2C and 5-HT7 receptors correlated positively correlated with 5-HIAA levels (Figure 5).
Figure 5.
Gene expression correlations of genes that have known roles in 5-HT homeostasis and signaling with midbrain 5-HT traits: (A) 5-HT levels, (B) 5-HIAA levels and (C) 5-HIAA/5-HT ratios. Dashed lines represent significance levels of 0.05, 0.01 and 0.001 (Spearman's rank test). Open bars represent positive correlations and filled bars denote negative correlations. Genes with multiple probes, designated by parentheses, are plotted separately.
To assess whether the gene expression networks linked to midbrain 5-HT homeostasis could act cell autonomously, we compared this list to a list of genes found to display anatomical correlation with either the pattern of Tph2 or Slc6a4 gene expression using the NeuroBlast tool (Hawrylycz et al. 2011) or with an embryonic raphe 5-HT neuron-specific gene list (Wylie et al. 2010). Out of the total of 341 genes nominated by NeuroBlast, 60 showed significant correlations with either 5-HT, 5-HIAA or 5-HT turnover rates (Suppl File 8). Out of the total of 354 genes enriched in embryonic 5-HT neurons, 64 correlated significantly with either 5-HT, 5-HIAA or 5-HT turnover rates (Suppl File 8). Among the 9 genes present in all three lists, 4 genes (Gch1, Maoa, S100a10, Slc18a2) are well known genes in 5-HT system. The 5 genes with unknown 5-HT-related functions are Dach2, Gng4, Grb10, Oprl1 and Vat1l.
Finally, given that the bulk of serotonergic projections from the midbrain innervate more anterior brain regions, we took this opportunity to identify genes whose midbrain expression might determine diencephalic serotonergic tone (Suppl File 9). In contrast to the midbrain, none of the well-characterized genes participating in 5-HT synthesis and metabolism significantly correlated to diencephalon serotonergic traits, suggesting a greater degree of local control over 5-HT traits in this projection area. The expression of three 5-HT receptors, however, 5-HT1D, 5-HT7 and 5-HT1B, exhibited significant positive correlations with these measures (5-HT1D: 5-HT levels, r = 0.65, P = 0.005; 5-HT7: 5-HIAA levels, r = 0.59, P = 0.01; 5-HT1B: 5-HIAA levels, r = 0.57, P = 0.02).
Relationship of midbrain 5-HT associated gene expression profiles to midbrain 5-HT QTLs
Of the 348 genes associated with mb5htq1, 2 and 3 (Figure 3), 43 were found to be among the genes whose expression was also found to be associated with midbrain 5-HT traits (Suppl File 10). Within this tabulation, the reader will find scores from visual inspection of expression patterns for genes expressed by raphe neurons, providing a further, prioritized list that can be used for future experiments.
Discussion
BXD RI strains constitute a powerful reference population with which to assess both genotype-phenotype and phenotype-phenotype relationships, as well as to nominate genomic loci that influence measured traits (Lu et al. 2008; Houtkooper et al. 2013). Surprisingly, neurochemical measures are comparatively limited in the GeneNetwork BXD database, including CNS and peripheral levels of 5-HT and 5-HIAA. It should be recognized at the outset that the conclusions we draw from our analyses are limited by the number of lines and animals per line utilized as well as the extent of variation in 5-HT traits exhibited by the parental C57BL/6J and DBA/2J parental lines. For genetic influences that impact 5-HT trait variation more broadly (e.g. after stress or in the context of other genetic variants), no associations can be inferred. Thus, with respect to biochemical, molecular and behavioral correlations pursued with the BXD resource, “absence of evidence does not mean evidence of absence”.
