Abstract
Studies performed in the liver in the 1960s led to the identification of lysosomes and the discovery of autophagy, the process by which intracellular proteins and organelles are degraded in lysosomes. Early studies in hepatocytes also uncovered how nutritional status regulates autophagy and how various circulating hormones modulate the activity of this catabolic process in the liver. The intensive characterization of hepatic autophagy over the years has revealed that lysosome-mediated degradation is important not only for maintaining liver homeostasis in normal physiological conditions, but also for an adequate response of this organ to stressors such as proteotoxicity, metabolic dysregulation, infection and carcinogenesis. Autophagic malfunction has also been implicated in the pathogenesis of common liver diseases, suggesting that chemical manipulation of this process might hold potential therapeutic value. In this Review—intended as an introduction to the topic of hepatic autophagy for clinical scientists—we describe the different types of hepatic autophagy, their role in maintaining homeostasis in a healthy liver and the contribution of autophagy malfunction to liver disease.
Introduction
Autophagy is the term (from the Greek words ‘auto’ meaning ‘self’ and ‘phagy’ meaning ‘to eat’) adopted to describe the series of molecular reactions that ultimately result in the degradation of intracellular components in lysosomes.1,2 Breakdown of proteins and organelles and the subsequent recycling of their essential constituents occur in all types of cells and organs with different purposes. It is intuitive to infer that degradation is an efficient method of cellular ‘cleaning’. Proteins and organelles bearing any type of abnormality due to faulty synthesis, or that become damaged by intracellular or extracellular insults, need to be removed before they become toxic.3 In this respect, autophagy is one of the most powerful systems for cellular clean-up, as lysosomal hydrolases can degrade all type of macromolecules (protein, lipids, nucleic acids and complex sugars).4 However, the functions of autophagy go far beyond the elimination of damaged cellular components and protein quality control. In fact, all intracellular proteins and organelles, even if they are not damaged, undergo some degree of continuous synthesis and degradation. This recycling allows for their renewal and prevents malfunction due to wear and tear. 3
An additional reason for degradation of macromolecular structures is to utilize their breakdown products as an internally derived source of energy.5,6 Autophagic degradation of macromolecular structures in lysosomes generates amino acids, free fatty acids and carbohydrates moieties that can be recycled for synthesis of new cellular components or further oxidized to generate ATP, mostly in conditions when nutrients are scarce. Both, degradation and recycling are particularly important in highly metabolically active organs such as the liver, in which periods of starvation as short as 4 hours are enough to induce maximal activation of autophagy.7 Interestingly, this starvation-induced upregulation of autophagy in the liver led to the initial identification of this pathway in mammals.7 Early studies in hepatocytes also provided detailed descriptions of the vesicles that participate in the sequestration and delivery of cytosolic material to lysosomes. Quantification of the changes in the number and size of these vesicles and in liver protein turnover led to the characterization of the regulatory effect of selected amino acids and hormones such as insulin and glucagon on hepatic autophagy.7–9 However, identification of the genes responsible for autophagy did not begin until 30 years later, when a detailed molecular dissection of this process was performed in yeast.10 Discovery of the genes controlling autophagy and the fact that most of them are conserved from yeast to mammals 11 has provided momentum for numerous studies connecting malfunctioning of the autophagic system with different human diseases such as cancer, neurodegeneration, myopathies, infectious conditions and metabolic disorders.1,2 Modulating autophagy for therapeutic purposes holds potential for the treatment of the plethora of disease states in which autophagic processes are dysregulated. In fact, pharmacological manipulation of autophagy has already begun testing in clinical trials for different types of cancers and for some severe neuronal and muscular degenerative diseases.
The liver, the organ in which autophagy research started, has also benefited from this enhanced understanding of autophagy, a process that is tightly connected to hepatic pathophysiology. This Review is intended as an introduction for clinical scientists to the topic of hepatic autophagy, its contribution to liver physiology and to the growing number of liver diseases linked to autophagy malfunction.
Autophagic pathways in the liver
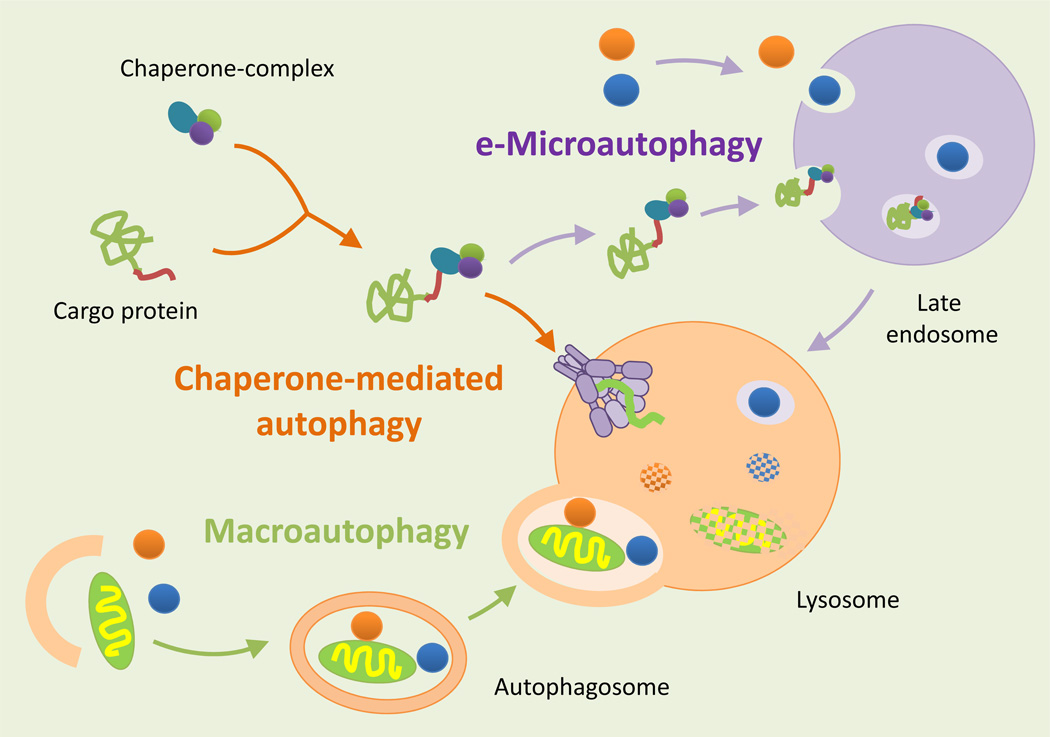
Hepatocytes, like most cells, have many ways to transport cytosolic material (cargo) to lysosomes for degradation. The identification of the mechanisms behind these different methods of delivery has helped define three co-existing types of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) (Figure 1). In many of these pathways, the autophagic process can be ‘constitutive’ (continuously active) or ‘inducible’ (activated in response to stimuli). In addition, cargo can be delivered to lysosomes in bulk (through random sequestration) or selectively (by individual targeting of each cargo molecule).
Figure 1. Autophagic pathways in the liver.
Three different types of autophagy co-exist in the liver. In macroautophagy, proteins, organelles, and cytosolic material are sequestered in a double-membraned vesicle (autophagosome) to be degraded by lysosomal hydrolases once the autophagosome fuses with lysosomes. In microautophagy, small vesicles form on the surface of late endosomes and are pinched off into the lumen to be degraded. By contrast, in CMA the substrate proteins cross the lysosomal membrane through a translocation complex instead of being delivered by vesicles. This type of transport requires the coordinate action of chaperones in the cytosolic side of the membrane that facilitate the unfolding of the substrate protein, and chaperones in the luminal side of the membrane that pull substrate proteins through the translocation complex one by one.
Macroautophagy
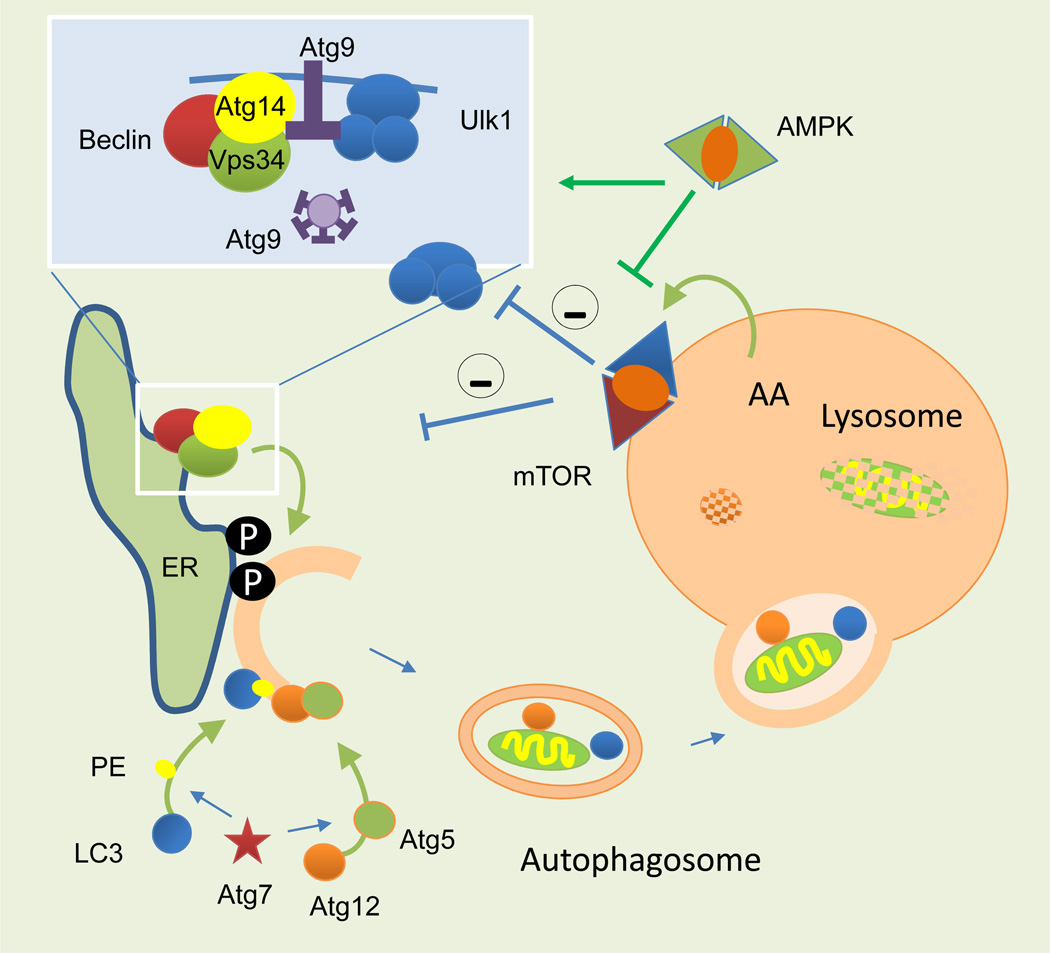
The distinctive step of macroautophagy is the formation of a double-membraned vesicle (autophagosome), which sequesters cytosolic material to be degraded by lysosomal hydrolases once the autophagosome fuses with lysosomes. Examples of types of macroautophagy that target specific organelles for degradation are mitophagy (mitochondria), pexophagy (peroxisomes), and lipophagy (lipid droplets). About 30 autophagy-related genes and their protein products (Atgs) organize into functional complexes that act co-ordinately at different steps of this process (reviewed elsewhere2,12). The temporal and spatial organization of the Atg complexes initiates autophagosome formation from sites such as the endoplasmic reticulum (ER), mitochondria, Golgi or the plasma membrane or endocytic system. Targeted autophagic degradation is possible owing to a subset of autophagy receptors, which link Atg proteins that initiate autophagosome formation to the surface of the designated cargo.13 For example, these receptors permit the identification and removal of one damaged mitochondrion from a whole pool of functional mitochondria14. Atg proteins also mediate the sealing of the autophagosome limiting membrane, known as the phagophore or isolation membrane, that encloses the cargo, and finally, Atg proteins also participate in the microtubule-dependent trafficking of the autophagosome and the ultimate fusion between autophagosomes and lysosomes.1,2 In certain conditions, autophagosomes might fuse instead with endosomes to create an intermediary compartment (amphisome) that then delivers cargo to lysosomes.15 The final step in macroautophagy, shared with the other autophagic pathways, is the degradation and recycling of breakdown products back into the cytosol. Several lysosomal membrane permeases involved in this recycling pathway have already been characterized in yeast, but the identification of mammalian homologues has just begun.16 The material freed from the lysosomes (amino acids, sugars and fatty acids) is sensed by regulatory complexes that modulate autophagy activity. For example, the mammalian target of rapamycin (mTOR), the best characterized negative regulator of macroautophagy, sits on the surface of lysosomes and is controlled by the efflux of select amino acids and glucose released from these compartments17 (Figure 2).
