Abstract
The development of a multicellular embryo from a single zygote is a complex and highly organized process that is far from understood. In higher plants, apical–basal patterning mechanisms are crucial to correctly specify root and shoot stem cell niches that will sustain and drive post-embryonic plant growth and development. The auxin-responsive AtWRKY23 transcription factor is expressed from early embryogenesis onwards and the timing and localization of its expression overlaps with the root stem cell niche. Knocking down WRKY23 transcript levels or expression of a dominant-negative WRKY23 version via a translational fusion with the SRDX repressor domain affected both apical–basal axis formation as well as installation of the root stem cell niche. WRKY23 expression is affected by two well-known root stem cell specification mechanisms, that is, SHORTROOT and MONOPTEROS–BODENLOS signalling and can partially rescue the root-forming inability of mp embryos. On the basis of these results, we postulate that a tightly controlled WRKY23 expression is involved in the regulation of both auxin-dependent and auxin-independent signalling pathways towards stem cell specification.
Keywords: Arabidopsis , embryogenesis, WRKY
Introduction
In the Arabidopsis root meristem, all cell types originate from stem cells that surround a small group of mitotically less active cells, designated as the quiescent centre (QC). The QC cells function as an organizing centre controlling the stem cell state of its neighbouring cells and derive from a single root founder cell, called the hypophysis, which is established during early embryogenesis [1]. Critical in this event is the action of the plant hormone auxin, which induces a transcriptional cascade through MONOPTEROS (MP) and BODENLOS (BDL). The MP gene encodes a member of the AUXIN RESPONSE FACTOR (ARF) family that can activate auxin-inducible gene expression, whereas BDL, a member of the AUX/IAA family, inhibits MP-mediated transcriptional induction in an auxin-dependent manner [1]. The combined action of MP and BDL promotes the transport of auxin from the embryo to the hypophysis and tightly regulates the expression of auxin-responsive transcription factors during embryogenesis [2, 3]. As such, auxin determines the expression domains of the PLETHORA (PLT) genes, members of the APETALA2 domain transcription factor family, redundantly required for the embryonic specification of the root stem cell niche [4, 5]. Accordingly, mp mutants, dominant-negative bdl mutants as well as multiple plt mutants all lack an embryonic root [5–7].
In addition, positioning and maintenance of the root stem cell niche also requires SHORT ROOT (SHR) and SCARECROW (SCR), members of the GRAS family of transcription factors [8, 9]. These transcription factors are not controlled by auxin but intriguingly, the combined action of PLT, SHR and SCR contributes to QC and stem cell patterning [4], indicating that both auxin-dependent, as well as -independent programs converge on the specification and maintenance of the root stem cell niche. Despite the identification of above-mentioned master regulators, there is only limited information about their downstream transcriptional networks. Moreover, molecular mechanisms linking auxin-dependent and -independent programs are currently unknown.
In this study, we identify AtWRKY23 as a novel player in the molecular pathway leading to root stem cell niche specification. We furthermore show that its spatiotemporal expression is fine-tuned by a positive auxin-dependent (BDL–MP) regulation and a repressing auxin-independent (SHR–SCR) control.