Our efforts revealed an ∼2 fold variation in the levels of 5-HT, 5-HIAA and 5-HT turnover in both brain and whole blood extracts across BXD strains, with the across vs within strain variation establishing a significant heritability of these measures. Consistent with these findings, human blood 5-HT levels have been found to exhibit high heritability (Weiss et al. 2005). Significant differences in blood 5-HT levels have also been reported in C57BL/6 and BALB/c mouse strains (Flood et al. 2012). 5-HT measures in the midbrain, where raphe cell bodies and their intrinsic projections reside, were significantly correlated with the same measures in the diencephalon, a region that receives ascending raphe projections. In contrast, BXD strains did not demonstrate a significant correlation between brain and blood 5-HT measures. These findings are consistent with observations obtained in ICR mice (Pietraszek et al. 1992) and suggest that basal CNS 5-HT homeostasis is greatly impacted by locally derived factors as opposed to determinants shared with blood 5-HT. Enterochromaffin cells utilize tryptophan hydroxylase 1 (TPH1) to synthesize 5-HT whereas TPH2 supports 5-HT synthesis neurons (Walther et al. 2003) and as such, 5-HT synthesis is likely under the control of distinct regulatory networks, not to mention those received in a tissue specific manner. Nonetheless, certain features of 5-HT homeostasis are shared between blood and brain. For example, the transporter responsible for packaging 5-HT in secretory vesicles for release in neurons (VMAT2, Slc18a2) is expressed by platelets, as is SERT, the chief mechanism of 5-HT clearance in the CNS. Additionally, the MAO isoform MAOB is present in both platelets and raphe neurons (Chen et al. 1993; Jahng et al. 1997). Despite the absence of evidence for molecular relationships dictating blood and brain 5-HT traits in our studies, we must consider that factors shared between these two measures may not vary across BXD strains or may be engaged in a context-dependent manner. Nonetheless, several CNS measures, including behavioral phenotypes, correlated with both brain and blood 5-HT traits. One conspicuous correlation with 5-HT turnover was found for brain weight, hippocampal weight and hippocampal stem cell number. Studies that manipulate 5-HT turnover independent of CNS 5-HT turnover are needed to determine whether these correlations reflect a result of 5-HT homeostasis per se versus a shared determinant that may in early life impact the trajectory of brain development (Mazer et al. 1997; Janusonis et al. 2004; Bonnin et al. 2007).
Consistent with our earlier report focused on phenotypes linked to SERT coding variation (Carneiro et al. 2009; Ye and Blakely 2011), we found many iron-related traits that significantly correlate with midbrain and diencephalon 5-HT levels. The associations of several phenotypic correlates with brain 5-HT pathways have been extensively studied and add credence to our analysis. For example, adult neurogenesis in the hippocampus significantly associated with midbrain 5-HT and 5-HIAA levels and is believed to play an important role in the chronic actions of SSRIs (Santarelli et al. 2003). Additionally, CNS 5-HT has been shown to be a strong regulator of bone formation and resorption (Yadav et al. 2009). More importantly, we identified several novel, or underappreciated phenotypes linked to CNS or blood 5-HT levels, including attributes of the visual, auditory and immune systems. Combined with the candidate gene approaches discussed above, these findings encourage increased attention to these systems and their control by central and peripheral 5-HT signaling. One caveat to these associations is the large number of phenotypes (∼3800) that we accessed for our 5-HT trait correlations. A Bonferroni correction for multiple comparisons requires that we achieve a P value of < 1.5×10-5. At this stringency level, none of the trait correlations tabulated would reach statistical significance. Thus, we offer the uncorrected tabulation as a hypothesis generator whose true significance must be assessed through other experiments.
In our studies, we analyzed 5-HT trait correlations for both males and females separately as well as in pooled samples where these measures exhibited significant correlations. Although the between gender comparisons required that we use fewer lines (24 out of 38 total) that had both male and female samples, we did find that males and female 5-HT traits from midbrain and blood were significantly correlated, whereas diencephalon traits lacked such a correlation. The latter observation suggests that gender-specific determinants of 5-HT homeostasis act more strongly in distal, forebrain projection areas. It will be interesting to re-examine these relationships using females at specific phases of the estrous cycle. Both Maoa and Maob genes, as well as the Htr2c gene that encodes the 5-HT2C receptor, are located on the X chromosome in mice and humans and this localization may contribute to the gender effects we observed, particularly, the lack of gender correlations in MAO-controlled 5-HT turnover measures. Alternatively, one or more of the gender specific QTLs for CNS 5-HT levels (see below) may be under strong gonadal hormone control. Such an idea is consistent with gender-specific modulation of human brain metabolism by fenfluramine (Anderson et al. 2004) and SSRI medications (Munro et al. 2012) as well as gender-dependent modulation of brain glucose consumption by SERT expression level (Dawson et al. 2009).
Both tissue and gender specificity was further evident in our studies identifying QTLs. For example, while a suggestive QTL on chromosome 10 (mb5htq3) for midbrain 5-HT levels as well as 5-HT turnover ratios also demonstrated suggestive correlation with diencephalon 5-HT levels (females) and 5-HT turnover (males), this locus was not identified as a QTL for blood 5-HT traits (either gender). Similarly, neither suggestive QTLs for blood 5-HT levels (bl5htq1 and bl5htq2) was identified as a QTL for the equivalent traits in midbrain or diencephalon.