Figure 2. Molecular components of macroautophagy.
Autophagosome formation starts with the recruitment of autophagy-related proteins (Atg) to specific cellular membranes (ER membrane depicted here) where modification of the membrane lipids allows for recruitment and assembly of additional Atgs. Conjugation of certain Atgs to the lipids and proteins in the forming membrane facilitates its elongation. Ultimately, it seals to form a double-membraned autophagosome vesicle. Complete degradation of the sequestered cargo occurs when lysosomes infuse their hydrolases into autophagosomes through membrane fusion. The amino acids resulting from the breakdown are released into the cytosol through permeases. This amino acid efflux activates the sensor kinase complex (mTOR) that sits on the lysosomal membrane and inhibits autophagosome induction. Other energy sensors such as AMPK repress the inhibitory effect of mTOR on autophagy. AA: amino acids; Atg: autophagy-related proteins; ER: endoplasmic reticulum; LC3: light chain 3 protein; mTOR: mammalian target of rapamycin; P: phosphorylation; PE: phosphatidylethanolamine.
Macroautophagy was originally thought to be activated only in response to stress, such as starvation. However, macroautophagy also has a role in the unstressed liver since genetic blockage of this pathway in hepatocytes leads to accumulation of damaged organelles and altered proteins even under basal conditions.18 Therefore, maintenance of hepatocyte homeostasis and protein quality control requires constitutive macroautophagy. Both constitutive and inducible macroautophagy share the same core Atg machinery but they are subjected to different regulation. For instance, constitutive macroautophagy is insensitive to the well-characterized inhibitory effects that mTOR, insulin-signalling, and selected amino acids exert on starvation-induced macroautophagy. 19 In most organs, different stimuli (ER stress, hypoxia, oxidative stress, DNA damage or mitochondria dysfunction) can activate macroautophagy through a broad array of signalling pathways that include JNK (c-Jun N-terminal kinase), CaMKK (calcium/calmodulin-dependent protein kinase kinase), LKB (liver kinase B), AKT (protein kinase B), Sirt1 (sirtuin 1), PERK (protein kinase RNA-like endoplasmic reticulum kinase), PDGF (platelet-derived growth factor), AMPK (AMP-activated protein kinase), and p53-mediated signalling (reviewed elsewhere20).
Microautophagy
Microautophagy was also originally described in hepatocytes as the internalization of cytosolic material in lysosomes by invaginations in the lysosomal membrane (Figure 1).4,21 These invaginations pinch off from the membrane as single-membraned vesicles that undergo degradation in the lysosomal lumen. In contrast to the high conservation of the genes involved in macroautophagy, yeast microautophagy genes are not conserved in mammals.22 Although this lack of conservation has called into question whether microautophagy occurs in mammals, studies support the idea that a microautophagy-like process takes place at late endosomes in mammalian cells, including hepatocytes23 (Figure 1). Endosomal microautophagy utilizes components of the ESCRT (endosomal sorting complexes required for transport) complex, known to participate in internalization and recycling of plasma membrane proteins through formation of these endosomal vesicles.24 Interestingly, ESCRT proteins and functional multivesicular bodies are also required to sustain macroautophagic activity.25 Both an in bulk and a selective form of this process mediated by the chaperone hsc70 have been described.23
Chaperone-mediated autophagy
The distinctive feature of CMA is that substrate proteins are recognized in the cytosol by a chaperone26 that brings them to lysosomes where they cross the membrane through a translocation complex instead of being delivered by vesicles (Figure 1).27 This type of transport requires the unfolding of the substrate protein so it can cross the lysosomal lipid bilayer, and the presence of chaperones on both sides of the membrane to pull substrate proteins through one by one. The lysosome-associated membrane protein type 2A (LAMP-2A)28 and the constitutively expressed chaperone hsc70, are essential players in this type of autophagy.29–31 CMA was initially identified in fibroblasts in culture but most of its molecular characterization has been performed in the liver. Upregulation of CMA occurs as part of the cellular response to various cellular insults such as prolonged nutrient deprivation, oxidative stress, and proteotoxicity.32,33
Autophagy in the healthy liver
The number of cellular functions known to be linked to the autophagic process has considerably expanded in the last decade as a result of the many studies using genetic and chemical approaches to block or upregulate autophagy. Comparison of transgenic mouse models with blockage of autophagy in different organs has permitted the identification of a subset of functions of autophagy shared by all cells, as well as organ-specific functions. In this section we describe two of the main general functions attributed to autophagy in the healthy liver.
Energetic cellular balance
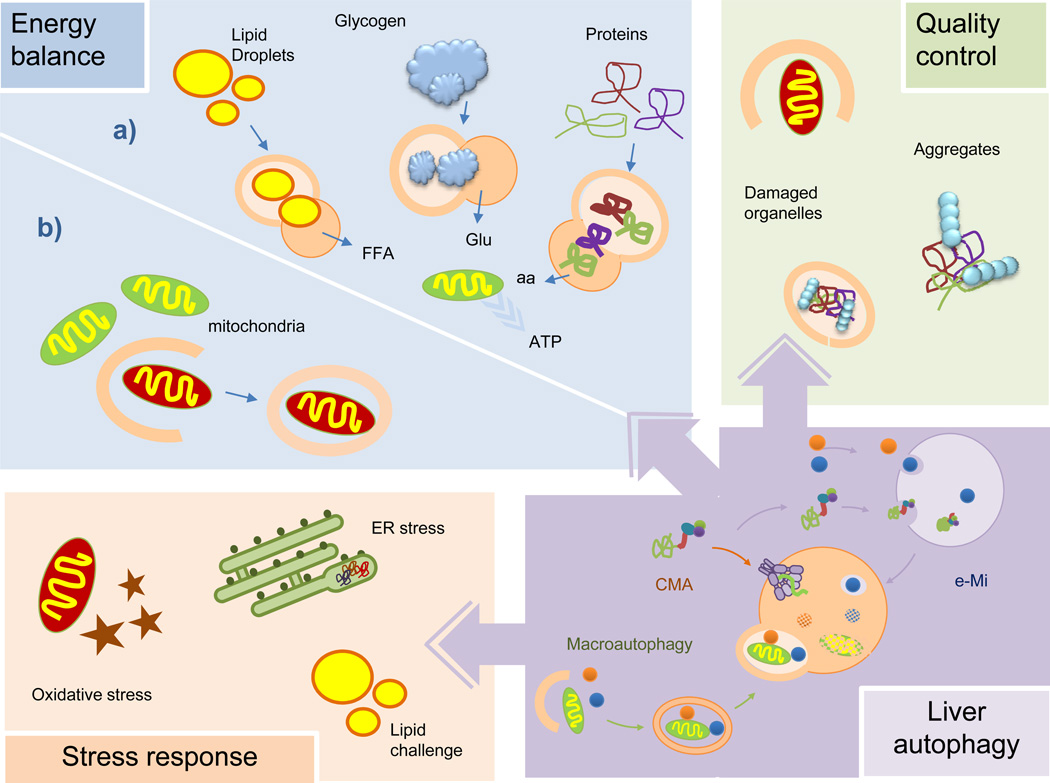
Autophagy helps maintain a positive energetic balance in the liver through different mechanisms: first, through breakdown of intracellular stores of macromolecules that can be utilized for cellular fuel; second, through control of key regulatory enzymes of cellular metabolism; and third, through preservation of mitochondrial homeostasis by efficient organelle turnover (Figure 3).
Figure 3. Functions of autophagy in liver physiology.
Autophagy maintains the energetic balance of the liver through turnover of different energy stores (a) and elimination of dysfunctional mitochondria (b). Autophagy eliminates altered proteins and organelles, as part of its function in cellular quality control, which otherwise will become toxic for hepatocytes. Liver autophagy is also activated for defense against hepatic damage resulting from oxidative stress, organelle stress (ER stress depicted here) or that induced by massive arrival of nutrients (lipotoxic challenge depicted here). aa: amino acids; ER: endoplasmic reticulum; FFA: free fatty acids; Glu: glucose.
Breakdown of intracellular energy stores
Hepatic macroautophagy is rapidly activated during the first 4–6 h of starvation. This increase in protein degradation replenishes the pool of intracellular amino acids and thus preserves protein synthesis under these conditions.21 The amino acids from the lysosomal breakdown can also enter the Krebs cycle and contribute to ATP generation34–36 or be utilized for gluconeogenesis.37 This autophagy-mediated adaptation to starvation is essential in neonates during the critical period between interruption of the transplacental supply of nutrients and lactation. In fact, mice lacking essential autophagy genes, although normal at birth, die in the first days of life owing to their inability to sustain high enough concentrations of amino acids to maintain ATP levels and the synthesis of essential proteins.38 Moreover, mice with specific deletion of essential autophagy genes in the liver have a marked inability to adapt to nutrient deprivation owing to failure of starvation-induced proteolysis and lipolysis.18 The energetic imbalance in these mice is further aggravated by the fact that mitochondria function, which is essential for beta-oxidation of lipids, is compromised due to the inefficient turnover of these organelles by autophagy.18
Activation of liver macroautophagy during starvation is promoted by the increase in circulating glucagon and the concomitant decrease in circulating levels of insulin and amino acids, both potent inhibitors of hepatic macroautophagy.7,9 Degradation of proteins by macroautophagy in the liver declines by 8 h into the starvation period and is followed by a gradual activation of CMA.39 Hepatic CMA activity reaches its maximum at 24 h after nutrient deprivation and remains activated for up to 3 days, serving as an internal source of amino acids.39 The mechanisms that trigger this activation of CMA in response to starvation are for the most part unknown. Signalling through the retinoic acid receptor alpha (RARα) is the only pathway described to modulate CMA activity so far.40 However, this pathway might not have an important role in the activation of CMA during starvation because the inhibition of CMA by retinoic acid signalling is insensitive to nutritional changes, and antagonists of RARα have an additive effect with starvation on CMA activation.40 Early studies analysed the possible stimulatory role of ketone bodies on CMA because ketones are generated in the liver during prolonged starvation.41 Although increased rates of CMA can be observed upon exposure of cultured cells to ketone bodies, these molecules do not mediate signalling but rather induce oxidation of proteins, which explains the increase in selective degradation through CMA.41 Given that activation of CMA by starvation does not require protein translation or synthesis, and the increase in the lysosomal levels of the receptor for this pathway during starvation is mainly the result of a decrease in the lysosomal turnover of this receptor,42 it is likely that starvation-mediated changes directly in the lysosomal compartment are responsible for CMA activation. In this respect, it would be interesting to elucidate if, as in the case of macroautophagy, nutrient sensing proteins at the lysosomal membrane are responsible for modulating the levels and organization of the CMA translocation complex.