Results and Discussion
Reduced WRKY23 activity and embryo development
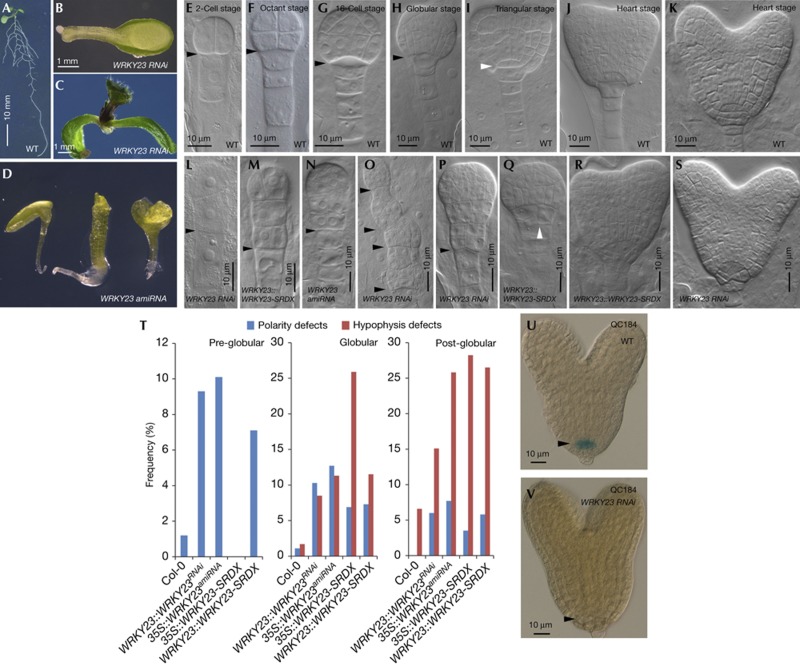
Recently, we showed that the WRKY23 transcriptional activator is involved in maintenance of root meristematic activity of Arabidopsis seedlings by regulating the local production of flavonols [10]. However, in the progeny of plants with a sub-optimal WRKY23 transcript level (WRKY23::WRKY23RNAi) or with a dominant repression of WRKY23’s targets (35 S::WRKY23-SRDX), a significant number of rootless and/or malformed seedlings could be observed shortly after germination [7.7% for WRKY23::WRKY23RNAi (n=504) and 77.6% for WRKY23::WRKY23-SRDX (n=165)] [10] (Fig 1A–C), suggesting a role for WRKY23 in meristem establishment during embryogenesis. Therefore, we evaluated our transgenic lines (at least two independent lines for each construct) for developmental defects during embryogenesis and generated new transgenic lines expressing the previously used WRKY23-SRDX construct and three different artificial microRNA constructs, all from the endogenous WRKY23 promoter (WRKY23::WRKY23-SRDX and WRKY23:: WRKY23amiRNA).
Figure 1.
Analysis of the effect of WRKY23 on embryo patterning and root meristem specification. (A) Ten-day-old Arabidopsis thaliana seedling. (B,C) WRKY23::WRKY23RNAi seedlings. (D) WRKY23:: WRKY23amiRNA seedlings. (E–K) Wild-type embryos. (L–S) WRKY23 knockdown and WRKY23-SRDX embryos. Black arrowheads mark boundary between apical and basal cell lineages. White arrowheads point to orientation of hypophysis division. (T) Quantification of polarity and hypophysis defects in lines with reduced WRKY23 levels or activity. (U,V) QC184-marker GUS expression in wild type (U) and in WRKY23::WRKY23RNAi embryos (V). amiRNA, artificial microRNA; WT, wild type.
During Arabidopsis embryogenesis, at the globular to early heart stage transition, the wild-type hypophysis divides horizontally in an upper lens-shaped cell and a lower tier cell. Both cells will subsequently divide vertically giving rise to the QC and the columella root cap, respectively [11] (Fig 1H–K). However, both WRKY23 knockdown and WRKY23-SRDX embryos fail to organize these essential divisions resulting in an abnormal cellular organization of the root meristem (referred to as hypophysis defects; Fig 1Q–T; supplementary Fig S1 and supplementary Table S1 online). To assess cell identity in the embryonic root stem cell niche of these aberrant embryos, we examined quiescent centre-specific QC184-marker expression. In wild-type embryos, this marker shows GUS activity only in the four QC cells present in the root stem cell niche (Fig 1U) [12]. In WRKY23::WRKY23RNAi embryos, however, no QC184 activity could be detected (100% of WRKY23::WRKY23RNAi embryos with a phenotype, n=7) (Fig 1V).
In about 9–10% of the WRKY23 knockdown embryos (n=183 for WRKY23::WRKY23RNAi and n=119 for WRKY23::WRKY23amiRNA) and about 7% of the WRKY23::WRKY23-SRDX embryos (n=267), defects could be observed in younger (pre-globular) embryos (compared with 1.2% in wild type, n=171) (Fig 1L–P, T, supplementary Fig S1 online and supplementary Table S1 online). These embryos failed to establish the proembryo (referred to as polarity defects; Fig 1E–G,L–P). In most cases, the defects in apical–basal patterning were confined to the lower region of the proembryo and frequently, two or more proembryos developed on top of each other (referred to as polarity defects; Fig 1O, supplementary Fig S1 and supplementary Table S1 online). Consistent with the above-described embryo phenotypes, the WRKY23::WRKY23amiRNA and WRKY23::WRKY23-SRDX lines produced malformed seedlings (Fig 1D, supplementary Fig S1 online), which is also in line with our previous observations of WRKY23::WRKY23RNAi and 35 S::WRKY23-SRDX seedlings [10].
Together, these data support a role for WRKY23 in defining the apical–basal boundary and the proper specification of the root pole during early embryonic patterning.