The strongest correlation with midbrain 5-HT levels was observed on SNP rs13480650, located within Pcdh15 gene coding for protocadherin 15 (PCDH15). PCDH15 is a cell adhesion molecule belonging to the nonclustered protocadherin subfamily in the protocadherin superfamily, playing important roles in the development of cochlea hair cells and has been associated with human Usher syndrome (Alagramam et al. 2001; Alagramam et al. 2001). Expression of a distinct cadherin gene cluster, protocadherin alpha, is required by serotonergic neuron to form proper projections (Katori et al. 2009), suggesting that multiple adhesive proteins may be critical for 5-HT neuron differentiation and signaling. Indeed, we recently validated that disruption of PCDH15 functions resulted in alterations in CNS 5-HT and SERT levels (Ye and Blakely, submitted). It is also worth noting that another cadherin gene, Cdh23, encoding cadherin 23 (CDH23 or octocadherin), locates within locus mb5htq2. CDH23 physically interacts with PCDH15 and this interaction is required for normal inner ear function (Lelli et al. 2010). Future studies that explore the interaction of CDH23 and PCDH15 in determining 5-HT homeostasis and signaling appear warranted.
In the genes identified from interval mapping of blood 5-HT levels, Nod2 (nucleotide-binding oligomerization domain containing 2) codes for Card15 (Caspase recruitment domain-containing protein 15) that is highly expressed in peripheral blood leukocytes (Strober and Watanabe 2011). Card15 is also known as inflammatory bowel disease protein 1 due to its association with inflammatory bowel disease (IBD)/Crohn's disease (Hugot et al. 2001; Ogura et al. 2001) and Blau syndrome (Miceli-Richard et al. 2001). Card15's potential regulation of 5-HT levels is also interesting given the role the protein plays in inducing immune responses to bacterial antigens, including LPS, and the presence of SERT (Marazziti et al. 1998) and 5-HT signaling (Prasad et al. 2005) in the immune system.
Assessment of midbrain gene expression patterns permitted an identification of transcriptional signatures linked to 5-HT traits. In the GeneNetwork BXD database, transcriptome profiles of multiple brain regions have been deposited, including hippocampus, striatum, amygdala, and the ventral tegmental area. However, midbrain mRNA expression profiles are lacking, precluding trait correlation analyses with expression variation derived from serotonin neurons. To remedy this situation, we pursued a microarray analysis of male midbrain mRNAs. Surprisingly, little correlation was found between 5-HT traits and the levels of expression of many genes known to control 5-HT homeostasis and signaling. Although at first surprising, this lack of correlation may simply reflect the significant control of these gene products by posttranslational mechanisms (e.g. kinase mediated regulation of Tph2 activity) that would not be detected in our mRNA analyses. We did uncover significant correlations with Gch1, Slc18a2, and Maoa, genes expressed by 5-HT neurons and that are involved in the biosynthesis, packaging and inactivation of the neurotransmitter. The network of genes associated with these molecules provides a window on other biological pathways that may rely on 5-HT signaling. One interesting gene that emerged from these networks is Hcrtr1, which encodes the hypocretin (orexin) receptor type 1. Orexin is a neuropeptide that is involved in the control of feeding behavior (de Lecea et al. 1998; Sakurai et al. 1998) and our NeuroBlast analysis suggests expression by 5-HT neurons. Agents that elicit 5-HT release diminish appetite (Curzon 1990) and Hcrtr1 null mice display behavior similar to that seen with antidepressant treatments (Scott et al. 2011). These connections, along with recent findings of variation in the HCRT1 gene that associate with unipolar depression, warrant further studies of the control of 5-HT signaling by orexin (Rainero et al. 2011). Finally, we identified multiple 5-HT receptors whose expression significantly correlated with midbrain and diencephalon 5-HT traits. Among the receptors, excitatory 5HT2A and inhibitory 5HT1B receptors showed positive correlations with both midbrain 5-HT and 5-HIAA levels, suggesting their potential roles in upregulating 5-HT synthesis. 5HT1B and 5HT7 receptors correlated with serotonergic traits measured in both brain regions, indicating their involvement in dictating global 5-HT tone in the brain. These findings, should they relate to relationships present in the human brain, may be of use in the development of better medications for the spectrum of 5-HT associated neuropsychiatric disorders.