Macroautophagy also persists in the liver after 8 h of starvation but there is a shift in the type of cargo sequestered by autophagosomes from proteins to lipids43 (Figure 3). Lipid droplets—cytosolic stores of triglycerides and cholesterol esters—are trapped inside autophagosomes and delivered to lysosomes for breakdown by the resident lysosomal lipases through a selective form of macroautophagy named macrolipophagy.43–45 Lysosome-mediated lipolysis is independent from the lipolytic process carried out by the cytosolic lipases and depends instead on intralysosomal lipases.46,47 Studies have shown that a whole transcriptional program is activated in the liver during starvation to prepare the autophagic/lysosomal system (including organelles and their enzymatic content) for the arrival and processing of the lipid cargo. 46,48 Although the relative contribution of lysosomal lipophagy to overall lipolysis under normal conditions might be discrete, lipophagy prevents massive accumulation of hepatic lipids in response to dietary lipid overload.43–45 Interestingly, mobilization of lipids by macroautophagy in the liver is not limited to hepatocytes. For instance, lipophagy is essential for the activation of hepatic stellate cells, a key event in liver fibrogenesis,49 which has led to the idea of transient chemical blockage of autophagy as a possible new therapeutic strategy for liver fibrosis 49,50.
Autophagy might also contribute to maintenance of the energetic balance in the liver through catabolism of glycogen, another major hepatic energy store51,52 (Figure 3). The selectivity of this process, known as glycophagy, has been reinforced by the discovery that the protein Stbd1 (starch-binding domain-containing protein 1) serves as the cargo-recognizing receptor that delivers glycogen to the autophagosome through interaction with the Atg protein GABARAPL1 (gamma-aminobutyric acid receptor-associated protein-like 1). Microautophagy also participates in delivery and degradation of glycogen in the vacuole in yeast,53 but it is unknown whether a similar process exists in mammals. More studies are also necessary to determine whether glycogenolysis, an essential process that provides glucose to extrahepatic tissues, is attributable to any type of autophagy.
Regulation of liver enzyme content
Besides active breakdown of energy stores, autophagy can contribute to the regulation of liver metabolism through the degradation of limiting enzymes in distinct metabolic pathways. CMA seems well suited for this task due to its ability to target single proteins for lysosomal degradation. In fact, key glycolytic enzymes have been shown to undergo proteolysis through this selective autophagic pathway.39,54 Degradation of regulators of cellular metabolism, such as p62, via macroautophagy55 has been proposed to also contribute to the transcriptional control of liver metabolism. In fact, most of the metabolic compromise in the mouse models of defective liver autophagy can be attributed to the accumulation of p62, because loss of autophagy in a p62 null animal presents with a very mild phenotype in comparison.56
Mitochondrial turnover
Proper functioning of mitochondria is key for the maintenance of the liver energetic balance. These organelles are most at risk of damage by reactive oxygen species, and often handle it by merging with the rest of the mitochondria network and diluting the burden.57 However, in cases of severe insult, altered mitochondria are actively excluded from fusing with the network and instead are delivered to lysosomes for complete degradation through a macroautophagy variant known as mitophagy58,59 (reviewed elsewhere14, 60). Changes at the membrane of the mitochondria are detected by cargo-recognizing molecules that bring along the essential components for the formation of the autophagosome57,59 (Figure 3). The molecular pair PINK and Parkin, the Bnip3-like protein NIX (also known as NIP3-like protein X), p62, and NBR-1 (next to BRCA1 gene 1 protein) have all been shown to contribute to mitophagy, although probably in response to different stimuli.59,61,62 For example, PINK and Parkin preferentially facilitate the degradation of depolarized mitochondria, but they are dispensable for starvation-induced mitophagy in which Bnip3 has a more prominent role.62 Mitophagy of damaged mitochondria is initiated through PINK1-dependent recruitment to the mitochondria of the ubiquitin ligase Parkin, which targets the protein mitofusin (involved in mitochondrial dynamics) for proteasome degradation.59 Parkin-induced degradation of mitofusin prevents accumulation of mitochondrial spheroids—the morphological representation of segregated damaged mitochondria often observed in liver injury.63 Lysosomes might directly fuse with the mitochondrial spheroids as an alternative mechanism for mitochondria degradation when autophagy is compromised.63 Although mitophagy is an important process in cellular quality control, it also mediates the normal turnover of healthy mitochondria and regulates the size of the mitochondria pool to accommodate, for example, changes in nutrient status.64
Cellular quality control
Autophagy has an important role in the clearance of abnormal or damaged hepatic proteins and organelles that would otherwise accumulate and lead to hepatotoxicity. Intracellular proteins with abnormal conformations resulting from faulty synthesis or posttranslational damage are frequent cargo of the different autophagic pathways in the liver. For example, although selective degradation of oxidized proteins is achieved in part through the proteasome, CMA is also activated in the liver after oxidative injury to mediate the removal of damaged proteins.32 Blockage of CMA renders cells more susceptible to oxidative damage,33 whereas genetic manipulations that prevent the age-dependent decrease of CMA in mouse liver markedly reduce the content of oxidized proteins in this organ.65 In fact, preservation of normal hepatic CMA activity until late in life in rodents improves overall hepatocyte homeostasis, maintains the ability of the liver to respond to stressors and keeps hepatic function in these old animals at levels comparable to those in young animals.65
The high levels of oxidative damage detected in the liver in mouse models of compromised autophagy highlights the importance of macroautophagy in the hepatocyte response against oxidative stress.18 Interestingly, hepatocytes have an antioxidant transcriptional program in place that is activated when autophagy is compromised to ameliorate the subsequent oxidative stress damage.66 The main regulator of this program, Nrf2, is under the control of an autophagy substrate, p62. Nfr2 is constitutively trapped by Keap1, which facilitates its degradation by the proteasome and maintains its transcriptional activity at a low level. When autophagy fails, p62 accumulates and its ability to bind Keap1 competitively prevents degradation of Nfr2, leading to the activation of the protective Nrf2- mediated program.67–69 However, the effect of Nrf2-mediated signalling in the liver might be more complex than initially anticipated because a study has shown that simultaneous elimination of Nrf2, Keap1 and Atg7 in mouse liver reduces the hepatic damage observed in Atg7-deficient animals.70
Although both macroautophagy71 and CMA32 become activated during oxidative stress, they are not redundant in their protective functions. In fact, the contribution of one pathway or the other becomes more prevalent depending on the target of the oxidative damage. Conditions with predominantly protein damage benefit from the selectivity of CMA, whereas organelle injury and accumulation of protein aggregates are handled for the most part by macroautophagy. Most of the anti-oxidant effect of macroautophagy has been attributed to its role in the removal of damaged or malfunctioning mitochondria through mitophagy, as described in the previous section (Figure 3).
Hepatic autophagy is also important for the maintenance of ER homeostasis. The very high rates of secretory protein synthesis sustained by hepatocytes impose continuous stress in the ER. Cells respond to ER proteotoxicity through the activation of the unfolding protein response (UPR), a local response that upregulates molecular chaperones, attenuates translation, and enhances degradation of misfolded proteins by the proteasome. Protein stress inside the ER lumen can also trigger macroautophagy,72,73 and this activation is in part mediated by transcription factors such as cJun74 but also by some of the factors that participate in the UPR75 (Figure 3). This coordinated activation of UPR and ER stressinduced autophagy might facilitate the disposal of misfolded proteins that cannot be fully degraded by the proteasome, as well as facilitate the selective degradation of portions of the ER to expedite recovery of ER homeostasis. 72,73,76 The ability of macroautophagy to minimize ER stress has been confirmed in vivo using genetic and dietary mouse models of obesity in which, as described in the following sections, severe compromise of autophagy is responsible for elevated ER stress, and restoration of normal levels of autophagy is sufficient to ameliorate this stress.77 However, considering the important contribution of the ER membrane to autophagosome biogenesis,78 the protective effect of macroautophagy against ER stress could have some limit, as conditions of chronic uncontrolled ER stress might have a negative effect on autophagic function.
In summary, degradation of hepatocyte organelles through autophagy serves both to eliminate those organelles that malfunction and thus prevent further cellular damage, and also to modulate the size and number of the pool of a particular organelle to adapt to intracellular and extracellular stress. Although the same basic structural autophagic components are shared in both cases, the signalling and modulatory pathways might be different depending on the purpose of the autophagic engagement.
Autophagy and liver disease
A growing number of liver pathologies have been associated with primary or secondary autophagy dysfunction. In some of these disorders, the consequences of the inadequacy of the autophagic process are closely linked to the role of autophagy in the maintenance of the energetic balance. In other instances, pathology arises because of the inability of hepatic autophagy to eliminate abnormal cellular components or structures damaged as a consequence of the underlying pathology. We have selected examples of liver pathologies in which autophagy malfunction contributes to loss of quality control, metabolic imbalance, or to the inability to carry out a stress response.
