Abstract
Driven by recent advances in medical imaging, image segmentation and numerical techniques, computer models of ventricular electrophysiology account for increasingly finer levels of anatomical and biophysical detail. However, considering the large number of model parameters involved parameterization poses a major challenge. A minimum requirement in combined experimental and modeling studies is to achieve good agreement in activation and repolarization sequences between model and experiment or patient data. In this study, we propose basic techniques which aid in determining bidomain parameters to match activation sequences. An iterative parameterization algorithm is implemented which determines appropriate bulk conductivities which yield prescribed velocities. In addition, a method is proposed for splitting the computed bulk conductivities into individual bidomain conductivities by prescribing anisotropy ratios.
1. Introduction
In-silico models of ventricular electrophysiology are widely recognized to be an invaluable adjunct to experimental in-vitro or in-vivo models. Recent advances in medical imaging, image segmentation and image-based finite element (FE) mesh generation [1], along with major advances in the numerical solution of model equations have led to the generation of micro-anatomically accurate and biophysically detailed ventricular models [2]. However, considering the large number of parameters in these models data assimilation and parameterization poses a major challenge.
A minimum requirement in such modeling studies which aim at making case-specific predictions on ventricular electrophysiology is that activation and repolarization sequences are carefully matched. Conduction velocity in the ventricles is orthotropic and may vary in space, thus profoundly influencing shape and location of activation isochrones. Moreover, while anisotropy ratios, on mathematical grounds, are of rather minor relevance when simulating impulse propagation in tissue, they play a prominent role when the stimulation of cardiac tissue via externally applied electric fields is studied.
In this study, we propose an automatic tuning procedure which iteratively refines bidomain bulk conductivities using observed conduction velocities in 1D cable simulations as input. We propose a method for splitting these bulk conductivities into individual bidomain conductivities by prescribing anisotropy ratios and using those experimental data afflicted with the smallest uncertainty.
2. Methods
2.1. Governing equations
The bidomain equations are considered to be among the most accurate descriptions of cardiac bioelectricity at a macroscopic size scale. In the elliptic-parabolic form they are given by
| (1) |
where ϕi and ϕe are intra- and extracellular potentials, respectively, Vm = ϕi – ϕe is the transmembrane voltage, σi and σe are the intra- and extracellular conductivity tensors, respectively, β is the membrane surface-to-volume ratio, Cm is the membrane capacitance per unit area, and Iion is the membrane ionic current density which depends on Vm and a set of state variables, η. At tissue boundaries, no flux boundary conditions are imposed on ϕi and ϕe.
In a 1D case where impulse propagation occurs along a thin strand of tissue aligned with an axis ζ, or in the case of a planar wave front moving along an axis ζ, the monodomain equation
| (2) |
is equivalent to the bidomain equation if the monodomain conductivity σmζ is chosen to be half the harmonic mean between intracellular and interstitial conductivities, that is,
| (3) |
suggesting that conduction velocity in a full 3D bidomain model at a given location can be matched closely by an equivalent monodomain model using a harmonic mean conductivity tensor.
2.2. Conduction velocity parameterization
Conduction velocity, Cv, is not a parameter in the bidomain equations and as such cannot be directly parameterized. However, assuming a continuously propagating planar wavefront along a given direction, ζ, space and time are related by which allows to replace spatial derivatives in Eq. (2) by temporal derivatives
| (4) |
Since membrane properties on the right hand side remain unchanged, Vm remains to be a solution of Eq. (4) as long as σmζ/Cv2/β is constant. Thus Cv is governed by the proportionality relation
| (5) |
Ideally, these parameters and their spatial variation would be measured accurately in-vivo for a given subject, however, this is difficult, if not impossible, to achieve. Even when considering ex-vivo measurements, the number of reports in the literature is scarce and the variation in measured values across these studies is vast. These uncertainties inevitably arise due to the significant degree of biological variation and the substantial errors in the measurement techniques themselves.
2.3. Iterative parameterization strategy
Modeling and technical uncertainties may also have an impact on model predictions as well. For instance, Cv also depends on the particular model used to describe cellular dynamics. Uncertainties due to technical factors such as discretization errors are of lesser concern since these are, in general, small (<5%) relative to the uncertainties in model parameters. Therefore, resorting to use overly expensive numerical schemes to minimize discretization errors is not a likely candidate strategy for improving the predictive power of computer simulations. The direct use of experimentally measured conductivity values, is not warranted neither when aiming to achieve good agreement with a specific experiment.