Having established a midbrain 5-HT associated transcriptome, we used this list to filter the genes identified in our QTL analysis. Genes in this combined list should derive from an impact of cells in the midbrain on 5-HT homeostasis. A caveat to this filtering is the possibility that genes driving variations in midbrain 5-HT could act through long-range connections (e.g. expression in frontal cortex neurons that project to the raphe) but we eliminated due to our use of a midbrain mRNA data set. Nonetheless, this analysis resulted in 43 genes (Suppl File 10). Interestingly, Pcdh15, which we noted above as containing the SNP that marks the peak of the most significant QTL in our study (mb5htq3), is among this list. Additionally, a bootstrap resampling analysis of mb5htq3 yields a location that solely includes the Pcdh15 gene (data not shown). Finally, in a separate study (Ye and Blakely, submitted) examining BXD determinants of SERT protein variation, we again identified Pcdh15 as a regulatory gene dictating CNS SERT protein levels. Together, these efforts point to an unexpected relationship between the cell adhesion protein and the development and/or function of 5-HT neurons, and further support the power of the BXD paradigm.
In summary, the current study reports an interrogation of BXD RI mouse strains at multiple levels (neurochemical trait, mRNA, phenotypes) to generate rational hypothesis that could explain the genetic control of 5-HT homeostasis and signaling in both central and peripheral tissues. Despite the caveats noted above, we observed strong genetic control of both 5-HT traits and gene expression profiles. In examining these determinants, we 1) found blood and brain 5-HT traits were controlled by different sets of genetic loci, 2) identified a group of phenotypic traits that correlate with 5-HT blood and/or brain traits, 3) revealed both known and novel genes that associate with 5-HT traits. Given the success of our studies, it is evident that an even more clear picture should emerge through the use of a larger family of recombinant lines, such as those present in the Collaborative Cross, an effort based on RI strains derived from 8 inbred and outbred lines, and where genetic variation similar to that of humans (Welsh et al. 2012). Finally, as more phenotypic data (such as protein expression data for 5-HT related genes) is deposited into the GeneNetwork database, our data sets will offer additional opportunities to dissect the complex actions of 5-HT in the brain and periphery, and hopefully provide clues to disorders associated with 5-HT dysfunction.
Supplementary Material
Acknowledgments
We acknowledge the expert support of Chris Svitek, Jane Wright, Qiao Han, Sarah Whitaker, Angela Steele, Tracy Moore-Jarrett, and Kathryn Lindler in general laboratory oversight. This work was supported by NIH Award MH096972 (RDB).
References
- Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, Woychik RP. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet. 2001;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- Alagramam KN, Yuan H, Kuehn MH, Murcia CL, Wayne S, Srisailpathy CR, Lowry RB, Knaus R, Van Laer L, Bernier FP, Schwartz S, Lee C, Morton CC, Mullins RF, Ramesh A, Van Camp G, Hageman GS, Woychik RP, Smith RJ. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Human molecular genetics. 2001;10:1709–1718. doi: 10.1093/hmg/10.16.1709. [DOI] [PubMed] [Google Scholar]
- Anderson AD, Oquendo MA, Parsey RV, Milak MS, Campbell C, Mann JJ. Regional brain responses to serotonin in major depressive disorder. J Affect Disord. 2004;82:411–417. doi: 10.1016/j.jad.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Arias B, Catalan R, Gasto C, Gutierrez B, Fananas L. 5-HTTLPR polymorphism of the serotonin transporter gene predicts non-remission in major depression patients treated with citalopram in a 12-weeks follow up study. Journal of clinical psychopharmacology. 2003;23:563–567. doi: 10.1097/01.jcp.0000095350.32154.73. [DOI] [PubMed] [Google Scholar]
- Ayada K, Tadano T, Endo Y. Gnawing behavior of a mouse in a narrow cylinder: a simple system for the study of muscle activity, fatigue, and stress. Physiology & behavior. 2002;77:161–166. doi: 10.1016/s0031-9384(02)00844-2. [DOI] [PubMed] [Google Scholar]