Alpha-1 antitrypsin deficiency
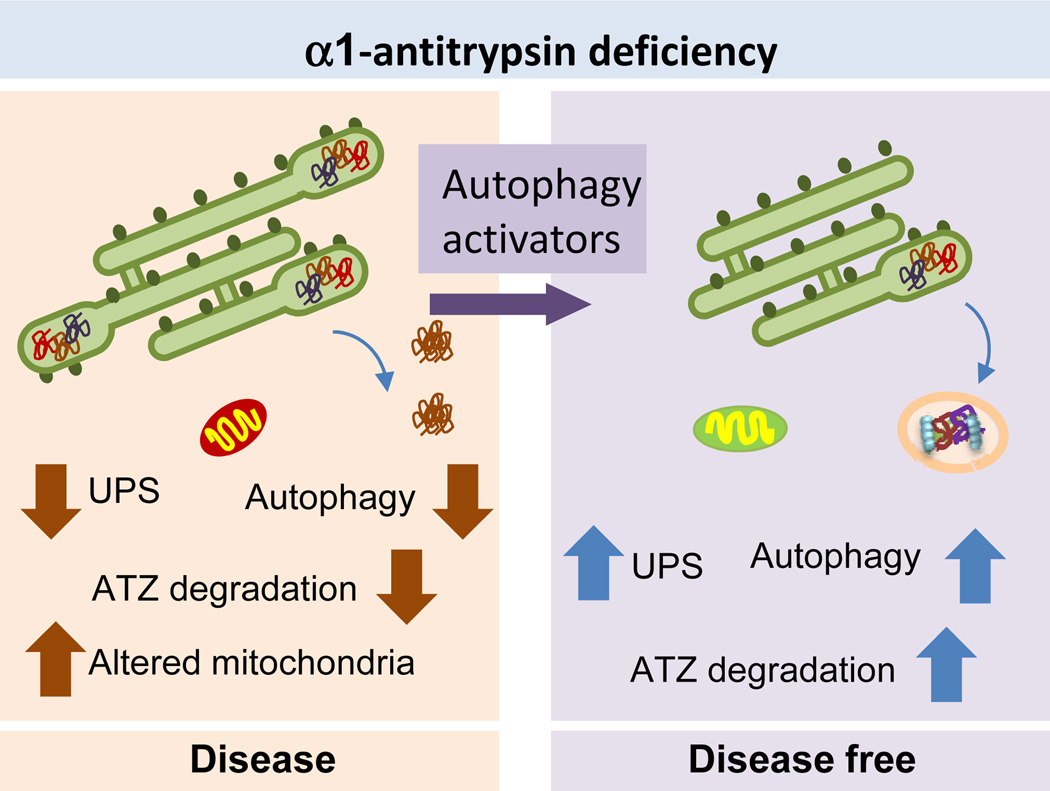
Failure of both proteasomal degradation and autophagy has been described in the liver-specific protein conformational disorder known as α1-antitrypsin deficiency. In this hereditary condition, a point mutation in the plasma protease inhibitor α1-antitrypsin, normally synthesized and secreted by the liver, causes trapping of the mutant protein (referred to as ATZ), in the ER in the form of insoluble protein inclusions.79 This proteotoxic insult ultimately progresses to chronic liver injury, hepatic fibrosis, and on occasion HCC (reviewed elsewhere80) (Figure 4a). Proteasomal degradation and autophagy are activated in response to the accumulation of ATZ in the ER and contribute to its degradation. 81,82 In fact, expression of ATZ in mouse models is sufficient to activate autophagy in vivo and, conversely, genetic blockage of macroautophagy leads to accumulation of ATZ intracellular inclusions in these animals.81 Constitutive upregulation of liver macroautophagy in this disease and the subsequent turnover of mutant α1-antitrypsin contribute to reduce toxicity, cellular damage and inflammatory responses in hepatocytes.81 However, liver damage can ultimately occur because the affected hepatocytes cannot further induce autophagy in response to stressors such as starvation, and they also exhibit alterations in mitophagy, which could explain the accumulation of abnormal mitochondria that is characteristic of the disease.82,83 Individual variability in the autophagic response of patients might explain the diversity of phenotypic expression and severity of liver disease in α1-antitrypsin deficiency and perhaps the predisposition to HCC.80
Figure 4. Liver autophagy and a-1-antitrypsin deficiency.
Left. Autophagy activity is reduced in patients with α1-antitrypsin deficiency. Failure to degrade the pathogenic form of the protein α1-antitrypsin (ATZ) results in its aggregation and accumulation as intracellular inclusions in hepatocytes. It also reduces turnover of damaged organelles, such as mitochondria, further contributing to hepatic damage. Right: Chemical enhancement of autophagy in the mouse model of this disease has been shown effective in reducing liver pathology by promoting the degradation of the accumulated protein, improving removal of the damaged organelles and restoring function of the ubiquitin proteasome system (UPS) also compromised in the disease.
The exciting finding that the autophagy-enhancing drug carbamazepine is able to decrease the number of ATZ-containing globules and reduce hepatic injury and fibrosis in mouse models of α1-antitrypsin deficiency supports the idea that pharmacological upregulation of autophagy might be of therapeutic value84 (Figure 4). Ongoing pilot studies are currently testing the efficacy of this autophagy activator in patients with α1-antitrypsin deficiency and end-stage liver failure for which the only current curative treatment is liver transplantation (http://clinicaltrials.gov/show/NCT01379469). The beneficial effect of enhancing autophagy in the α1-antitrypsin deficient liver has been further validated in studies with the transcription factor EB (TFEB), a master regulatory gene that controls a transcriptional program capable of upregulating autophagy and the lysosomal system.48 Gene transfer of Tfeb rescues the α1-antitrypsin deficient phenotype in mice by increasing the degradation of the oligomeric ATZ protein through autophagy.85 Although most current efforts are focused on α1-antitrypsin deficiency, pharmacologic or genetic activation of autophagy has therapeutic potential for protecting against hepatic damage caused by primary or secondary accumulation of other toxic pathogenic proteins in the liver. For example, autophagy has been shown to mediate the elimination of hepatic Mallory bodies,86 which are inclusions of keratin intermediate filaments that form in livers affected by chronic conditions such as alcoholic hepatitis, cirrhosis, Wilson’s disease, and hepatocellular carcinoma (HCC).
Toxic injury to the liver
Autophagy is an essential component of the cellular response to numerous stressors. Of relevance for the liver are the oxidative and hypoxic stress that underlie the basis of common types of hepatic injury such as those inflicted by ischaemia–reperfusion or by hepatotoxic drugs including alcohol. Although autophagy is activated as a protective mechanism in most of these instances, compromised autophagic activity, often as consequence of the effect of the aggressor on autophagy, aggravates the course of disease. Repairing the autophagic defect or enhancing the autophagic capacity of the liver might have curative value in these conditions.
Ischaemia–reperfusion injury
Ischaemia–reperfusion injury, a common complication of liver surgery and transplantation, is a result of the insult to hepatocytes during the ischaemic period and the subsequent damage from the activation of pro-inflammatory pathways and elevation of oxidative stress. Initial studies reported contradictory data on the protective or damaging effect of autophagy during ischaemia–reperfusion injury.87–90 Part of the confusion originates from the fact that read-outs of the autophagic process in studies with animal models in vivo and in patients are often limited to electron microscopy analysis or measurement of Atg proteins levels, which provide information on the steady-state of autophagy but functional measurements are lacking. For example, an increase in the number of autophagosomes observed by electron microscopy or in the levels of integral autophagosome Atg proteins could be a result of enhanced induction of autophagy but could also be the result of compromised clearance of autophagosomes. Thus, general agreement exists that multiple factors have to be taken into consideration in the interplay of autophagy with ischaemia–reperfusion injury. The effect of autophagy blockage or upregulation in ischaemia–reperfusion injury can be influenced by the ischaemic time, temperature, method of organ preservation, previous liver conditions and even aging. In the case of liver surgery, a single ischaemic event does not seem sufficient to induce autophagy, but ischaemic preconditioning (brief periods of vascular occlusion shown to be protective against ischaemia–reperfusion injury) enhances autophagic upregulation and decreases necrosis.87 Autophagy involvement is different in the case of transplants owing to the low temperature utilized to preserve the donor’s liver. The switch from cold to warm temperature during reperfusion is associated with an increase in autophagic vacuoles in the transplanted liver,88 which could be in part triggered by the lack of nutrients in the preserving solution.89 Claims of the deleterious effect of autophagy under these conditions are based on the hepatic protection observed upon treatment with phosphoinositide (PI) 3-kinase inhibitors or inhibitors of lysosomal proteolysis.90 However, these compounds have effects beyond autophagy, and in the case of lysosomal proteolysis inhibitors, a study with chloroquine found that although beneficial in the early period, it aggravates liver damage in later ischaemia–reperfusion stages.91 The idea of a beneficial effect of autophagy in liver transplantation is gaining more support because upregulation of autophagy is a common feature of interventions shown to be protective against ischaemia–reperfusion injury such as pretreatment with proteasome inhibitors,92 AMPK93 and CaMKIV (calcium/calmodulin-dependent protein kinase IV)94 activators or lithium.95 In addition, general consensus exists that upregulation of autophagy has a beneficial effect in cases in which autophagy in the donor liver is reduced to start with, for example in steatotic rodent and human liver grafts,93,96 and in experimental animal models when using livers from old mouse donors,97 or mouse livers subjected to pretreatments that reduce autophagic activity such as hydrogen sulphide.98 Although further investigation is required to understand the basis for autophagic protection against hepatic ischaemia–reperfusion injury, the ability of autophagy to contribute amino acids, to remove the damage caused by anoxia and to reduce generation of reactive oxygen species by elimination of mitochondria by mitophagy are all likely mechanisms
Alcohol abuse
Owing to the role of the liver in xenobiotic metabolism, hepatocytes are exposed to many chemicals that at high toxic concentrations often interfere with hepatic lysosomal protein degradation.99 For example, the liver is the organ that sustains the highest level of damage during alcohol abuse. Hepatocytes have been show to upregulate autophagy in response to alcohol exposure.100 The autophagic increase is selective for lipophagy and mitophagy, which interestingly target the two organelles most affected by the alcohol intoxication exposure, that is lipid droplets and mitochondria.100 However, reduced autophagic activity upon exposure to alcohol has also been reported in other studies,101,102 which suggests that different factors, such as the duration of the alcoholic intoxication, might modulate the effect on autophagy. For example, although autophagic activation might occur in response to the oxidative stress resulting from alcohol catabolism in hepatocytes,103 primary and secondary oxidants generated during this process have been described to upregulate autophagosome synthesis but prevent their clearance because of their toxic effect on lysosomes.104 The inhibitory effect of alcohol on autophagy can also be secondary to the inhibition of AMPK after chronic ethanol consumption105 or to the increase in hepatic lipids that occurs as result of SREBP-induced lipogenesis106 Considering these inhibitory effects of long-term alcohol consumption on autophagy, chemical upregulation of this pathway might be of use against alcohol-induced liver damage.