Auxin response in WRKY23RNAi embryos
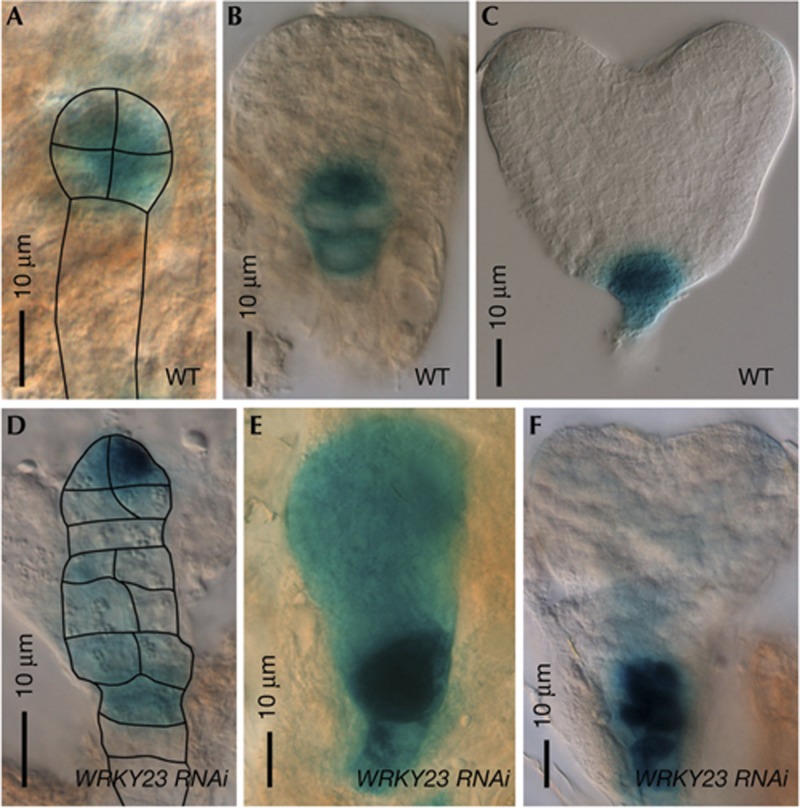
Determination of the apical–basal axis during embryogenesis is preceded by the active organization of auxin response maxima [13]. In wild type, the auxin response maximum, visualized by activity of the synthetic auxin-responsive promoter DR5, is first confined to the apical cell and the derived proembryo and later to the hypophysis (Fig 2A,B). However, in WRKY23::WRKY23RNAi globular embryos, DR5::GUS activity was diffuse and could be detected throughout the entire embryo (100% of WRKY23::WRKY23RNAi embryos with a phenotype, n=5) (Fig 2D,E). Also at later stages during which strong auxin response can be seen at the wild-type root stem cell niche (Fig 2C), WRKY23::WRKY23RNAi embryos showed aberrant DR5 expression patterns (100% of WRKY23::WRKY23RNAi embryos with a phenotype, n=12) (Fig 2F; supplementary Fig S2 online). These results indicate that defects seen in WRKY23 knockdown embryos might be due to defects in auxin distribution. This is further supported by analyses of PIN FORMED1 (PIN1) protein localization in 35S::WRKY23-SRDX embryos (33%, n=18), which deviated from the wild type (n=7) (supplementary Fig S2 online). For example, ectopic PIN1 below the QC could explain the DR5 expression pattern, namely more diffuse and in the suspensor. Therefore, intact WRKY23 expression and WRKY23 activity appear to be important to maintain correct auxin flow and response.
Figure 2.
Auxin response maxima in embryos with reduced WRKY23 levels. (A–F) DR5-visualized auxin response in wild type (WT) (A–C) and WRKY23::WRKY23RNAi embryos (D,F).
We previously showed that WRKY23 regulates the biosynthesis of quercetin, a flavonol known to act as an endogenous auxin transport inhibitor [10, 14]. To analyse whether the observed WRKY23 knockdown and WRKY23-SRDX embryo phenotypes could be attributed to a lack of flavonols, the flavonol-deficient tt4-8 mutant (n=176) and the quercetin-deficient tt7-5 (n=316) and tt7-6 mutants (n=327) were analysed during embryogenesis. However, no difference could be observed between wild type (n=203 for Ws and n=155 for Col-0) and the tt mutant embryos (supplementary Table S2 online). This suggests that dependent on the developmental stage, WRKY23 might regulate different transcriptional networks resulting in disrupted auxin flow and response: a flavonol-dependent control on root meristem maintenance and a flavonol-independent mechanism during Arabidopsis embryo development.