- Barnes NM, Sharp T. A review of central 5-HT receptors and their function. Neuropharmacology. 1999;38:1083–1152. doi: 10.1016/s0028-3908(99)00010-6. [DOI] [PubMed] [Google Scholar]
- Barondes SH. Thinking about Prozac. Science. 1994;263:1102–1103. doi: 10.1126/science.8108727. [DOI] [PubMed] [Google Scholar]
- Barr LC, Goodman WK, McDougle CJ, Delgado PL, Heninger GR, Charney DS, Price LH. Tryptophan depletion in patients with obsessive-compulsive disorder who respond to serotonin reuptake inhibitors. Archives of general psychiatry. 1994;51:309–317. doi: 10.1001/archpsyc.1994.03950040053007. [DOI] [PubMed] [Google Scholar]
- Barton DA, Esler MD, Dawood T, Lambert EA, Haikerwal D, Brenchley C, Socratous F, Hastings J, Guo L, Wiesner G, Kaye DM, Bayles R, Schlaich MP, Lambert GW. Elevated brain serotonin turnover in patients with depression: effect of genotype and therapy. Arch Gen Psychiatry. 2008;65:38–46. doi: 10.1001/archgenpsychiatry.2007.11. [DOI] [PubMed] [Google Scholar]
- Bonnin A, Goeden N, Chen K, Wilson ML, King J, Shih JC, Blakely RD, Deneris ES, Levitt P. A transient placental source of serotonin for the fetal forebrain. Nature. 2011;472:347–350. doi: 10.1038/nature09972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin A, Torii M, Wang L, Rakic P, Levitt P. Serotonin modulates the response of embryonic thalamocortical axons to netrin-1. Nat Neurosci. 2007;10:588–597. doi: 10.1038/nn1896. [DOI] [PubMed] [Google Scholar]
- Carneiro AM, Airey DC, Thompson B, Zhu CB, Lu L, Chesler EJ, Erikson KM, Blakely RD. Functional coding variation in recombinant inbred mouse lines reveals multiple serotonin transporter-associated phenotypes. Proc Natl Acad Sci U S A. 2009;106:2047–2052. doi: 10.1073/pnas.0809449106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, Taylor A, Poulton R. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Chen K, Wu HF, Shih JC. The deduced amino acid sequences of human platelet and frontal cortex monoamine oxidase B are identical. Journal of neurochemistry. 1993;61:187–190. doi: 10.1111/j.1471-4159.1993.tb03554.x. [DOI] [PubMed] [Google Scholar]
- Cook EH, Leventhal BL. The serotonin system in autism. Current opinion in pediatrics. 1996;8:348–354. doi: 10.1097/00008480-199608000-00008. [DOI] [PubMed] [Google Scholar]
- Cross S, Kim SJ, Weiss LA, Delahanty RJ, Sutcliffe JS, Leventhal BL, Cook EH, Jr, Veenstra-Vanderweele J. Molecular genetics of the platelet serotonin system in first-degree relatives of patients with autism. Neuropsychopharmacology. 2008;33:353–360. doi: 10.1038/sj.npp.1301406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Mombereau C, Vassout A. The tail suspension test as a model for assessing antidepressant activity: review of pharmacological and genetic studies in mice. Neuroscience and biobehavioral reviews. 2005;29:571–625. doi: 10.1016/j.neubiorev.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Curzon G. Serotonin and appetite. Ann N Y Acad Sci. 1990;600:521–530. doi: 10.1111/j.1749-6632.1990.tb16907.x. discussion 530-521. [DOI] [PubMed] [Google Scholar]
- Dawson N, Ferrington L, Olverman HJ, Harmar AJ, Kelly PA. Sex influences the effect of a lifelong increase in serotonin transporter function on cerebral metabolism. Journal of neuroscience research. 2009;87:2375–2385. doi: 10.1002/jnr.22062. [DOI] [PubMed] [Google Scholar]
- de Lecea L, Kilduff TS, Peyron C, Gao X, Foye PE, Danielson PE, Fukuhara C, Battenberg EL, Gautvik VT, Bartlett FS, 2nd, Frankel WN, van den Pol AN, Bloom FE, Gautvik KM, Sutcliffe JG. The hypocretins: hypothalamus-specific peptides with neuroexcitatory activity. Proc Natl Acad Sci U S A. 1998;95:322–327. doi: 10.1073/pnas.95.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado PL, Charney DS, Price LH, Aghajanian GK, Landis H, Heninger GR. Serotonin function and the mechanism of antidepressant action. Reversal of antidepressant-induced remission by rapid depletion of plasma tryptophan. Archives of general psychiatry. 1990;47:411–418. doi: 10.1001/archpsyc.1990.01810170011002. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Siegmund D. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics. 1999;151:373–386. doi: 10.1093/genetics/151.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood ZC, Engel DL, Simon CC, Negherbon KR, Murphy LJ, Tamavimok W, Anderson GM, Janusonis S. Brain growth trajectories in mouse strains with central and peripheral serotonin differences: relevance to autism models. Neuroscience. 2012;210:286–295. doi: 10.1016/j.neuroscience.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Gershon MD. 5-Hydroxytryptamine (serotonin) in the gastrointestinal tract. Current opinion in endocrinology, diabetes, and obesity. 2013;20:14–21. doi: 10.1097/MED.0b013e32835bc703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjerris A, Sorensen AS, Rafaelsen OJ, Werdelin L, Alling C, Linnoila M. 5-HT and 5-HIAA in cerebrospinal fluid in depression. J Affect Disord. 1987;12:13–22. doi: 10.1016/0165-0327(87)90056-5. [DOI] [PubMed] [Google Scholar]
- Hawrylycz M, Ng L, Page D, Morris J, Lau C, Faber S, Faber V, Sunkin S, Menon V, Lein E, Jones A. Multi-scale correlation structure of gene expression in the brain. Neural Netw. 2011;24:933–942. doi: 10.1016/j.neunet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Hegmann JP, Possidente B. Estimating genetic correlations from inbred strains. Behav Genet. 1981;11:103–114. doi: 10.1007/BF01065621. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Jahng JW, Houpt TA, Wessel TC, Chen K, Shih JC, Joh TH. Localization of monoamine oxidase A and B mRNA in the rat brain by in situ hybridization. Synapse. 1997;25:30–36. doi: 10.1002/(SICI)1098-2396(199701)25:1<30::AID-SYN4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Janusonis S, Gluncic V, Rakic P. Early serotonergic projections to Cajal-Retzius cells: relevance for cortical development. J Neurosci. 2004;24:1652–1659. doi: 10.1523/JNEUROSCI.4651-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Archives of general psychiatry. 2011;68:444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katori S, Hamada S, Noguchi Y, Fukuda E, Yamamoto T, Yamamoto H, Hasegawa S, Yagi T. Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J Neurosci. 2009;29:9137–9147. doi: 10.1523/JNEUROSCI.5478-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelli A, Kazmierczak P, Kawashima Y, Muller U, Holt JR. Development and regeneration of sensory transduction in auditory hair cells requires functional interaction between cadherin-23 and protocadherin-15. J Neurosci. 2010;30:11259–11269. doi: 10.1523/JNEUROSCI.1949-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenders JW, Eisenhofer G, Abeling NG, Berger W, Murphy DL, Konings CH, Wagemakers LM, Kopin IJ, Karoum F, van Gennip AH, Brunner HG. Specific genetic deficiencies of the A and B isoenzymes of monoamine oxidase are characterized by distinct neurochemical and clinical phenotypes. J Clin Invest. 1996;97:1010–1019. doi: 10.1172/JCI118492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller CR, Hamer DH, Murphy DL. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- Lu L, Wei L, Peirce JL, Wang X, Zhou J, Homayouni R, Williams RW, Airey DC. Using gene expression databases for classical trait QTL candidate gene discovery in the BXD recombinant inbred genetic reference population: mouse forebrain weight. BMC Genomics. 2008;9:444. doi: 10.1186/1471-2164-9-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JJ, McBride PA, Brown RP, Linnoila M, Leon AC, DeMeo M, Mieczkowski T, Myers JE, Stanley M. Relationship between central and peripheral serotonin indexes in depressed and suicidal psychiatric inpatients. Archives of general psychiatry. 1992;49:442–446. doi: 10.1001/archpsyc.1992.01820060022003. [DOI] [PubMed] [Google Scholar]
- Marazziti D, Rossi A, Giannaccini G, Baroni S, Lucacchini A, Cassano GB. Presence and characterization of the serotonin transporter in human resting lymphocytes. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 1998;19:154–159. doi: 10.1016/S0893-133X(97)00204-2. [DOI] [PubMed] [Google Scholar]