Paracetamol overdose
Hepatic toxicity caused by overdoses with paracetamol is another clinically relevant condition in which the therapeutic value of modulating autophagy has been explored. Treatment with rapamycin has been shown to reduce the liver injury caused by this drug, suggesting a protective role for autophagy.44 The severe damage that paracetamol’s reactive metabolites cause in mitochondria is probably the trigger for the observed activation of Parkin-dependent mitophagy.63,107
NAFLD and metabolic syndrome
Multiple levels of interaction have been described between liver autophagic function and NAFLD. The characteristic accumulation of hepatic lipids in this condition is often a consequence of increased lipid influx in this organ, enhanced de novo synthesis of hepatic lipids and reduced mobilization and utilization of liver lipid stores.108 The high prevalence of NAFLD in individuals with type 2 diabetes or with morbid obesity suggests common pathogenic mechanisms for all these conditions. The fact that alterations in autophagy have been described in NAFLD, obesity and diabetes has led to proposals that autophagic failure might underlie the basis of the metabolic syndrome and justifies the current interest in exploring the use of chemical modulators of autophagy in its treatment.108–109
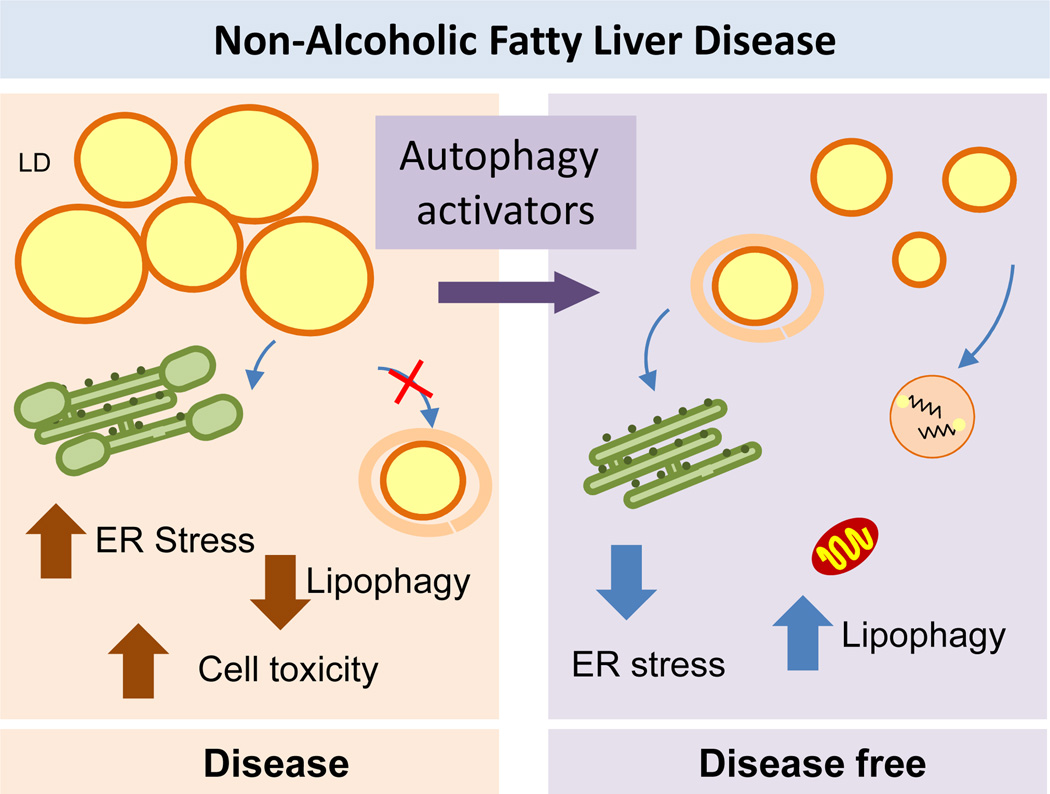
As in other diseases, the interplay between NAFLD and autophagy might change as the disease progresses, and the failure of autophagy can be a cause or a consequence of the disease. The role of macroautophagy in the mobilization of hepatic lipids43 supports a protective role of autophagy against NAFLD and explains why hepatocytes upregulate autophagy in response to dietary lipid challenges as a mechanism of defence against lipotoxicity.43,110 In fact, studies in mouse models overexpressing TFEB have demonstrated that this factor protects against steatosis induced by a high-fat diet in an autophagy dependent manner.46–48 In addition to the protective effect of lipid mobilization by lipophagy, autophagy might also be beneficial against the ER-stress associated with lipotoxicity. However, the defensive effect of autophagy in this condition might have limitations as studies have attributed hepatic steatosis and compromised turnover of lipids in models of NAFLD, at least partially, to a reduction of autophagic activity.77,113 Failure of autophagy in NAFLD might be explained by observations that chronic exposure to a high-fat diet or acute exposure to a cholesterol-enriched diet actually inhibits autophagy.43,110 Upregulation or inhibition of autophagy in response to a lipid challenge might be dependent not only on the intensity and duration of the challenge but also on the types of lipids. For example, hepatocyte exposure to unsaturated fatty acid induces autophagy whereas exposure to saturated fatty acids suppresses autophagy and induces apoptosis.45 However, the compromise of autophagy in hepatosteatosis might still be reversible, because injection of TFEB in mice being fed a high-fat diet stops the development of obesity in an autophagy-dependent manner.48 Furthermore, the same intervention in a genetic mouse model of obesity improves the metabolic syndrome phenotype,48 which suggests that activation of autophagy under these conditions might not only have preventive value but also could be utilized in the treatment of the metabolic disorder, at least in the case of autophagy induced by TFEB.
Several mechanisms might be responsible for the autophagic failure in NAFLD (Figure. 5). Lipid-enriched diets have been shown to impose changes in the membrane lipid composition of autophagosomes and lysosomes that reduce their fusogenic capacity, thus inhibiting completion of the autophagic process.110 These diet-induced changes in the lysosomal membrane also have a negative effect on CMA, as they reduce the stability of the CMA receptor in lysosomes.111 Interestingly, changes in the lipid composition of the lysosomal membrane upon dietary challenges are remarkably similar to those observed during physiological aging, a condition also associated with reduced CMA,112 which underscores the importance of environmental influences and nutritional choices on liver autophagic function. Impairment of autophagy in NAFLD might also occur at the level of induction owing to changes in cellular signalling. Autophagy in the liver is normally suppressed by insulin signalling,9 which intuitively suggests that conditions with insulin resistance, such as NAFLD and obesity, should lead to augmented hepatic autophagy. However, experimental evidence has revealed suppression of liver autophagy and mitophagy in obesity mouse models with insulin resistance and hyperinsulinemia through a FoxO1-dependent mechanism.113 Reduced autophagy under these conditions is in part a consequence of decreased expression of essential Atgs, which interestingly occurs whether the hyperinsulinemia is normalized or not.77,113 Surprisingly, restoration of normal hepatic autophagic function in the context of obesity is sufficient to improve hepatic insulin action and restore peripheral glucose tolerance.77 This unexpected turn of events, by which autophagy modulates sensitivity to its own inhibitor, insulin, seems to originate from the ability of this pathway to attenuate ER stress, which has been proven to contribute to insulin insensitivity.77
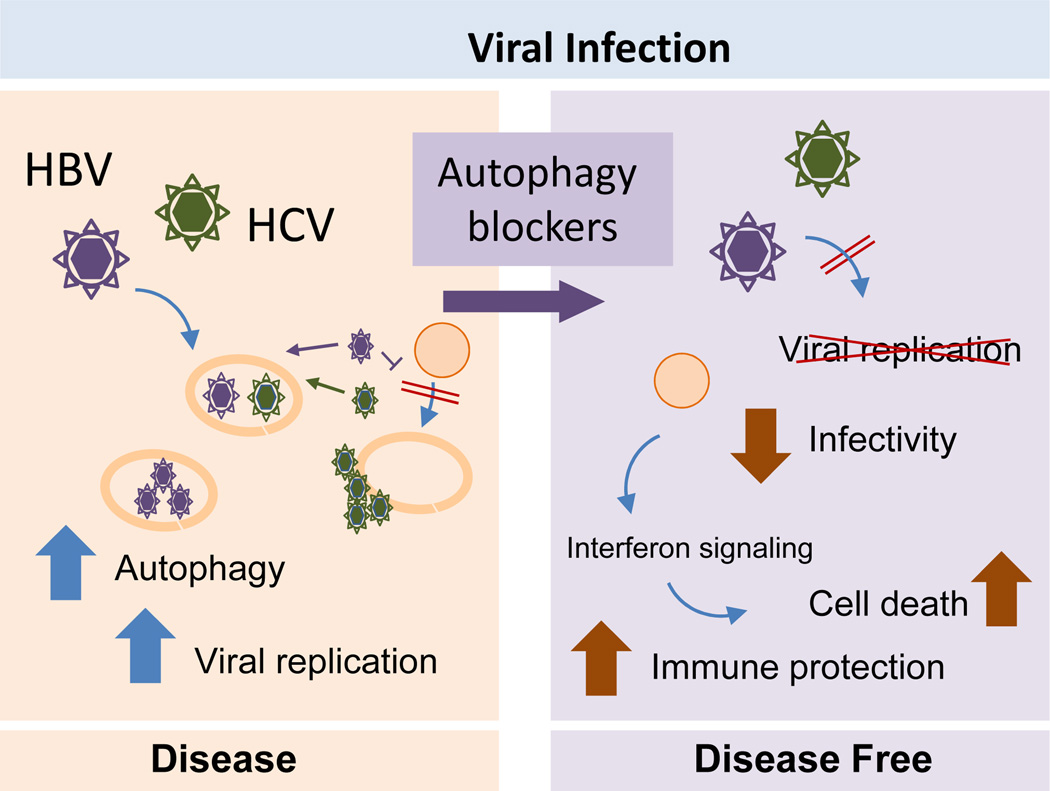
Figure 5. Hick-jacking of the autophagic system during liver viral infection.
Left: Hepatitis B (HBV) and C (HCV) virus induce formation of autophagosomes but prevent the degradation of these compartments by lysosomes. This allows virus to replicate inside or in the surface of autophagosomes. Right: Autophagy blockage has proven to be an effective anti-viral treatment against HBV and HCV by limiting viral replication and activating the host immune response.
When considering the course of NAFLD, it is still a matter of debate whether the compromised autophagic function in the liver is a consequence of obesity and changes in insulin sensing or whether hepatic autophagy impairment strikes earlier on in the metabolic syndrome, and is followed by hepatic steatosis, lipotoxicity and metabolic dysregulation. Examples of both scenarios have been documented.43–45,111 As described before, the secondary compromise of autophagy by dietary lipids is now well supported, 43,110,111 but it is plausible that, for example in the case of the metabolic syndrome associated with aging, the primary defect in autophagic function associated with age could be the trigger for inadequate lipid handling in the liver and the subsequent metabolic dysfunction. Distinguishing between cause and consequence might not always be possible, but the identification of this bidirectional interplay between autophagy and lipids in the liver supports the notion that breaking this vicious cycle, either by enhancing autophagy function or reducing the inhibitory effect of the metabolic imbalance (lipids and insulin) on this pathway, should help in these situations.
The possible therapeutic value of modulating autophagy in these abnormal metabolic conditions has not been clinically tested yet but drugs such as carbamazepine and rapamycin have shown promising effects in the reduction of liver steatosis induced in rodents by either dietary lipids or alcohol (Figure 5).114 Whether the therapeutic benefit is in part or fully attributable to their stimulatory effect on autophagy requires future investigation. Furthermore, the improvement of the metabolic conditions in mouse models of diet-induced obesity subjected to caloric-restriction or exercise has been correlated with the positive effect of both interventions on autophagy.109
Viral hepatitis
Autophagy aids in the removal of pathogens (a process called xenophagy) by working in conjunction with the innate immune system. However, several microorganisms have evolved unique mechanisms to circumvent, suppress, or exploit autophagic machinery to ensure their own survival and replication (reviewed elsewhere115–117). For instance, HSV-1 and HIV-1 block autophagy to abrogate their degradation through this pathway by blunting autophagosome formation or interrupting autophagosome–lysosome fusion, respectively118,119 (Figure 6). By contrast, pathogens such as polio and dengue activate autophagy in order to enhance their own replication.120
Figure 6. Autophagy failure in the pathogenesis of NAFLD.
Left: The failure of autophagy detectable in NAFLD could be the cause for the accumulation of lipids in this organ or a consequence, since although autophagy has an active role in the mobilization of intracellular lipids, abnormal increase in the lipid content has been shown to exert an inhibitory effect on autophagy. Defective autophagy and lipotoxicity may contribute to ER stress. Right: Activation of autophagy reduces steatosis in NAFLD by promoting lipid droplets (LD) mobilization and reducing ER stress.