WRKY23 expression overlaps with root stem cell niche
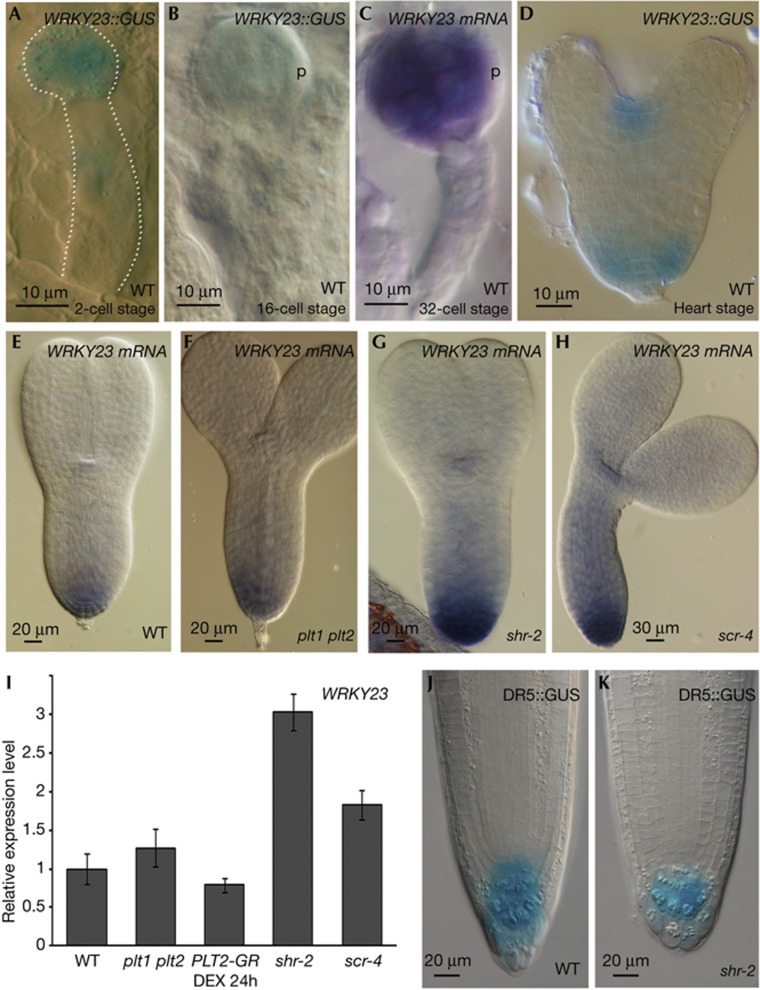
Detailed WRKY23 expression analysis, using WRKY23::GUS and whole-mount mRNA in situ hybridization revealed specific expression patterns during embryo development. WRKY23 expression could be detected in the apical cells of a two-cell stage embryo (Fig 3A) and as embryogenesis proceeded, WRKY23 transcripts were detected in the proembryo (with lower expression in the protoderm layer) until the globular stage (Fig 3B–C; supplementary Fig S3 online). During the transition from globular to heart stage, WRKY23 expression became restricted to the developing shoot apical meristem and root stem cell niche (Fig 3D). These experiments show that WRKY23 has a dynamic spatiotemporal expression pattern, which overlaps with the root stem cell niche. This pattern of WRKY23 expression is maintained during post-embryonic development, both in root and shoot apical meristems [10] (supplementary Fig S3 online).
Figure 3.
WRKY23 expression in embryo and primary root tips. (A–H) WRKY23 expression in wild type (A–E), plt1plt2 (F), shr-2 (G) and scr-4 (H) embryos visualized through WRKY23::GUS (A,B,D) and whole-mount mRNA in situ hybridization (C,E–H). (I) qRT–PCR on WRKY23 in plt1plt2, DEX treated PLT2::PLT2-GR, shr-2 and scr-4 3-day-old seedlings. (J,K) DR5::GUS activity in 3-day-old wild type (J) and shr-2 root tips (K). DEX, dexamethasone; mRNA, messenger RNA; PLT, PLETHORA; qRT–PCR, quantitative real-time polymerase chain reaction; SCR, SCARECROW; SHR, SHORT ROOT; WT, wild type.