- Mazer C, Muneyyirci J, Taheny K, Raio N, Borella A, Whitaker-Azmitia P. Serotonin depletion during synaptogenesis leads to decreased synaptic density and learning deficits in the adult rat: a possible model of neurodevelopmental disorders with cognitive deficits. Brain Res. 1997;760:68–73. doi: 10.1016/s0006-8993(97)00297-7. [DOI] [PubMed] [Google Scholar]
- Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- Munro CA, Workman CI, Kramer E, Hermann C, Ma Y, Dhawan V, Chaly T, Eidelberg D, Smith GS. Serotonin modulation of cerebral glucose metabolism: sex and age effects. Synapse. 2012;66:955–964. doi: 10.1002/syn.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Paulmann N, Grohmann M, Voigt JP, Bert B, Vowinckel J, Bader M, Skelin M, Jevsek M, Fink H, Rupnik M, Walther DJ. Intracellular serotonin modulates insulin secretion from pancreatic beta-cells by protein serotonylation. PLoS biology. 2009;7:e1000229. doi: 10.1371/journal.pbio.1000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietraszek MH, Takada Y, Yan D, Urano T, Serizawa K, Takada A. Relationship between serotonergic measures in periphery and the brain of mouse. Life Sci. 1992;51:75–82. doi: 10.1016/0024-3205(92)90221-a. [DOI] [PubMed] [Google Scholar]
- Pineyro G, Castanon N, Hen R, Blier P. Regulation of [3H]5-HT release in raphe, frontal cortex and hippocampus of 5-HT1B knock-out mice. Neuroreport. 1995;7:353–359. [PubMed] [Google Scholar]
- Pollock BG, Ferrell RE, Mulsant BH, Mazumdar S, Miller M, Sweet RA, Davis S, Kirshner MA, Houck PR, Stack JA, Reynolds CF, Kupfer DJ. Allelic variation in the serotonin transporter promoter affects onset of paroxetine treatment response in late-life depression. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2000;23:587–590. doi: 10.1016/S0893-133X(00)00132-9. [DOI] [PubMed] [Google Scholar]
- Prasad HC, Zhu CB, McCauley JL, Samuvel DJ, Ramamoorthy S, Shelton RC, Hewlett WA, Sutcliffe JS, Blakely RD. Human serotonin transporter variants display altered sensitivity to protein kinase G and p38 mitogen-activated protein kinase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11545–11550. doi: 10.1073/pnas.0501432102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainero I, Ostacoli L, Rubino E, Gallone S, Picci LR, Fenoglio P, Negro E, Rosso C, De Martino P, De Marchi M, Furlan PM, Pinessi L. Association between major mood disorders and the hypocretin receptor 1 gene. J Affect Disord. 2011;130:487–491. doi: 10.1016/j.jad.2010.10.033. [DOI] [PubMed] [Google Scholar]
- Rand M, Reid G. Source of ‘serotonin’ in serum. Nature. 1951;168:385. doi: 10.1038/168385b0. [DOI] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, Griem A, Kovacs M, Ott J, Merikangas KR. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA : the journal of the American Medical Association. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Yamagata T, Matsumoto A, Shiba Y, Nagashima M, Taniguchi S, Jimbo E, Momoi MY. MAOA/B deletion syndrome in male siblings with severe developmental delay and sudden loss of muscle tonus. Brain Dev. 2013 doi: 10.1016/j.braindev.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, Williams SC, Richardson JA, Kozlowski GP, Wilson S, Arch JR, Buckingham RE, Haynes AC, Carr SA, Annan RS, McNulty DE, Liu WS, Terrett JA, Elshourbagy NA, Bergsma DJ, Yanagisawa M. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92:573–585. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S, Weisstaub N, Lee J, Duman R, Arancio O, Belzung C, Hen R. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301:805–809. doi: 10.1126/science.1083328. [DOI] [PubMed] [Google Scholar]
- Schain RJ, Freedman DX. Studies on 5-hydroxyindole metabolism in autistic and other mentally retarded children. J Pediatr. 1961;58:315–320. doi: 10.1016/s0022-3476(61)80261-8. [DOI] [PubMed] [Google Scholar]
- Scott MM, Marcus JN, Pettersen A, Birnbaum SG, Mochizuki T, Scammell TE, Nestler EJ, Elmquist JK, Lutter M. Hcrtr1 and 2 signaling differentially regulates depression-like behaviors. Behav Brain Res. 2011;222:289–294. doi: 10.1016/j.bbr.2011.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serretti A, Kato M, De Ronchi D, Kinoshita T. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Molecular psychiatry. 2007;12:247–257. doi: 10.1038/sj.mp.4001926. [DOI] [PubMed] [Google Scholar]