HBV infection
In the case of the liver, considerable attention has been paid to the interplay between autophagic degradation and viral hepatitis. HBV —a hepatotropic DNA virus associated with acute viral hepatitis and chronic liver diseases such as cirrhosis and HCC121—induces autophagy in cells in culture, transgenic mice and in livers from infected patients.122–124 Paradoxically, despite the increased number of autophagosomes produced, protein breakdown is not augmented, suggesting that HBV induces the early autophagy stages but halts autophagosome maturation and clearance (Figure 6).122,123 HBV-mediated induction of autophagy has been attributed so far to the action of two different viral proteins, the hepatitis B virus X protein (HBx) and the small surface protein (SHBs).122,123.125 The multifunctional regulatory protein HBx has been shown to activate the kinase activity of the autophagy initiation complex122 and also to induce the expression of Beclin-1, the most important modulator of that complex.125 In the case of the SHBs protein, activation of autophagy might be secondary to its ability to induce ER stress.123 How precisely autophagy contributes to enhancing HBV viral replication is still unclear, but most studies support an effect on DNA replication and viral envelopment 122,123 The inability of HBV to replicate in vivo in mouse models with compromised macroautophagy has provided rationale for the claim that pharmacological inhibition of autophagy might be a reasonable adjuvant therapeutic strategy to treat patients infected with HBV.124
HCV infection
Autophagic changes have also been observed in rodent liver and human hepatocyte in culture upon infection by the RNA virus HCV, which shares with HBV its hepatotropism and ability to cause acute hepatitis and chronic liver disease.126 Independent of HCV genotype, a consistent increase in the number of autophagosomes is observed in HCV-infected hepatocytes.127–129 Although reports are conflicting about whether fusion of autophagosomes with lysosomes is required or not for the beneficial effects of autophagy on viral RNA replication, consensus exists on the capability of HCV to drive autophagic activation. Several HCV non-structural proteins interact with Atg proteins and can directly induce autophagosome formation (Figure 6).130,131 Interestingly, besides upregulation of core components of the autophagy machinery, HCV infection also induces expression of selective autophagy effectors such as PINK1 and Parkin to specifically drive mitophagy.132 As with HBV, some HCV proteins can induce autophagy in infected cells through activation of the UPR in the ER.129,133,134 In fact, suppression of the ER stress signals in this context diminishes HCV replication, demonstrating that the induction of autophagy caused by HCV depends on an intact ER stress response. 129,133
The precise mechanism of how autophagy promotes HCV replication remains elusive, but several studies have suggested that autophagic vacuoles might act as a membranous web—a complex of viral proteins, replicating RNA and altered cell membranes—that serves as a transient scaffold for efficient translation of viral RNA135 (Figure 6). Autophagosomes might also facilitate viral replication by sequestering negative regulators or conversely by attracting viral proteins needed for virion propagation.128,136 For example, pro-viral autophagy proteins Atg5, Atg12, and Beclin-1 are required for delivery of incoming viral RNA to the translation apparatus for initiation of HCV replication.128 Similarly, Atg5 has been shown to interact with HCV NS5B, a viral protein necessary to establish the membrane-bound replication complex that catalyzes RNA synthesis.136 Interestingly, this interaction is transient, emphasizing the importance of autophagic involvement during the onset of viral infection but not necessarily to sustain it.128,136 Lastly, part of the beneficial effect of blockage of autophagosome clearance in viral replication might be accomplished by preventing degradation of lipid droplets, which are known to be a favourite site for assembly and production of HCV virions.137
Blockade of liver autophagy could become a therapeutic option for HCV treatment as silencing of different autophagy proteins has been shown experimentally to be effective in inhibiting viral replication, decreasing intracellular HCV levels and extracellular infectivity. 128,136 Furthermore, suppression of autophagy in infected cells induces severe cytoplasmic vacuolation and cell death, underscoring the importance of autophagy in minimizing toxic vacuole formation.134 Part of the beneficial effect of suppression of macroautophagy in the context of HCV infection might be attributable to enhancement of the host’s innate immune system133,138 (Figure 6). Inhibition of autophagy in HCV-infected hepatocytes activates interferon signalling and induces apoptosis.133,138 Overall, given that autophagy seems to be important in the early stages of infection, autophagy blockers might be best-suited for immediate treatment upon HCV exposure.
Hepatocellular carcinoma
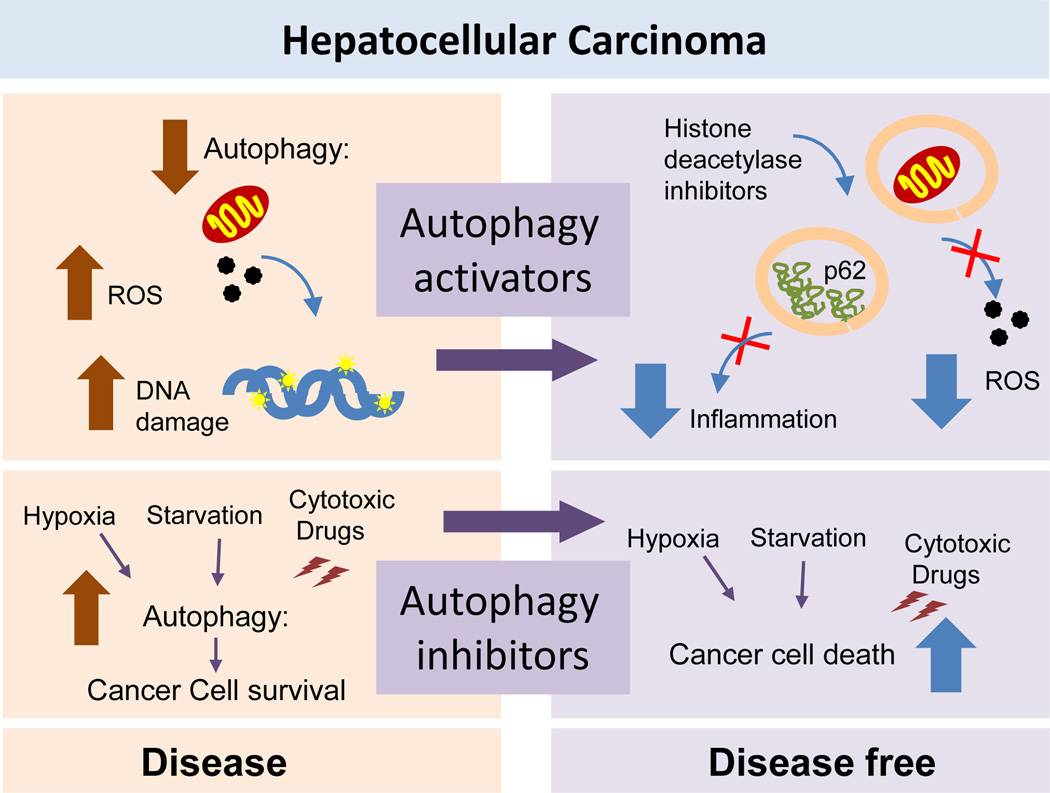
Early studies of macroautophagy in cancer emphasized its tumour-suppressive function on the basis that autophagy was reduced in different types of liver cancer139 and that in normal cells autophagy helps mitigate damage during genotoxic and proteotoxic stress, assuring chromosomal stability and reducing chances of malignant transformation140,141 (and reviewed elsewhere142–145). The fact that several tumour suppressor proteins induce macroautophagy whereas some oncogenic proteins inhibit this process also reinforced the anti-oncogenic role of autophagy.143,145 In the case of the liver, the idea that reduced levels of autophagy will favour cell proliferation fitted well with the early findings that autophagy and overall protein catabolism were inhibited in the regenerating liver following partial hepatectomy to promote the switch from a catabolic to an anabolic state.146 However, later studies have revealed that the role of autophagy in tumorogenesis is context-dependent (Figure 7). Some cancer cells can upregulate autophagic activity to enhance their survival in the hostile tumour microenvironment, thus promoting tumour growth142–145. This protumorigenic effect occurs in part because macroautophagy efficiently removes the deleterious products caused by endogenous damage (hypoxia-induced reactive oxygen species) or exogenous damage (cytostatic or cytotoxic drugs),147 and in part because autophagy assists these cells to maintain a positive energetic balance in the face of increasing metabolic demands.55, 148
Figure 7. The complex interplay between autophagy and hepatic carcinogenesis.
Left: Autophagy is protective against malignant transformation by reducing the damage generated during genotoxic and proteotoxic stress. However, once carcinogenesis occurs, cancer cells may downregulate or upregulate autophagy in a contex-dependent manner. Thus, in basal conditions most hepatocarcinoma cells have reduced autophagy, which may favour additional DNA damage and increase oxidative stress. In contrast, in response to cytotoxic treatments or as result of the poor nutritional conditions of the center of a solid tumor, cancer cells upregulate autophagy with defensive purposes. Right: Both activation and inhibition of autophagy have shown anti-tumoral effect. Activation of autophagy is protective by reducing levels of intracellular damage and improving genome stability, reducing the inflammatory response associated to p62 accumulation and as co-adjuvant of drugs that require activation of autophagy. Autophagy inhibition renders cancer cells more vulnerable to different stressors limiting their proliferation and survival.
In the case of HCC, the most prevalent form of liver cancer, most studies support a tumour-suppressive function of autophagy. HCC cell lines and tumour biopsies often exhibit decreased expression of autophagy markers.149 In fact, there is an inverse correlation between the aggressiveness of the HCC and autophagic activity that links diminished autophagy with poorer prognosis.149 Likewise, expression of Atgs such as beclin-1 after curative resection has been shown to correlate with disease-free survival in patients.149 Experimental evidence also supports the protective effect of liver autophagy against carcinogenesis, as mice with heterozygous disruption of Beclin-1 are prone to developing spontaneous malignancies including HCC as well as accelerated HBV-induced carcinomas.150 Other studies have demonstrated that autophagy exerts a pro-carcinogenic effect in HCC by protecting hepatic cancer cells from damage resulting, for example, from hypoxic stress,147,151 or by inducing expression of factors that stimulate cell invasion and metastasis (Figure 7).152 The precise mechanisms that lead to autophagic alterations in HCC have yet to be uncovered, as are the conditions and/or the timings that determine whether and when tumour cells upregulate or downregulate macroautophagy. However, the fact that some tumour suppressors and oncogenes directly modulate autophagy and that several signalling pathways regulating autophagy closely overlap with those that regulate tumorigenesis (reviewed elsewhere142,143,153) supports the existence of continuous feedback between both processes. For example, the signalling adaptor p62 has risen as one of the favourite candidates in the regulation of the interplay between carcinogenesis and autophagy in light of the previously mentioned effect of p62 on the activation of the Nrf2-dependent transcriptional program.67,154 Increase in p62 levels and induction of Nrf2 have been described in >25% of human HCC biopsies, and experimental blockage of p62 in HCC cell lines is effective at reducing cell growth.154
The potential of chemical modulation of macroautophagy as antioncogenic therapy is undergoing testing in clinical trials for several types of cancers (Figure 7). One of the requirements before pharmacologically manipulating autophagy in HCC is to gain a clear understanding of whether autophagy is depressed or upregulated at the time of the intervention. IN additional, current anti-HCC treatments have different effects on macroautophagy, which might determine the selection of inhibitors or activators of this pathway as co-adjuvant therapeutics. For example, sorafenib, common in the treatment of advanced HCC, showed improved efficacy in causing cell death in vitro and in vivo when used in combination with pharmacological inhibitors of autophagy.155 In fact, induction of autophagy by this drug has been proposed to contribute to the acquired drug resistance in patients with HCC.156 Inhibition of autophagy also increases the efficacy of antiangiogenic agents against HCC.157 However, blockage of autophagy might not be a universal approach as a co-adjuvant agent in HCC therapy given that other drugs, such as histone deacetylase inhibitors, actually stimulate autophagy and depend on its activation to cause cytotoxicity and death in cancer cells.158
Other current limitations to the generalized use of macroautophagy modulators in HCC therapeutics is the rather limited number of available compounds. Rapamycin and other derivatives acting on the mTOR complex, the most commonly used activators of autophagy, have shown antitumor efficacy in advanced HCC in a phase II study,159 but their additional autophagy-independent effects on processes such as protein translation and cellular growth highlights the need for more potent and selective autophagy activators. Similar limitations apply to chloroquine, an agent that disrupts lysosomal acidification, and that is the leading compound used in the clinic to block autophagic flux. An emerging alternative to chemical modulation of autophagy in the treatment of HCC could be gene therapy to target microRNAs described to upregulate or downregulate autophagy.160
A role for CMA in cancer cell metabolism has also been identified, in this case by contributing to maintaining high rates of glycolysis in transformed cells.161 CMA activity has been shown to be constitutively active in different types of cancer, including HCC, regardless of the status of macroautophagy.161,162 The fact that genetic inhibition of CMA in human cancer cell lines or directly in tumours of human cancer cells in mice effectively reduces tumour growth and metastasis161 supports the possible antioncogenic therapeutic value of negative modulators of this pathway.