Previous studies have highlighted SHR, SCR and PLT transcription factors as essential players mediating root pole establishment during embryogenesis and maintenance of the post-embryonic root meristem [4, 9]. Therefore, we investigated to what extent WRKY23 might be influenced by or interact with these pathways. In plt1plt2 double mutants as well as in the inducible 35S::PLT2-GR lines, WRKY23 expression did not change significantly compared with the corresponding controls pointing towards a PLT-independent mechanism (Fig 3E,F,I). Using mRNA in situ hybridization in embryos and qRT–PCR analysis on primary root tips revealed higher levels of WRKY23 transcript in shr and scr mutants (Fig 3E,G–I), suggesting a repressive effect of the SHR–SCR pathway on WRKY23 expression. However, this effect is likely not direct, as WRKY23 is absent from the transcriptomics-flagged SHR-regulated genes [15, 16] (data not shown). To check if the upregulation of WRKY23 in the shr background could be due to altered auxin distribution or auxin response, expression of auxin-responsive genes and DR5::GUS activity in shr-2 root tips were analysed. Although no obvious difference could be observed in the DR5-visualized auxin response between shr and wild-type roots (Fig 3J,K), the strong upregulation of WRKY23 in shr-2 (∼10-fold) was associated with a mild increase in the expression level of a selection of auxin-responsive genes (∼three to fivefold) (supplementary Fig S4 online). However, although auxin strongly induced WRKY23 in a wild-type root tip, this auxin-mediated upregulation of WRKY23 occurred in cell layers outside the SHR-expression domain (supplementary Fig S5 online). In addition, the SHR–SCR-mediated effect on WRKY23 is not mediated by auxin, as neither SHR nor SCR appear to be auxin inducible (supplementary Fig S6 online). On the basis of these results, we cannot fully exclude that the repressing action of the SHR–SCR pathway on WRKY23 expression is an indirect effect of affected auxin signalling; but, our data support that this is likely not the case. Further genetic analyses will be required to convincingly conclude that WRKY23 functions downstream of SHR–SCR in the same pathway or that SHR–SCR function upstream of WRKY23 in the same pathway.
Genetic interaction between MP–BDL and WRKY23
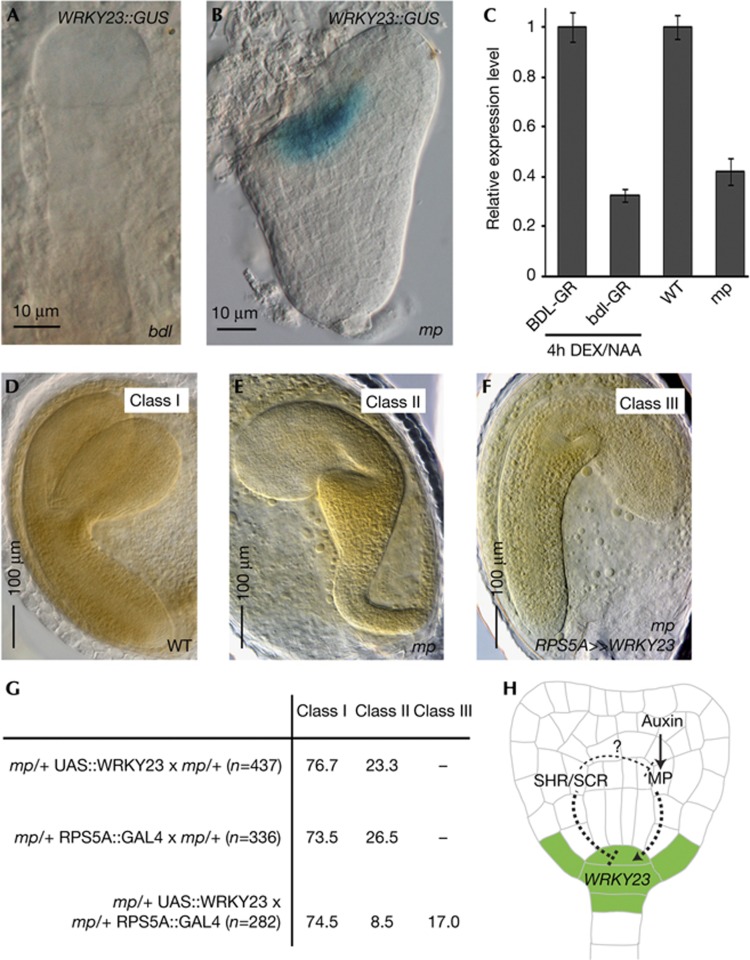
We previously showed that WRKY23 acts downstream of SLR–ARF7–ARF19, key auxin signalling components during lateral root development [10, 17]. However, the establishment of a functional root meristem during embryogenesis largely depends on the transcriptional activator ARF5/MP and its inhibitor IAA12/BDL, as reflected in the rootless phenotypes of their respective mutants [6, 7, 18]. Remarkably, expression patterns of MP and BDL in young embryos overlap with WRKY23 expression, namely in the proembryo excluding the protoderm [2, 7, 19]. Moreover, the morphological defects observed in the WRKY23 knockdown and WRKY23-SRDX lines are very similar to those seen in mp and bdl mutants. Therefore, we checked WRKY23 expression in mpB4149 and bdl mutant backgrounds. We found that the typical WRKY23::GUS activity in the central cells of a 16-cell stage proembryo could not be detected in bdl embryos (Fig 4A, compared with Fig 3B). To independently confirm the BDL control on WRKY23 expression, we made use of seedlings expressing a dexamethasone-inducible, stabilized bdl version [3]. After a 4-h dexamethasone and auxin co-treatment of 35S::bdl-GR seedlings (using 35S::BDL-GR as a control), it was observed that the auxin inducibility of WRKY23 is affected in the absence of a functional BDL pathway (Fig 4C).