- Stamford JA, Davidson C, McLaughlin DP, Hopwood SE. Control of dorsal raphe 5-HT function by multiple 5-HT(1) autoreceptors: parallel purposes or pointless plurality? Trends in neurosciences. 2000;23:459–465. doi: 10.1016/s0166-2236(00)01631-3. [DOI] [PubMed] [Google Scholar]
- Steinbusch HW. Distribution of serotonin-immunoreactivity in the central nervous system of the rat-cell bodies and terminals. Neuroscience. 1981;6:557–618. doi: 10.1016/0306-4522(81)90146-9. [DOI] [PubMed] [Google Scholar]
- Strober W, Watanabe T. NOD2, an intracellular innate immune sensor involved in host defense and Crohn's disease. Mucosal Immunol. 2011;4:484–495. doi: 10.1038/mi.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan GM, Oquendo MA, Huang YY, Mann JJ. Elevated cerebrospinal fluid 5-hydroxyindoleacetic acid levels in women with comorbid depression and panic disorder. The international journal of neuropsychopharmacology / official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2006;9:547–556. doi: 10.1017/S1461145705006231. [DOI] [PubMed] [Google Scholar]
- Taylor BA, Bedigian HG, Meier H. Genetic studies of the Fv-1 locus of mice: linkage with Gpd-1 in recombinant inbred lines. J Virol. 1977;23:106–109. doi: 10.1128/jvi.23.1.106-109.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BA, Wnek C, Kotlus BS, Roemer N, MacTaggart T, Phillips SJ. Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm Genome. 1999;10:335–348. doi: 10.1007/s003359900998. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Thompson R, Haley CS. Confidence intervals in QTL mapping by bootstrapping. Genetics. 1996;143:1013–1020. doi: 10.1093/genetics/143.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DJ, Peter JU, Bashammakh S, Hortnagl H, Voits M, Fink H, Bader M. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003;299:76. doi: 10.1126/science.1078197. [DOI] [PubMed] [Google Scholar]
- Walther DJ, Peter JU, Winter S, Holtje M, Paulmann N, Grohmann M, Vowinckel J, Alamo-Bethencourt V, Wilhelm CS, Ahnert-Hilger G, Bader M. Serotonylation of small GTPases is a signal transduction pathway that triggers platelet alpha-granule release. Cell. 2003;115:851–862. doi: 10.1016/s0092-8674(03)01014-6. [DOI] [PubMed] [Google Scholar]
- Watson C, Paxinos G. Chemoarchitectonic atlas of the mouse brain. London: Academic; 2010. [Google Scholar]
- Weiss LA, Abney M, Parry R, Scanu AM, Cook EH, Jr, Ober C. Variation in ITGB3 has sex-specific associations with plasma lipoprotein(a) and whole blood serotonin levels in a population-based sample. Human genetics. 2005;117:81–87. doi: 10.1007/s00439-004-1250-3. [DOI] [PubMed] [Google Scholar]
- Welsh CE, Miller DR, Manly KF, Wang J, McMillan L, Morahan G, Mott R, Iraqi FA, Threadgill DW, de Villena FP. Status and access to the Collaborative Cross population. Mamm Genome. 2012;23:706–712. doi: 10.1007/s00335-012-9410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whibley A, Urquhart J, Dore J, Willatt L, Parkin G, Gaunt L, Black G, Donnai D, Raymond FL. Deletion of MAOA and MAOB in a male patient causes severe developmental delay, intermittent hypotonia and stereotypical hand movements. Eur J Hum Genet. 2010;18:1095–1099. doi: 10.1038/ejhg.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie CJ, Hendricks TJ, Zhang B, Wang L, Lu P, Leahy P, Fox S, Maeno H, Deneris ES. Distinct transcriptomes define rostral and caudal serotonin neurons. J Neurosci. 2010;30:670–684. doi: 10.1523/JNEUROSCI.4656-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav VK, Oury F, Suda N, Liu ZW, Gao XB, Confavreux C, Klemenhagen KC, Tanaka KF, Gingrich JA, Guo XE, Tecott LH, Mann JJ, Hen R, Horvath TL, Karsenty G. A serotonin-dependent mechanism explains the leptin regulation of bone mass, appetite, and energy expenditure. Cell. 2009;138:976–989. doi: 10.1016/j.cell.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Blakely RD. Natural and engineered coding variation in antidepressant-sensitive serotonin transporters. Neuroscience. 2011;197:28–36. doi: 10.1016/j.neuroscience.2011.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.