Abstract
We synthesized a panel of unnatural base pairs whose pairing depends on hydrophobic and packing forces and identify dTPT3-dNaM, which is PCR amplified with a natural base pair-like efficiency and fidelity. In addition, the dTPT3 scaffold is uniquely tolerant of attaching a propargyl amine linker, resulting in the dTPT3PA-dNaM pair, which is amplified only slightly less well. The identification of dTPT3 represents significant progress towards developing an unnatural base pair for the in vivo expansion of an organism's genetic alphabet and for a variety of in vitro biotechnology applications where it is used to site-specifically label amplified DNA, and it also demonstrates for the first time that hydrophobic and packing forces are sufficient to mediate natural-like replication.
The four letter natural genetic alphabet is conserved throughout nature and is based on the complementary shape and hydrogen-bonding (H-bonding) of the natural nucleotides. Since its original proposal by Benner in 1990,1 the development of unnatural base pairs has become an active area of research.2-7 Early efforts focused on novel pairs that form via orthogonal H-bonding patterns, and progress along this route continues.8 As an alternative approach, inspired in part by Kool's demonstration that H-bonds are not an absolute requirement for replication,9 our group2,10-12 and the Hirao group13-17 have demonstrated that hydrophobic and packing forces are also sufficient to mediate unnatural base pair replication. In particular, we have developed a class of unnatural base pairs, exemplified by d5SICS-dNaM (Fig. 1A), that when incorporated into DNA are efficiently replicated without sequence bias,2,18 and efficiently transcribed into RNA.19,20 However, neither d5SICS-dNaM, nor any of the other unnatural base pairs reported to date, are replicated with natural base pair-like efficiencies or fidelities. In addition to potentially compromising the potential uses of these unnatural base pairs for both in vivo and in vitro applications, this raises the fundamental question of whether hydrophobicity and packing forces are truly sufficient to mediate replication with natural-like efficiencies and fidelities.
Figure 1.
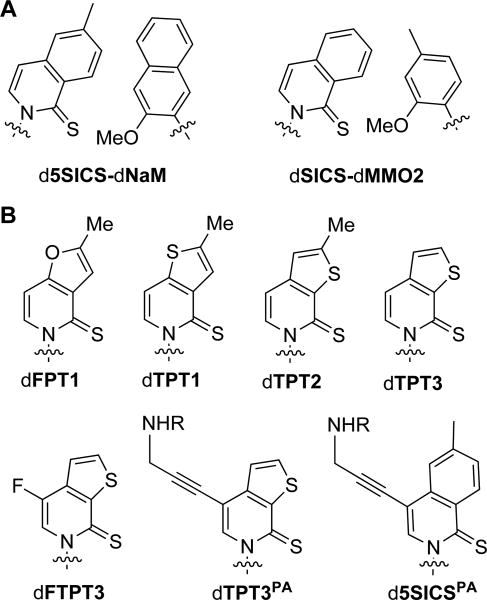
(A) Previously identified unnatural base pairs. (B) Analogs used in this study, with d5SICSPA shown for comparison.20 R=COCHCl2. Sugar and phosphate backbone are omitted for clarity.
The d5SICS-dNaM pair was identified from the optimization of dSICS-dMMO2 (Fig. 1A), which was identified from a screen of 3600 candidate unnatural base pairs.11 Early optimization efforts focused on improving dSICS as a partner for dMMO2, eventually yielding d5SICS. After progress stalled, our efforts turned to the optimization of dMMO2 as a partner for d5SICS, which eventually yielded dNaM.2,12,18 Although the discovery of dNaM represented a significant improvement in replication, continued optimization efforts again stalled,12,18,21,22,23 implying that further optimization of dNaM as a partner for d5SICS was unlikely. Thus, our focus turned to the optimization of d5SICS as a partner for dNaM.
