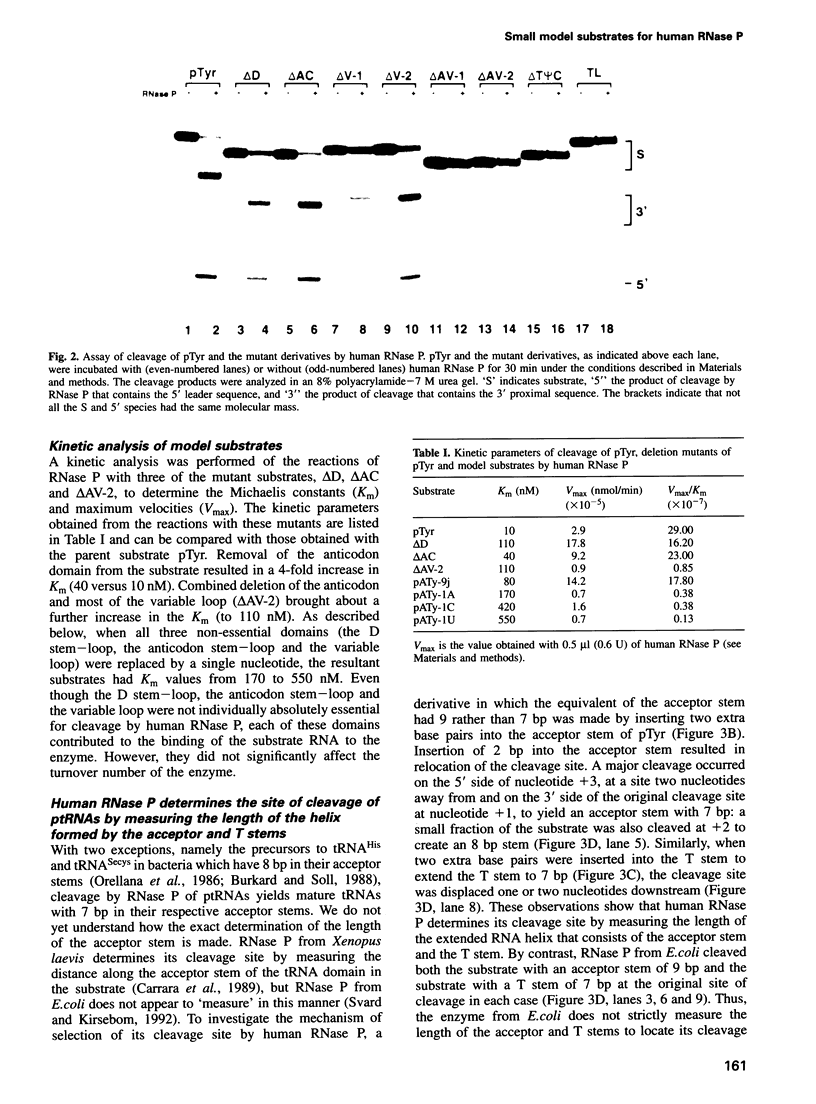
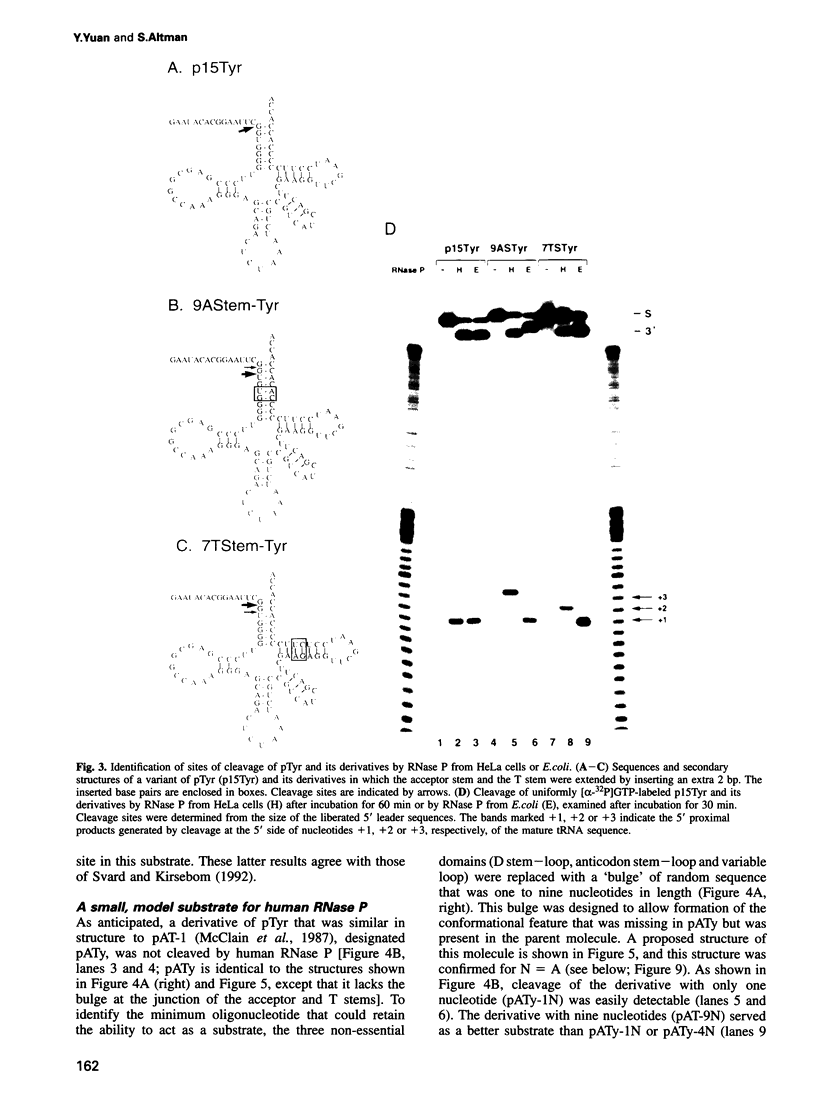
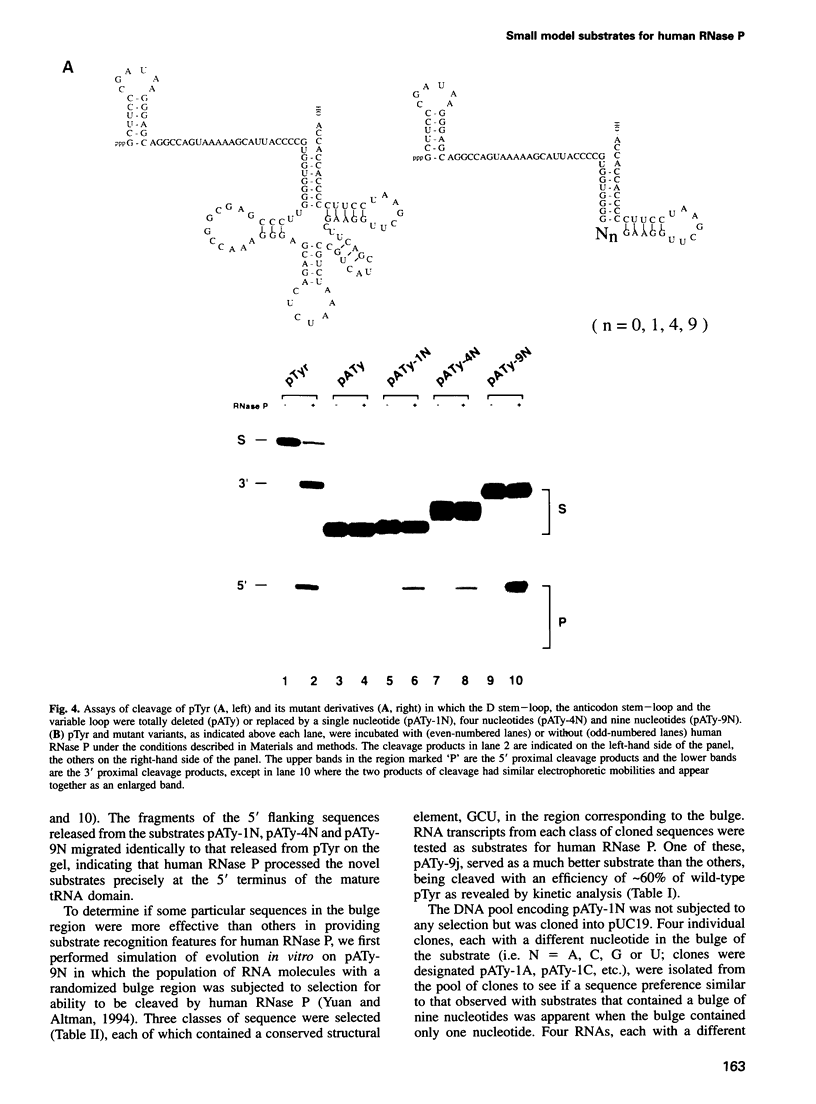
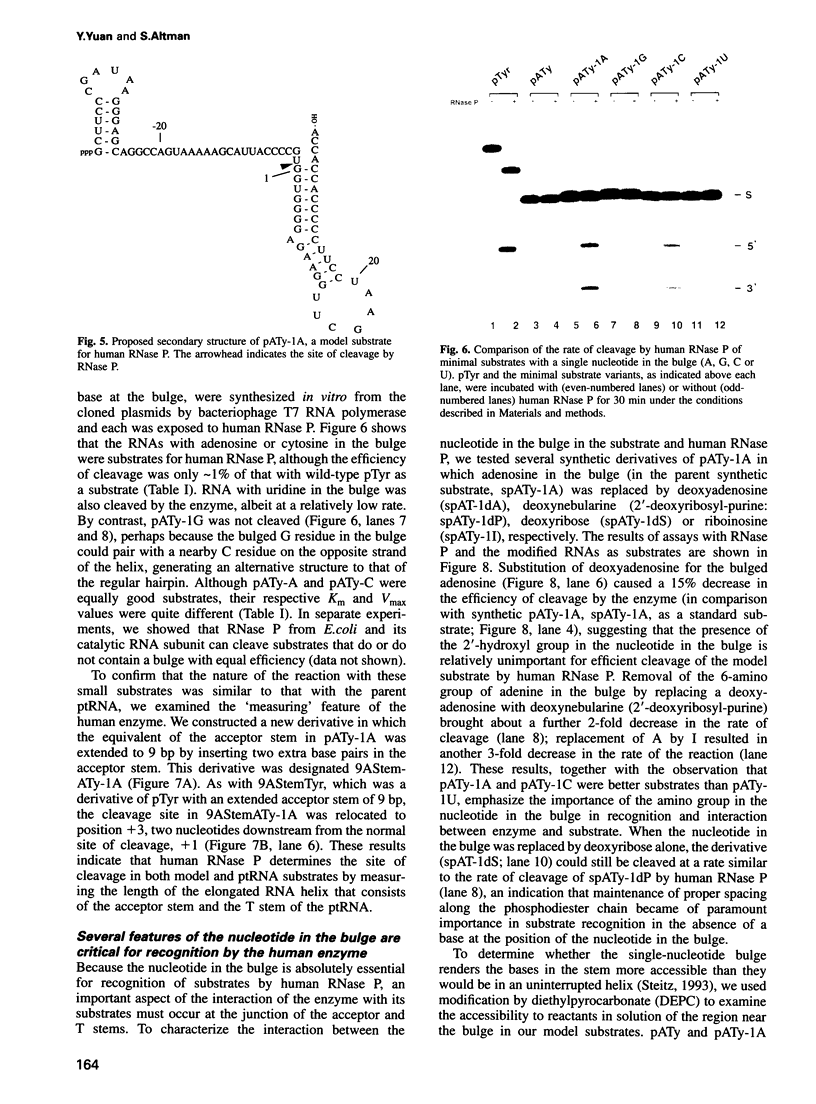
Abstract
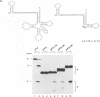
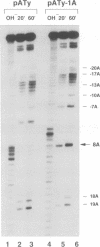
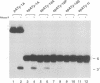
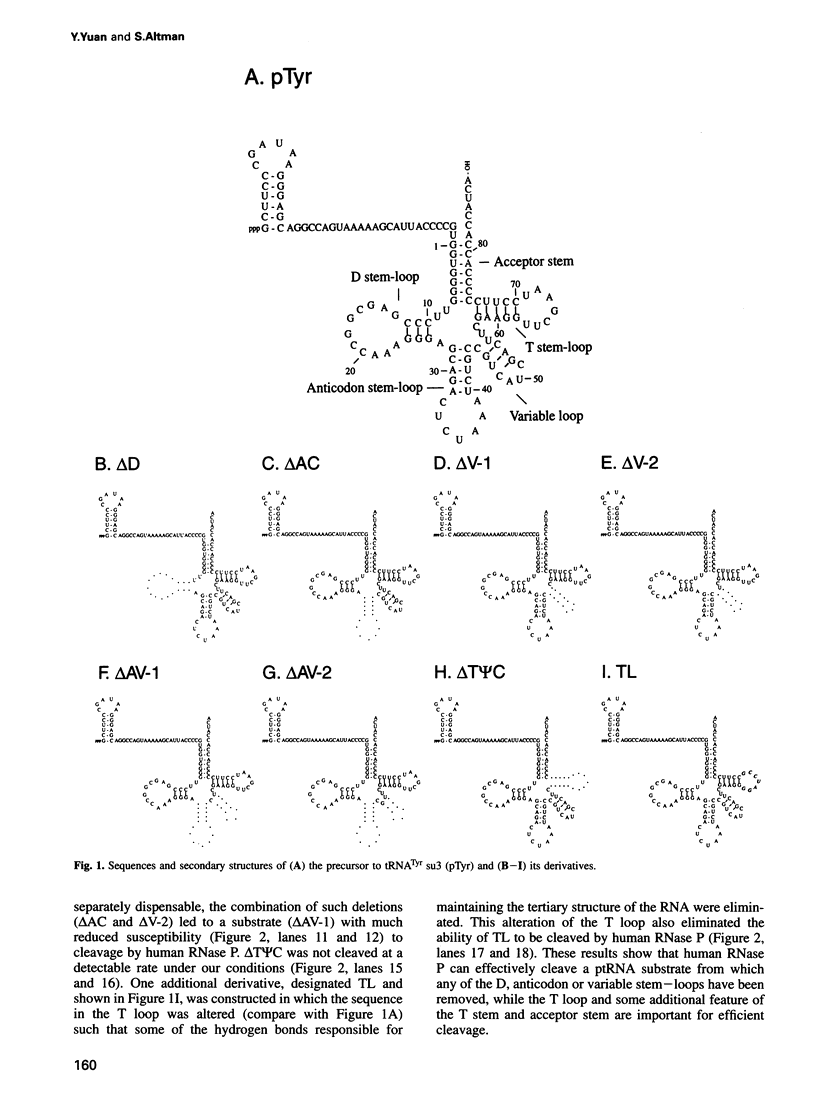
RNase P from HeLa cells can efficiently cleave tRNA precursor molecules in vitro but cannot cleave potential substrates from which the D, anticodon and variable loops and stems of the tRNA moiety have all been removed. However, molecules from which the latter subdomains have been removed individually do serve as substrates. We show here that molecules that contain only a 5' leader sequence, the acceptor stem and the T stem and loop of the tRNA domain, and a bulge as small as one nucleotide downstream from nucleotide 7 in the tRNA sequence at the junction of the two stems, can serve as substrates for human RNase P. The identity of the nucleotide in the bulge is important in determining both the efficiency of