To find a balanced trade-off, we propose a strategy which relies on prescribing Cv directly. This is based on the consideration that Cv is a quantity which is much easier, more robustly and more accurately measurable in-vivo than tissue conductivities. The proposed strategy assumes that Cv predicted by a given computer simulation setup, , can be represented as a function
| (6) |
which depends on the main factors conductivity along an axis ζ, σmζ, surface-to-volume ratio, β, the chosen model of cellular dynamics, Iion, and technical factors such as spatial discretization, Δζ, and others such as the used spatio-temporal discretization method, convergence criteria and error tolerances, the influence of which shall be summarized by ξ. In most practical scenarios, ξ, Iion and Δζ are parameters defined by users in the course of selecting a simulation software, an ionic model and a provided mesh to describe the geometry. Thus, only two free parameters, σmζ and β, are left which can be tuned to achieve a close match between the pre-specified conduction velocity, Cv, and the velocity, , predicted by the simulation.
In ventricular models Cv is orthotropic, thus necessitating to find parameters along each of the three eigenaxes. Among the two parameters available for fitting , β scales conduction velocity isotropically along all axes ζ. Using (5) and (2), and keeping β at a chosen default value, one can find unique monodomain conductivities along all axes ζ, which yield the prescribed conduction velocities, , by iteratively refining conductivities σmζ based on measured in simple 1D cable simulations. The iterative update scheme we propose is given as
| Algorithm 1 Iterative conductivity tuning |
|---|
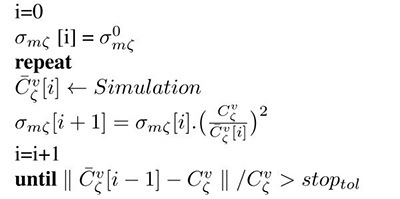
|
where is an arbitrary initial guess which is used during the first simulation run.
2.4. Compensation of discretization effects
When using bidomain parameters as chosen based on the proposed strategy, Cv must be independent of the choice of spatial discretization, Δζ. The influence of Δζ is studied in a 1D strand model of 1cm length in which Δζ is varied between 1μm up to 400μm. σmζ is iteratively tuned at the 1μm grid to yield a Cv of 0.6m/s, 0.4m/s and 0.2m/s, respectively. At each discretization and for each σmζ, propagation is initiated at the left hand side end of the cable and as well as the spatial extent of the wavefront, ΔX ζ, is measured at its center. Plots are constructed to show as a function of Δζ. Finally, each simulation is repeated, replacing σmζ, as fitted for the 1μm grid, by , as computed by the iterative tuning loop described in algorithm 1.
2.5. Obtaining bidomain conductivities
Although anisotropy ratios, on mathematical grounds, are of rather minor relevance when simulating impulse propagation in tissue, when the stimulation of cardiac tissue via externally applied electric fields are studied, anisotropy ratios play a prominent role. For this case additional constraints are required to adjust the anisotropy ratio α while keeping conduction velocities unaltered. Using the definition to characterize differences in conductivities between intracellular and extracellular space along an axis ζ, and to characterize differences in conductivity within the same space, but between an axis ζ = t|n relative to the longitudinal axis, ζ = l, we may express the anisotropy ratio εl,ζas
Note that we present quantities relative to the longitudinal axes where ζ is then either t or n.
To determine the four individual bidomain conductivities we chose the desired anisotropy ratio εlt and the longitudinal conductivity σil as these quantities show the smallest variance in the literature [3-5]. Using these constraints the bidomain conductivities are determined using (3) as
| (7) |
where * denotes quantities which were determined by iterative simulations, as outlined in Algorithm 1, and primed quantities are chosen values. Solving equations (7) for αl and αζ allows to find the bidomain conductivities
| (8) |
| (9) |
In the transversely orthotropic case, Eq. (7) is solved with ζ = t, whereas in the orthotropic case Eq. (7) is solved once more with ζ = n.
3. Results
3.1. Monodomain-bidomain equivalence
While the exact equivalence between bidomain and monodomain model using harmonic mean conductivities does not hold in 3D, along the principal eigenaxes the agreement between the models is quite good (Fig. 1). This close agreement suggests that the equivalent monodomain model can be used within the Algorithm 1, yielding a set of conductivities which enforce the prescribed Cv also when using a 3D bidomain model.
Figure 1.
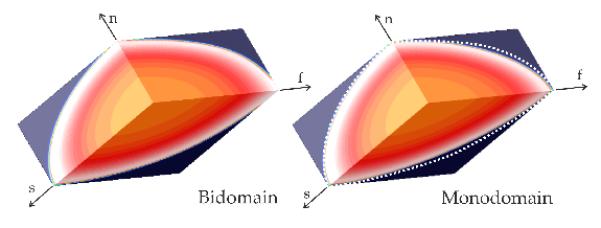
Comparison between bidomain and an equivalent monodomain model in a 3D slab model. White dashed lines in the Monodomain panel indicate position of the wavefront in a bidomain setting for the same instant in time. f, s and n are orthotropic main axes.