Structural studies in duplex DNA demonstrated that d5SICS-dNaM forms via cross-strand intercalation.23,24 This results in a structure that is more similar to a mispair than a correct pair, making the efficient replication of the unnatural base pair difficult to understand. However, recent studies have shown not only that pairing of dNaM with d5SICSTP within a polymerase active site induces the polymerase to undergo the open-to-closed structural transition characteristic of normal synthesis, but also that within the closed complex, dNaM and d5SICSTP pair in a Watson-Crick like fashion.24 This mutually induced fit mechanism is likely responsible for the efficient incorporation of the unnatural triphosphate, but structure-activity relationship (SAR) data suggested that after translocation within the polymerase binding site, in preparation for incorporation of the next triphosphate, the nascent unnatural base pair again returns to an intercalated state, making de-intercalation a requirement for continued DNA synthesis.12 Thus, we reasoned that further optimization might be possible with d5SICS analogs that conform to the SAR rules governing efficient base pair synthesis (via triphosphate incorporation) and extension (via incorporation of the next triphosphate), but that are less prone to intercalate. As SAR data has clearly demonstrated that the thio N-glycoside of d5SICS is required for both synthesis and extension, we reasoned that modifying and/or reducing the aromatic surface of the second ring might represent a route to favor de-intercalation.
To explore the reduction of aromatic surface area of the d5SICS scaffold, as well as to systematically vary nucleobase dipole moment and polarizability, we synthesized the furano and thieno substituted pyridine-2(1H)-thione heterocycles dFPT1, dTPT1, and dTPT2 (Fig. 1B, Supporting Information). Briefly, the nucleobase analogs were synthesized via an intermolecular Curtius rearrangement, and then coupled to (2R,5R)-5-chloro-2-(((4-methylbenzoyl)oxy)methyl) tetrahydrofuran-3-yl 4-methylbenzoate, resulting in a mixture of anomeric nucleosides, with the pure β-anomer obtained by column chromatography. After sulfonylation and toluyl deprotection, the free nucleosides were converted to triphosphates under Ludwig conditions, and purified by anion exchange chromatography and reverse phase HPLC.
In previous work, we employed steady-state kinetics to analyze both unnatural base pair synthesis and extension. However, the chemical steps underlying d5SICS-dNaM synthesis and extension are sufficiently efficient that under steady-state conditions product formation is limited by dissociation,25 rendering the steady-state kinetics data less helpful for the optimization of processive replication. Thus, we analyzed the new derivatives using a pre-steady state assay.22 The assay is based on determining, under a fixed set of conditions, the amount of a 23mer primer that is extended by addition of the unnatural triphosphate opposite its cognate nucleotide in a 45mer template by the Klenow fragment of E. coli DNA polymerase I (Kf). The efficiency of unnatural base pair synthesis is characterized by measuring the percent incorporation (%inc) at a given concentration of the unnatural and next correct triphosphate (dCTP), determined from the ratio [24mer+25mer]/[23mer+ 24mer+25mer]. The efficiency of extension is characterized by measuring the percent extension (%ext) at a given concentration of the next correct triphosphate (dCTP) and saturating concentrations of unnatural triphosphate, determined from the ratio [25mer]/[24mer+25mer]. Under the conditions selected, d5SICSTP is incorporated opposite dNaM with a %inc of 57, and the resulting d5SICS-dNaM pair is extended with a %ext of only 15. The derivatives showed a wide range of behaviors (Table 1). dFPT1TP is incorporated less efficiently, but dFPT1-dNaM is then extended more efficiently, while the opposite is true for dTPT1TP. However, dTPT2TP is incorporated more efficiently than d5SICSTP , and the subsequently formed dTPT2-dNaM pair is also extended more efficiently than d5SICS-dNaM.
Table 1.
Kinetic and PCR amplification data.