Conclusions
The relevance of autophagy in liver physiology goes beyond the initially reported role as a supplier of amino acids during starvation. New roles for autophagy encompass a complex array of functions including global regulatory effects on liver metabolism, essential roles in protein and organelle quality control, and active defence against liver pathogens. The complexity of hepatic autophagy stems from not only its diverse roles in physiologic and pathophysiologic processes but also from the multiple mechanisms that contribute to its regulation.
Despite the general agreement on the beneficial effects of autophagy activation in liver physiology and in the protection against liver diseases, manipulation of this pathway with therapeutic purposes might not be as clear-cut as anticipated. In almost every liver pathology, blockage of autophagy can have opposite effects depending on differences in the type, duration and intensity of the pathogenic insult and on the way in which hepatocytes react to this insult. When autophagy manipulation is considered as adjuvant to other therapeutic approaches, the effect of these other interventions on the autophagic pathway might also dictate the choice of activators or inhibitors of autophagy. The array of compounds available to modulate autophagy is still relatively limited and most of them target the same autophagy steps (frequently initiation), whereas trafficking, fusion and degradation are still poor drugable targets. The development of disease-specific autophagy modulators requires a more complete understanding of the different signalling pathways that converge on autophagy and how they differentially regulate basal versus inducible or selective versus nonselective autophagy. In fact, although blockage of autophagy has been shown to be effective in reducing liver damage in different diseases, complete prolonged inhibition of autophagy would eventually have a detrimental effect on this organ, and consequently drugs that target only inducible autophagy but preserve constitutive autophagy would be desirable in those chronic settings. Such drugs would enable us to better target the specific autophagy pathway altered in the disease of interest without perturbing the global autophagic process in hepatocytes.
Review Criteria.
A PubMed search of English language papers published between 1960 and 2012 was conducted using the terms: “autophagy” and “liver” in combination with the following terms: ”liver disease”, ”energy”, “NAFLD”, “transplantation”, “hepatitis”, “hepatocellular carcinoma”, “steatosis”, “alcohol”, “ischemia”, “transplantation”, “mitophagy”, “alpha-1 antitrypsin”, “acetaminophen”. Retrieved abstracts were read and relevant publications were acquired as full text. Some of the papers in the current reference list were identified from the reference section of the initial leads and then acquired as abstracts or full text.
Key points.
Autophagy contributes to the maintenance of the energetic balance, participates in hepatocyte quality control and aids in the defence against exogenous pathogens and conditions that cause cellular toxicity in the liver
The different autophagic pathways that coexist in the liver are in dynamic intercommunication with one another, allowing for a coordinated response to maintain homeostasis
Failure to properly execute the autophagic program renders hepatocytes vulnerable to stressors and unable to accommodate the extreme energetic demands of this organ
Autophagy malfunction underlies the pathogenesis of common liver diseases including metabolic disorders, protein conformational diseases, viral infection and carcinogenesis
Chemical modulation of autophagy has proven to be beneficial in certain liver pathologies, although manipulation of autophagy needs to be customized according to the status of the autophagic pathway in each disease state
Acknowledgments
Work in our laboratory is supported by grants from the National Institutes of Health AG21904, AG031782, DK098408, a Hirschl/Weill-Caulier Career Scientist Award and the generous support of R. and R. Belfer. JLS was supported by NIH/NIA T32NS007098, T32-GM007288 and F30AG046109.
Biographies
Jaime L. Schneider obtained her undergraduate degree from Northwestern University with a major in Biological Sciences. She worked as a technician at the Feinberg School of Medicine at Northwestern University in the Department of Pulmonary and Critical Care Medicine. Currently, Jaime is in the MD/PhD training program (MSTP) at the Albert Einstein College of Medicine. She is working on characterizing the role of Chaperone-Mediated Autophagy in vivo in Dr. Ana Maria Cuervo’s laboratory for her thesis dissertation.
Ana Maria Cuervo obtained her medical degree from the U. Valencia, Spain and received training in biochemistry and cell biology with emphasis on autophagy during her PhD and her postdoctoral training under the mentoring of Drs. E. Knecht and J.F. Dice, respectively. She is currently a Professor at the Albert Einstein College of Medicine, where her group continues studies on the molecular characterization of autophagic pathways in mammals and the contribution of autophagy malfunctioning to disease and aging.
Footnotes
Financial Disclosure
The authors declare no competing interest.
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Biochim Biophys Acta. 2012;1824:3–13. doi: 10.1016/j.bbapap.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 4.De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 6.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–C16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature. 1977;270:174–176. doi: 10.1038/270174a0. [DOI] [PubMed] [Google Scholar]
- 9.Mortimore G, Mondon CE. Inhibition by insulin of valine turnover in liver. J Biol Chem. 1970;245:2375–2383. [PubMed] [Google Scholar]
- 10.Klionsky DJ, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 11.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. Journal of Biological Chemistry. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 12.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 13.Novak I, Dikic I. Autophagy receptors in developmental clearance of mitochondria. Autophagy. 2011;7:301–303. doi: 10.4161/auto.7.3.14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Klionsky DJ. Permeases recycle amino acids resulting from autophagy. Autophagy. 2007;3:149–150. doi: 10.4161/auto.3631. [DOI] [PubMed] [Google Scholar]
- 17.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto A, Cremona M, Rothman J. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortimore GE, Lardeux BR, Adams CE. Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. J Biol Chem. 1988;263:2506–2512. [PubMed] [Google Scholar]
- 22.Sakai Y, Koller A, Rangell L, Keller G, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris. Identification of specific steps and morphological intermediates. J Cell Biol. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahu R, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 25.Rusten TE, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 26.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 27.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 29.Kaushik S, Cuervo AM. Chaperones in autophagy. Pharmacol Res. 2012;66:484–493. doi: 10.1016/j.phrs.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23:184–189. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiffin R, Christian CJ, Knecht E, Cuervo AM. Activation of Chaperone-mediated Autophagy during Oxidative Stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Nat Acad Sci USA. 2006;103:5905–5910. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsui Y, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 35.Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal. 2010;3:ra31. doi: 10.1126/scisignal.2000911. [DOI] [PubMed] [Google Scholar]
- 36.Lum J, et al. Growth Factor Regulation of Autophagy and Cell Survival in the Absence of Apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 37.Ezaki J, et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–736. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 39.Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 40.Anguiano J, et al. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol. 2013 doi: 10.1038/nchembio.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn PF, Dice JF. Ketone bodies stimulate chaperone-mediated autophagy. J Biol Chem. 2005;280:25864–25870. doi: 10.1074/jbc.M502456200. [DOI] [PubMed] [Google Scholar]
- 42.Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 43.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding WX, et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mei S, et al. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Ther. 2011;339:487–498. doi: 10.1124/jpet.111.184341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol. 2013 doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Settembre C, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013 doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thoen LF, et al. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55:1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 50.Hernandez-Gea V, et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Jiang S, et al. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J Biol Chem. 2010;285:34960–34971. doi: 10.1074/jbc.M110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takikita S, et al. The values and limits of an in vitro model of Pompe disease: the best laid schemes o' mice an' men. Autophagy. 2009;5:729–731. doi: 10.4161/auto.5.5.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lv L, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 57.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glick D, et al. BNip3 regulates mitochondrial function and lipid metabolism in the liver. Mol Cell Biol. 2012;32:2570–2584. doi: 10.1128/MCB.00167-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ding WX, et al. Parkin and mitofusins reciprocally regulate mitophagy and mitochondrial spheroid formation. J Biol Chem. 2012;287:42379–42388. doi: 10.1074/jbc.M112.413682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nature Medicine. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsumoto N, et al. Comprehensive proteomics analysis of autophagy-deficient mouse liver. Biochem Biophys Res Commun. 2008;368:643–649. doi: 10.1016/j.bbrc.2008.01.112. [DOI] [PubMed] [Google Scholar]
- 67.Jain A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lau A, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 70.Taguchi K, et al. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scherz-Shouval R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogata M, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fuest M, et al. The transcription factor c-Jun protects against sustained hepatic endoplasmic reticulum stress thereby promoting hepatocyte survival. Hepatology. 2012;55:408–418. doi: 10.1002/hep.24699. [DOI] [PubMed] [Google Scholar]
- 75.Kouroku Y, et al. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 76.Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- 77.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell metabolism. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- 80.Perlmutter DH, Silverman GA. Hepatic fibrosis and carcinogenesis in alpha1-antitrypsin deficiency: a prototype for chronic tissue damage in gain-of-function disorders. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kamimoto T, et al. Intracellular inclusions containing mutant alpha1-antitrypsin Z are propagated in the absence of autophagic activity. J Biol Chem. 2006;281:4467–4476. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- 82.Qu D, Teckman JH, Omura S, Perlmutter DH. Degradation of a mutant secretory protein, alpha1-antitrypsin Z, in the endoplasmic reticulum requires proteasome activity. J Biol Chem. 1996;271:22791–22795. doi: 10.1074/jbc.271.37.22791. [DOI] [PubMed] [Google Scholar]
- 83.Teckman JH, An JK, Loethen S, Perlmutter DH. Fasting in alpha1-antitrypsin deficient liver: consultative activation of autophagy. Am J Physiol. 2002;283:G1156–G1165. doi: 10.1152/ajpgi.00041.2002. [DOI] [PubMed] [Google Scholar]
- 84.Hidvegi T, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 85.Pastore N, et al. Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in alpha-1-anti-trypsin deficiency. EMBO Mol Med. 2013 doi: 10.1002/emmm.201202046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harada M. Autophagy is involved in the elimination of intracellular inclusions, Mallory-Denk bodies, in hepatocytes. Med Mol Morphol. 2010;43:13–18. doi: 10.1007/s00795-009-0476-5. [DOI] [PubMed] [Google Scholar]