the cleavage and the conformation of the substrate and/or the enzyme-substrate complex. We also show that the human enzyme locates the appropriate site for cleavage of its substrates in part by 'measuring' the length of the helices in the acceptor and T stems in both model and natural substrates.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartkiewicz M., Gold H., Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989 Apr;3(4):488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- Burkard U., Söll D. The unusually long amino acid acceptor stem of Escherichia coli selenocysteine tRNA results from abnormal cleavage by RNase P. Nucleic Acids Res. 1988 Dec 23;16(24):11617–11624. doi: 10.1093/nar/16.24.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara G., Calandra P., Fruscoloni P., Doria M., Tocchini-Valentini G. P. Site selection by Xenopus laevis RNAase P. Cell. 1989 Jul 14;58(1):37–45. doi: 10.1016/0092-8674(89)90400-5. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990 Aug 10;62(3):407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science. 1984 Jan 20;223(4633):285–286. doi: 10.1126/science.6199841. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Lumelsky N., Altman S. Specific interactions in RNA enzyme-substrate complexes. Science. 1989 Dec 22;246(4937):1578–1584. doi: 10.1126/science.2480641. [DOI] [PubMed] [Google Scholar]
- Kahle D., Wehmeyer U., Krupp G. Substrate recognition by RNase P and by the catalytic M1 RNA: identification of possible contact points in pre-tRNAs. EMBO J. 1990 Jun;9(6):1929–1937. doi: 10.1002/j.1460-2075.1990.tb08320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Orellana O., Cooley L., Söll D. The additional guanylate at the 5' terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol Cell Biol. 1986 Feb;6(2):525–529. doi: 10.1128/mcb.6.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Douthwaite S., Garrett R. A., Noller H. F. A "bulged" double helix in a RNA-protein contact site. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7331–7335. doi: 10.1073/pnas.78.12.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Svärd S. G., Kirsebom L. A. Several regions of a tRNA precursor determine the Escherichia coli RNase P cleavage site. J Mol Biol. 1992 Oct 20;227(4):1019–1031. doi: 10.1016/0022-2836(92)90518-o. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. Major groove accessibility of RNA. Science. 1993 Sep 17;261(5128):1574–1577. doi: 10.1126/science.7690496. [DOI] [PubMed] [Google Scholar]
- Wu H. N., Uhlenbeck O. C. Role of a bulged A residue in a specific RNA-protein interaction. Biochemistry. 1987 Dec 15;26(25):8221–8227. doi: 10.1021/bi00399a030. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Altman S. Selection of guide sequences that direct efficient cleavage of mRNA by human ribonuclease P. Science. 1994 Mar 4;263(5151):1269–1273. doi: 10.1126/science.8122108. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Hwang E. S., Altman S. Targeted cleavage of mRNA by human RNase P. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8006–8010. doi: 10.1073/pnas.89.17.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]