3.2. Conduction velocity parameterization
Using the iterative algorithm 1, conductivities were chosen to arrive at the prescribed velocities of =0.6, 0.4 and 0.2m/s for planar wave fronts traveling along the axes ζ = f, s, n, respectively. Fig. 2A shows how simulated velocities are affected when increasing grid resolution Δζ stepwise from 1μm to 400μm. Numerical errors of 5% incurred at different spatial resolutions of Δζ =275μm, 180μm and 90μm for and , respectively. As expected, scaling of simulated velocities with the prescribed velocity, i.e. , reveals that spatial discretization errors are mainly governed by the ratio Δζ/ΔX ζ(Fig. 2B). Convergence experiments were repeated for =0.6m/s and all discretizations Δζ, but equations were re-parameterized using the automatic parameterization strategy (APS) as described in algorithm 1. While conductivities varied in the range between −6 to +24%, wave fronts propagated with the exact prescribed Cv, independently of Δζ.
Figure 2.

A) Influence of spatial discretization Δζ upon predicted conduction velocities for propagation along the axes f, s and n. B) Deviations in relative velocity, depend on the ratio Δζ to spatial extent of wave front, ΔXζ. Using APS, predicted Cv (black dotted trace) is independent of Δζ, at the cost of minor variations of the chosen conductivity value, σmf (pink trace)
3.3. Choice of individual conductivities
Considering a transversely isotropic scenario, the analytic expressions 9 were used to obtain individual conductivities given three different anisotropy ratios: 1) unequal anisotropy, with εl,t = 4.5, 2) equal anisotropy, with εl,t = 1, and 3) inverse anisotropy, with εl,t = 1/4.5. The propagation patterns and the virtual electrode propagation (VEP) are shown in Fig. 3. Note that during normal propagation differences in anisotropy ratio do not manifest in the activation patterns. However, a different VEP is induced for each anisotropy ratio.
Figure 3.

Anisotropy ratios are chosen without affecting conduction velocity. Upper panels show activation sequences for three different anisotropy ratios. Lower panels show the induced VEPs in response to a strong hyperpolarizing point stimulus for the same given anisotropy ratios.
4. Discussion
In this study an iterative parameterization strategy is proposed that allows to find appropriate tissue conductivities which result then in the prescribed Cv along the orthotropic eigenaxes f, s and n of the tissue, as well as prescribed anisotropy ratios. Computationally cheap 1D equivalent monodomain models can be used in the parameterization loop to determine conductivities which are also suited for bidomain simulations. It is worth noting that parameter variations due to the automatic parameterization strategy, between −6% up to +24% around the nominal values, are well below the experimentally measured variability. The effectiveness of the methods is demonstrated for spatial discretizations of up to 400μm, however, for even coarser discretizations this may not be the case anymore. However, in line with the current trend towards anatomically detailed ventricular models, finer spatial resolution down to average discretizations of around ≈ 100μm are becoming standard to resolve geometric details. With such finer spatial steps Δζ, relative discretization of wave fronts Δζ/ΔX ζ is less than 0.3 and necessary modifications of conductivity values are below 6%.
Acknowledgment
This research is supported by Austrian Science Fund FWF grant F3210-N18.
References
- [1].Prassl AJ, Kickinger F, Ahammer H, et al. Automatically Generated, Anatomically Accurate Meshes for Cardiac Electrophysiology Problems. IEEE Trans Biomed Eng. 2009;56:1318–1330. doi: 10.1109/TBME.2009.2014243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Plank G, Burton RAB, Hales P, et al. Generation of histo-anatomically representative models of the individual heart: tools and application. Philos Trans A Math Phys Eng Sci. 2009 Jun;367(1896):2257–2292. doi: 10.1098/rsta.2009.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Clerc L. Directional Differences of Impulse Spread in Trabecular Muscle from Mammalian Heart. J Physiol. 1976;255:335–346. doi: 10.1113/jphysiol.1976.sp011283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Roberts DE, Hersh LT, Scher AM. Influence of cardiac fiber orientation on wavefront voltage, conduction velocity, and tissue resistivity in the dog. Circ Res. 1979;44(5):701–12. doi: 10.1161/01.res.44.5.701. [DOI] [PubMed] [Google Scholar]
- [5].Roberts DE, Scher AM. Effect of tissue anisotropy on extra-cellular potential fields in canine myocardium in situ. Circulation Research. 1982;50:342–51. doi: 10.1161/01.res.50.3.342. [DOI] [PubMed] [Google Scholar]