| Presteady-state kinetics | OneTaq PCR (48 cycles) | Taq PCR (20 cycles) | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| dXTP | %inca | %extb | amplification ×1012 | retention, % | fidelity, % | amplification ×103 | retention, % | fidelity, % |
| 5SICS | 57.0 ± 0.2 | 15.1 ± 1.1 | 9.4 | 96.3 ± 1.7 | 99.91 ± 0.04 | 7.7 | 86.7 ± 1.0 | 98.90 ± 0.01 |
| FPT1 | 7.2 ± 0.2 | 32.0 ± 1.5 | 9.7 | 62 ± 2 | 98.89 ± 0.09 | 10.9 | 42.0 ± 1.1 | 93.73 ± 0.18 |
| TPT1 | 28.7 ± 0.5 | 8.8 ± 0.2 | 13.3 | 60 ± 7 | 98.8 ± 0.3 | 6.7 | 33.76 ± 0.03 | 91.81 ± 0.01 |
| TPT2 | 65.7 ± 0.5 | 34.5 ± 0.5 | 16.6 | 96.8 ± 0.9 | 99.90 ± 0.02 | 9.9 | 93.05 ± 0.04 | 99.46 ± 0.01 |
| TPT3 | 72.3 ± 0.5 | 49.8 ± 1.3 | 12.9 | > 99 | >99.98 | 11.7 | 95.6 ± 1.7 | 99.66 ± 0.13 |
| FTPT3 | 66.3 ± 0.5 | 33.8 ± 0.2 | 6.5 | 97.9 ± 1.0 | 99.95 ± 0.02 | 5.0 | 98 ± 3 | 99.9 ± 0.2 |
| TPT3PA | 68.3 ± 0.4 | 31.5 ± 0.7 | 4.7 | 98.6 ± 1.2 | 99.97 ± 0.03 | 3.5 | 85 ± 4 | 98.7 ± 0.4 |
| 5SICSPA | 7.0 ± 0.2 | 5.5 ± 0.1 | 9.2 | 45 ± 2 | 98.16 ± 0.12 | 6.4 | -c | -c |
Incorporation assay conditions: 40 nM unnatural triphosphate, 2 μM dCTP, 10 s.
Extension assay conditions: 10 μM unnatural triphsophate, 2 mM dCTP, 10 s.
Unnatural base pair was lost during amplification.
Based on this data we further explored the dTPT2 scaffold with dTPT3 (Fig. 1B). With this analog, removal of the methyl group was expected to further modify the tendency of the nucleobase to cross-strand intercalate, and it was synthesized by adaptation of the dTPT2 synthesis (Supporting Information). Interestingly, both dTPT3TP incorporation opposite dNaM, and extension of the resulting unnatural pair is more efficient than with dTPT2TP (Table 1).
To more fully evaluate replication, the previously reported template D6,2,26 containing d5SICS or a d5SICS analog paired opposite dNaM was amplified by PCR (in this template, the unnatural base pair is flanked on each side by three randomized natural nucleotides, Supporting Information). Two sets of PCR reactions were explored (Table 1), one employing 48 cycles with OneTaq polymerase, a commercially available mixture of exonuclease-negative Taq polymerase and exonuclease-positive DeepVent polymerases, and one employing 20 cycles of amplification with exonuclease-negative Taq alone, to explore the contribution of proofreading. Efficiency was determined by monitoring amplification level, and fidelity (defined as unnatural base pair retention per doubling) was determined from the percentage of the amplified DNA that retained the unnatural base pair, which was determined from the relative peak intensities of a sequencing chromatogram (Supporting Information and Malyshev, et al.26). For comparison, with OneTaq natural DNA is amplified 5.5 × 1013-fold and DNA containing d5SICS-dNaM is amplified 9.4 × 1012-fold, and 96.3% of the DNA retained the unnatural pair, corresponding to a fidelity of 99.91%. Under these conditions DNA containing dFPT1-dNaM or dTPT1-dNaM is amplified ∼1 × 1013-fold, but with fidelity that is slightly reduced relative to d5SICS-dNaM (∼98.8%). However, amplification of DNA containing dTPT2-dNaM proceeded more efficiently than d5SICS-dNaM, and with an indistinguishable fidelity. Amplification of DNA containing dTPT3-dNaM again proceeded more efficiently than d5SICS-dNaM, and remarkably, with no mispaired sequences detected. With a lower limit of detection estimated to be one mispaired sequence in 102 (corresponding to 99% retention), this places a lower limit of 99.98% on the fidelity of dTPT3-dNaM replication.