- 87.Domart MC, et al. Concurrent induction of necrosis, apoptosis, and autophagy in ischemic preconditioned human livers formerly treated by chemotherapy. J Hepatol. 2009;51:881–889. doi: 10.1016/j.jhep.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 88.Lu Z, et al. Participation of autophagy in the degeneration process of rat hepatocytes after transplantation following prolonged cold preservation. Arch Histol Cytol. 2005;68:71–80. doi: 10.1679/aohc.68.71. [DOI] [PubMed] [Google Scholar]
- 89.Kim JS, et al. Impaired autophagy: A mechanism of mitochondrial dysfunction in anoxic rat hepatocytes. Hepatology. 2008;47:1725–1736. doi: 10.1002/hep.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gotoh K, et al. Participation of autophagy in the initiation of graft dysfunction after rat liver transplantation. Autophagy. 2009;5:351–360. doi: 10.4161/auto.5.3.7650. [DOI] [PubMed] [Google Scholar]
- 91.Fang H, Liu A, Dahmen U, Dirsch O. Dual role of chloroquine in liver ischemia reperfusion injury: reduction of liver damage in early phase, but aggravation in late phase. Cell Death Dis. 2013;4:e694. doi: 10.1038/cddis.2013.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Padrissa-Altes S, Zaouali MA, Bartrons R, Rosello-Catafau J. Ubiquitin-proteasome system inhibitors and AMPK regulation in hepatic cold ischaemia and reperfusion injury: possible mechanisms. Clin Sci (Lond) 2012;123:93–98. doi: 10.1042/CS20110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zaouali MA, et al. AMPK involvement in endoplasmic reticulum stress and autophagy modulation after fatty liver graft preservation: a role for melatonin and trimetazidine cocktail. J Pineal Res. 2013;55:65–78. doi: 10.1111/jpi.12051. [DOI] [PubMed] [Google Scholar]
- 94.Evankovich J, et al. Calcium/calmodulin-dependent protein kinase IV limits organ damage in hepatic ischemia-reperfusion injury through induction of autophagy. Am J Physiol Gastrointest Liver Physiol. 2012;303:G189–G198. doi: 10.1152/ajpgi.00051.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu A, Fang H, Dahmen U, Dirsch O. Chronic lithium treatment protects against liver ischemia/reperfusion injury in rats. Liver Transpl. 2013;19:762–7472. doi: 10.1002/lt.23666. [DOI] [PubMed] [Google Scholar]
- 96.Degli Esposti D, et al. Ischemic preconditioning induces autophagy and limits necrosis in human recipients of fatty liver grafts, decreasing the incidence of rejection episodes. Cell Death Dis. 2011;2:e111. doi: 10.1038/cddis.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang JH, et al. Autophagy suppresses age-dependent ischemia and reperfusion injury in livers of mice. Gastroenterology. 2011;141:2188–2199 e6. doi: 10.1053/j.gastro.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang D, et al. The role of AKT1 and autophagy in the protective effect of hydrogen sulphide against hepatic ischemia/reperfusion injury in mice. Autophagy. 2012;8:954–962. doi: 10.4161/auto.19927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kovacs AL, Seglen PO. Inhibition of hepatocytic protein degradation by inducers of autophagosome accumulation. Acta Biol Med Ger. 1982;41:125–130. [PubMed] [Google Scholar]
- 100.Ding WX, Li M, Yin XM. Selective taste of ethanol-induced autophagy for mitochondria and lipid droplets. Autophagy. 2011;7:248–249. doi: 10.4161/auto.7.2.14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Noh BK, et al. Restoration of autophagy by puerarin in ethanol-treated hepatocytes via the activation of AMP-activated protein kinase. Biochem Biophys Res Commun. 2011;414:361–366. doi: 10.1016/j.bbrc.2011.09.077. [DOI] [PubMed] [Google Scholar]
- 102.Yang L, Wu D, Wang X, Cederbaum AI. Cytochrome P4502E1, oxidative stress, JNK, and autophagy in acute alcohol-induced fatty liver. Free Radic Biol Med. 2012;53:1170–1180. doi: 10.1016/j.freeradbiomed.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thomes PG, et al. Proteasome activity and autophagosome content in liver are reciprocally regulated by ethanol treatment. Biochem Biophys Res Commun. 2012;417:262–267. doi: 10.1016/j.bbrc.2011.11.097. [DOI] [PubMed] [Google Scholar]
- 104.Thomes PG, et al. Multilevel regulation of autophagosome content by ethanol oxidation in HepG2 cells. Autophagy. 2013;9:63–73. doi: 10.4161/auto.22490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sid B, Verrax J, Calderon PB. Role of AMPK activation in oxidative cell damage: Implications for alcohol-induced liver disease. Biochem Pharmacol. 2013;86:200–209. doi: 10.1016/j.bcp.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 106.Wu D, Wang X, Zhou R, Cederbaum A. CYP2E1 enhances ethanol-induced lipid accumulation but impairs autophagy in HepG2 E47 cells. Biochem Biophys Res Commun. 2010;402:116–122. doi: 10.1016/j.bbrc.2010.09.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ni HM, Bockus A, Boggess N, Jaeschke H, Ding WX. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology. 2012;55:222–232. doi: 10.1002/hep.24690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Koliaki C, Roden M. Hepatic energy metabolism in human diabetes mellitus, obesity and non-alcoholic fatty liver disease. Mol Cell Endocrinol. 2013 doi: 10.1016/j.mce.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 109.Cui M, Yu H, Wang J, Gao J, Li J. Chronic Caloric Restriction and Exercise Improve Metabolic Conditions of Dietary-Induced Obese Mice in Autophagy Correlated Manner without Involving AMPK. J Diabetes Res. 2013;2013:852754. doi: 10.1155/2013/852754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rodriguez-Navarro JA, et al. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2012;109:E705–E714. doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 113.Liu HY, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lin CW, et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58:993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sir D, Ou JH. Autophagy in viral replication and pathogenesis. Mol Cells. 2010;29:1–7. doi: 10.1007/s10059-010-0014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Orvedahl A, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 119.Kyei GB, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kudchodkar SB, Levine B. Viruses and autophagy. Rev Med Virol. 2009;19:359–378. doi: 10.1002/rmv.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rautou PE, et al. Autophagy in liver diseases. J Hepatol. 2010;53:1123–1134. doi: 10.1016/j.jhep.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 122.Sir D, et al. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci U S A. 2010;107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li J, et al. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol. 2011;85:6319–6333. doi: 10.1128/JVI.02627-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tian Y, Sir D, Kuo CF, Ann DK, Ou JH. Autophagy required for hepatitis B virus replication in transgenic mice. J Virol. 2011;85:13453–13456. doi: 10.1128/JVI.06064-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tang H, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- 126.Rosen HR. Clinical practice. Chronic hepatitis C infection. The New England journal of medicine. 2011;364:2429–2438. doi: 10.1056/NEJMcp1006613. [DOI] [PubMed] [Google Scholar]
- 127.Ait-Goughoulte M, et al. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sir D, et al. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Su WC, et al. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol. 2011;85:10561–10571. doi: 10.1128/JVI.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shrivastava S, Bhanja Chowdhury J, Steele R, Ray R, Ray RB. Hepatitis C virus upregulates Beclin1 for induction of autophagy and activates mTOR signaling. J Virol. 2012;86:8705–8712. doi: 10.1128/JVI.00616-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim SJ, Syed GH, Siddiqui A. Hepatitis C virus induces the mitochondrial translocation of Parkin and subsequent mitophagy. PLoS Pathog. 2013;9:e1003285. doi: 10.1371/journal.ppat.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Taguwa S, et al. Dysfunction of autophagy participates in vacuole formation and cell death in cells replicating hepatitis C virus. J Virol. 2011;85:13185–13194. doi: 10.1128/JVI.06099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gosert R, et al. Identification of the Hepatitis C Virus RNA Replication Complex in Huh-7 Cells Harboring Subgenomic Replicons. Journal of Virology. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guevin C, et al. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology. 2010;405:1–7. doi: 10.1016/j.virol.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McLauchlan J. Lipid droplets and hepatitis C virus infection. Biochim Biophys Acta. 2009;1791:552–559. doi: 10.1016/j.bbalip.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 138.Shrivastava S, Raychoudhuri A, Steele R, Ray R, Ray RB. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology. 2011;53:406–414. doi: 10.1002/hep.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kisen GO, et al. Reduced autophagic activity in primary rat hepatocellular carcinoma and ascites hepatoma cells. Carcinogenesis. 1993;14:2501–2505. doi: 10.1093/carcin/14.12.2501. [DOI] [PubMed] [Google Scholar]
- 140.Mathew R, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liang X, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 142.Chen HY, White E. Role of autophagy in cancer prevention. Cancer Prev Res (Phila) 2011;4:973–983. doi: 10.1158/1940-6207.CAPR-10-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morselli E, et al. Anti- and pro-tumor functions of autophagy. Biochim Biophys Acta. 2009;1793:1524–1532. doi: 10.1016/j.bbamcr.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 146.Pfeifer U. Inhibited autophagic degradation of cytoplasm during compensatory growth of liver cells after partial hepatectomy. Virchows Arch B Cell Pathol Incl Mol Pathol. 1979;30:313–333. doi: 10.1007/BF02889111. [DOI] [PubMed] [Google Scholar]
- 147.Song J, et al. Autophagy in hypoxia protects cancer cells against apoptosis induced by nutrient deprivation through a Beclin1-dependent way in hepatocellular carcinoma. J Cell Biochem. 2011;112:3406–3420. doi: 10.1002/jcb.23274. [DOI] [PubMed] [Google Scholar]
- 148.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ding ZB, et al. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008;68:9167–9175. doi: 10.1158/0008-5472.CAN-08-1573. [DOI] [PubMed] [Google Scholar]
- 150.Qu X. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. Journal of Clinical Investigation. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Toshima T, et al. Autophagy enhances hepatocellular carcinoma progression by activation of mitochondrial beta-oxidation. J Gastroenterol. 2013 doi: 10.1007/s00535-013-0835-9. [DOI] [PubMed] [Google Scholar]
- 152.Li J, et al. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis. 2013;34:1343–1351. doi: 10.1093/carcin/bgt063. [DOI] [PubMed] [Google Scholar]
- 153.Wang RC, Levine B. Autophagy in cellular growth control. Febs Letters. 2010;584:1417–1426. doi: 10.1016/j.febslet.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Inami Y, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shi YH, et al. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy. 2011;7:1159–1172. doi: 10.4161/auto.7.10.16818. [DOI] [PubMed] [Google Scholar]
- 156.Gauthier A, Ho M. Role of sorafenib in the treatment of advanced hepatocellular carcinoma: An update. Hepatol Res. 2013;43:147–154. doi: 10.1111/j.1872-034X.2012.01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guo XL, et al. Inhibition of autophagy enhances anticancer effects of bevacizumab in hepatocarcinoma. J Mol Med (Berl) 2013;91:473–483. doi: 10.1007/s00109-012-0966-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu YL, et al. Autophagy potentiates the anti-cancer effects of the histone deacetylase inhibitors in hepatocellular carcinoma. Autophagy. 2010;6:1057–1065. doi: 10.4161/auto.6.8.13365. [DOI] [PubMed] [Google Scholar]
- 159.Decaens T, et al. Phase II study of sirolimus in treatment-naive patients with advanced hepatocellular carcinoma. Dig Liver Dis. 2012;44:610–616. doi: 10.1016/j.dld.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 160.Xu Y, et al. miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2013;29:2019–2024. doi: 10.3892/or.2013.2338. [DOI] [PubMed] [Google Scholar]
- 161.Kon M, et al. Chaperone-mediated autophagy is required for turmor growth. Sci. Trans. Med. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Saha T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy. 2012;8 doi: 10.4161/auto.21654. [DOI] [PMC free article] [PubMed] [Google Scholar]