Due to the absence of proofreading, amplification with only Taq polymerase is more likely to differentiate the unnatural base pairs. Under the conditions employed with only Taq, natural DNA is amplified 2.8 × 104-fold and DNA containing d5SICS-dNaM is amplified 7.7 × 103-fold and with a fidelity of 98.90%. DNA containing dFPT1-dNaM or dTPT1-dNaM is amplified with significantly lower fidelity. However, DNA containing dTPT2-dNaM, and especially dTPT3-dNaM, is amplified with greater efficiency and fidelity. In fact, dTPT3-dNaM is amplified with a fidelity that is only slightly less than that observed with OneTaq. Clearly, based on the pre-steady state kinetics and both PCR conditions, dTPT2 and especially dTPT3 are significantly better optimized as partners for dNaM than d5SICS or any of the other analogs examined.
In principle, an unnatural base pair should allow for the site-specific inclusion of different functionalities into DNA, either pre- or post-amplification,14,27 for biophysical, SELEX (Systematic Evolution of Ligands by Exponential Enrichment)28,29 or nanomaterial applications.30 To explore the potential of the dTPT3 scaffold for such applications, we synthesized dFTPT3TP and dTPT3PATP (Fig. 1B), with the former installing an NMR active nuclei for biophysical characterization31,32 and the latter bearing a linker for the site-specific modification of the amplified DNA. Both analogs were obtained from the toluyl-protected nucleoside of dTPT3 (Supporting Information). Briefly, dFTPT3 was obtained in via fluorination, sulfonation, benzoyl deprotection, and phosphorylation, to yield the corresponding triphosphate. dTPT3PATP was obtained via iodination, sulfonylation, coupling with the dichloro acetyl propargylamine, deprotection, and phosphorylation.
Both fluoro and propargyl amine substituents have similar effects on the pre-steady state kinetics, slightly reducing the %inc opposite dNaM, and more substantially reducing the %ext (Table 1). However, %inc and %ext for both analogs remain significantly greater than those for d5SICS. Interestingly, DNA containing either dFTPT3-dNaM or dTPT3PA-dNaM is PCR amplified with nearly the same fidelity as is dTPT3-dNaM and remarkably, both are amplified as well or better than DNA containing d5SICS-dNaM (Table 1). In fact, with Taq alone, dFTP3-dNaM is better replicated than any previously reported unnatural base pair. The ability of the dTPT3 scaffold to support the propargyl amine linker is particularly remarkable as dTPT3PA-dNaM is replicated significantly better than d5SICSPA-dNaM (Fig. 1B), which previously was the most efficiently amplified linker-bearing unnatural base pair.20 With OneTaq, the fidelity is 99.97% versus 98.16% per doubling for dTPT3PA-dNaM and d5SICSPA-dNaM, respectively. Dramatically, with Taq alone, d5SICSPA-dNaM is lost during amplification while dTPT3PA-dNaM is amplified with a fidelity of 98.7%.
Finally, to explore the use of dTPT3PA for the site-specific labeling of DNA, we examined the amplification, deprotection, and labeling of a 134mer DNA containing a centrally positioned dTPT3PA-dNaM or d5SICSPA-dNaM. Each template was amplified ∼500-fold, deprotected, coupled to NHS-PEG4-biotin, and analyzed by gel shift with streptavidin (Fig. 2A and B). With d5SICSPA-dNaM, 72% of the DNA was shifted, while for dTPT3PA-dNaM, 80% of the DNA was shifted. To explore site-specific labeling at potentially more challenging positions, dTPT3PA-dNaM or d5SICSPA-dNaM was incorporated into a 60mer DNA at the first, ninth, and eleventh position, resulting in duplexes 1–3, respectively (Fig. 2C). With d5SICSPA-dNaM, 6%, 56%, and 84% of the amplified DNA was shifted, while with dTPT3PA-dNaM, 72%, 81%, and 94% was shifted. Clearly, the more efficient amplification of dTPT3PA-dNaM, especially when it is located near the end of a template, results in a significantly greater efficiency of site-specific labeling. This should facilitate many applications, including those requiring that both ends of the same DNA strand are modified, which is otherwise challenging.
Figure 2.
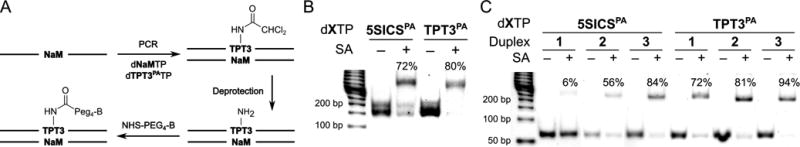
Determination of post-amplification DNA labeling efficiency via streptavidin (SA) gel shift. (A) Labeling strategy (illustrated for dTPT3PA-dNaM; B = biotin). (B) Labeling with unnatural base pair centrally located in 134mer duplex. The faster and slower migrating bands corresponds to dsDNA, and the 1:1 complex between dsDNA and streptavidin, respectively. (C) Labeling with unnatural base pair positioned at first, ninth, or eleventh positions, corresponding to duplexes 1–3, respectively.
In summary, we found that contraction of the distal phenyl ring of d5SICS to a methyl thieno ring significantly increases replication, and that subsequent removal of the methyl group increases it still further. These effects are clearly dependent on the detailed structure of the nucleobase. Moreover, it is noteworthy that dTPT3 represents a rare example among predominantly hydrophobic unnatural base pairs where a reduction in hydrophobicity facilitates replication. A simple explanation for this SAR is that when appropriately positioned, the sulfur atom increases intrastrand packing interactions, while the methyl group favors interstrand packing interactions. Thus, both addition of the sulfur and removal of the methyl group are expected to stabilize the transition state for incorporation, and also favor de-intercalation of the nascent unnatural base pair, which our model predicts would facilitate extension.12 The robust accommodation of substituents within the dTPT3 scaffold is also consistent with this model, since only when the pair adopts a Watson-Crick-like structure is a substituent at the position explored predicted to be disposed into the major groove, where it should be relatively non-perturbative.
Regardless of the precise physical underpinnings of its optimized replication, the identification of dTPT3-dNaM represents a milestone in our effort to expand the genetic alphabet. Under standard PCR conditions, DNA containing dTPT3-dNaM is amplified by OneTaq with an efficiency that is only 4-fold lower than DNA containing just the natural base pairs, and with a fidelity in excess of 99.98%. This fidelity corresponds to an error rate of 10-4 per nucleotide, which overlaps with the 10-4 to 10-7 error rate of fully natural DNA with commonly used PCR systems.33 With Taq, the efficiency is only 2.5-fold lower than that of a natural base pair, and the fidelity is 99.7%. This fidelity corresponds to an error rate of 10-3, which is similar to that observed with the Taq-mediated amplification of natural DNA.34 This efficient and high fidelity replication should facilitate efforts to expand the genetic alphabet in vivo and create the first semisynthetic organism with increased potential for information storage and retrieval. Moreover, the ability of the dTPT3 scaffold to accommodate a fluoro or protected propargyl amine linker substituent and still be efficiently replicated when paired with dNaM suggests that it is a particularly robust scaffold and should make possible a number of in vitro applications. Finally, the data demonstrate for the first time that hydrophobic and packing forces are sufficient to mediate the replication of unnatural base pairs with truly naturallike efficiencies and fidelities.
Supplementary Material
Acknowledgments
Funding Sources: This work was supported by the NIH (GM 060005).
Footnotes
Supporting Information. Synthesis, characterization, pre-steady state kinetic data, PCR assay and analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Piccirilli JA, Krauch T, Moroney SE, Benner SA. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 2.Malyshev DA, Dhami K, Quach HT, Lavergne T, Ordoukhanian P, Torkamani A, Romesberg FE. Proc Natl Acad Sci U S A. 2012;109:12005–12010. doi: 10.1073/pnas.1205176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minakawa N, Ogata S, Takahashi M, Matsuda A. J Am Chem Soc. 2009;131:1644–1645. doi: 10.1021/ja807391g. [DOI] [PubMed] [Google Scholar]
- 4.Chelliserrykattil J, Lu H, Lee AH, Kool ET. ChemBioChem. 2008;9:2976–2980. doi: 10.1002/cbic.200800339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brotschi C, Mathis G, Leumann CJ. Chem Eur J. 2005;11:1911–1923. doi: 10.1002/chem.200400858. [DOI] [PubMed] [Google Scholar]
- 6.Kaul C, Muller M, Wagner M, Schneider S, Carell T. Nat Chem. 2011;3:794–800. doi: 10.1038/nchem.1117. [DOI] [PubMed] [Google Scholar]
- 7.Yamashige R, Kimoto M, Takezawa Y, Sato A, Mitsui T, Yokoyama S, Hirao I. Nucleic Acids Res. 2012;40:2793–2806. doi: 10.1093/nar/gkr1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Chen F, Alvarado JB, Benner SA. J Am Chem Soc. 2011;133:15105–15112. doi: 10.1021/ja204910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran S, Ren RX, Rumney S, Kool ET. J Am Chem Soc. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMinn DL, Ogawa AK, Wu Y, Liu J, Schultz PG, Romesberg FE. J Am Chem Soc. 1999;121:11585–11586. [Google Scholar]
- 11.Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. J Am Chem Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo YJ, Hwang GT, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2009;131:3246–3252. doi: 10.1021/ja807853m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirao I. Curr Opin Chem Biol. 2006;10:622–627. doi: 10.1016/j.cbpa.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 14.Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. Nat Methods. 2006;3:729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 15.Hirao I, Mitsui T, Kimoto M, Yokoyama S. J Am Chem Soc. 2007;129:15549–15555. doi: 10.1021/ja073830m. [DOI] [PubMed] [Google Scholar]
- 16.Kimoto M, Kawai R, Mitsui T, Yokoyama S, Hirao I. Nucleic Acids Res. 2009;37:e14. doi: 10.1093/nar/gkn956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitsui T, Kitamura A, Kimoto M, To T, Sato A, Hirao I, Yokoyama S. J Am Chem Soc. 2003;125:5298–5307. doi: 10.1021/ja028806h. [DOI] [PubMed] [Google Scholar]
- 18.Lavergne T, Malyshev DA, Romesberg FE. Chem Eur J. 2012;18:1231–1239. doi: 10.1002/chem.201102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo YJ, Matsuda S, Romesberg FE. J Am Chem Soc. 2009;131:5046–5047. doi: 10.1021/ja9006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo YJ, Malyshev DA, Lavergne T, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2011;133:19878–19888. doi: 10.1021/ja207907d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo YJ, Romesberg FE. ChemBioChem. 2009;10:2394–2400. doi: 10.1002/cbic.200900413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavergne T, Degardin M, Malyshev DA, Quach HT, Dhami K, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2013;135:5408–5419. doi: 10.1021/ja312148q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malyshev DA, Pfaff DA, Ippoliti SI, Hwang GT, Dwyer TJ, Romesberg FE. Chem Eur J. 2010;16:12650–12659. doi: 10.1002/chem.201000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Betz K, Malyshev DA, Lavergne T, Welte W, Diederichs K, Dwyer TJ, Ordoukhanian P, Romesberg FE, Marx A. Nat Chem Biol. 2012;8:612–614. doi: 10.1038/nchembio.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 26.Malyshev DA, Seo YJ, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2009;131:14620–14621. doi: 10.1021/ja906186f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Lavergne T, Malyshev DA, Zimmermann J, Adhikary R, Dhami K, Ordoukhanian P, Sun Z, Xiang J, Romesberg FE. Chem Eur J. 2013;19:14205–14209. doi: 10.1002/chem.201302496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keefe AD, Cload ST. Curr Opin Chem Biol. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 29.Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I. Nat Biotechnol. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 30.Fritzsche W, Bier F. International Symposium on DNA-Based Nanodevices. American Institute of Physics Conference Proceedings; 2008. [Google Scholar]
- 31.Barhate NB, Barhate RN, Cekan P, Drobny G, Sigurdsson ST. Org Lett. 2008;10:2745–2747. doi: 10.1021/ol800872a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitevski-LeBlanc JL, Prosser RS. Prog Nucl Magn Reson Spectrosc. 2012;62:1–33. doi: 10.1016/j.pnmrs.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 33.New England Biolabs. PCR Reagents Version 3.0. Ipswich, MA: p. 3. [Google Scholar]
- 34.Tindall KR, Kunkel TA. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.