Figure 4.
WRKY23 expression and activity upon perturbation of the MP–BDL module. (A,B) WRKY23::GUS in bdl (A) and mpB4149embryo (B). (C) qRT–PCR on auxin-treated, inducible bdl roots as well as in untreated mp seedlings compared with their respective controls. Plants were grown for 5 days after germination and then treated for 4 h with DEX and NAA in liquid medium. (D–G) Activation of UAS::WRKY23 by RPS5A::GAL4 driver line in mpB4149 background The embryos observed in the progeny of mp/+ UAS::WRKY23 × mp/+ RPS5A::GAL4 fall into three classes: class I resembles wild type (D), class II resembles mp (E) and class III appears a partial rescue of mp (F). (G) Quantitative data in percentage of the partial rescue of mp by WRKY23. (H) Graphical summary of control on WRKY23 expression during embryogenesis. BDL, BODENLOS; DEX, dexamethasone; MP, MONOPTEROS; qRT–PCR, quantitative real-time polymerase chain reaction; SCR, SCARECROW; SHR, SHORT ROOT; WT, wild type.
In the progeny of heterozygous mp plants, homozygous mp embryos can only be morphologically distinguished at early heart stage by the defective division of the hypophysis [2]. At this stage, no WRKY23 expression could be detected in the basal pole of mpB4149mutant embryos (Fig 4B, compared with Fig 3D), which might be in line with a repression of WRKY23 in mp seedlings (Fig 4C). However, the observed loss of WRKY23 expression could be an indirect effect of the absence of root identity in mp embryos. Therefore, to genetically analyse whether WRKY23 is part of the MP–BDL signalling pathway defining the root pole during embryogenesis, a UAS::WRKY23 construct was transactivated by the RPS5A::GAL4 driver line in the mpB4149 background. Following a Mendelian segregation, mpB4149 heterozygous plants produced 76.2% (mp/+UAS::WRKY23 × mp/+; n=437) or 73.5% (mp/+RPS5A::GAL4 × mp/+; n=336) wild-type looking embryos (class I) and 23.3% (mp/+UAS::WRKY23 × mp/+) or 26.5% (mp/+ RPS5A::GAL4 × mp/+) mutant embryos (class II, lacking a proper root and hypocotyl and often displaying 1 cotyledon). Interestingly, although the 1:4 segregation was maintained in RPS5A»WRKY23 × mpB4149 embryos, we could morphologically divide the mutant embryos into two classes: characteristic homozygous mpB4149 embryos lacking a root and hypocotyl (class II, 8.5%, n=282) and embryos that are less affected in their basal parts (class III, seeming restoration of the root, 17.0%, n=282) (Fig 4D–G). These embryos still show defects at the root pole but clearly have a more root-like structure compared with the control crosses (Fig 4F). As WRKY23 can partially rescue mpB4149 embryos, it is likely that WRKY23 acts downstream of MP. However, given the absence of unambiguous, genetic experimental evidence that can discriminate between epistasis and bypass suppression, WRKY23 can also function in a pathway parallel to that in which MP functions.
Conclusion
Our findings suggest a central role for WRKY23 at the onset of plant development independent of its previous reported regulation of flavonol biosynthesis. Correct expression of WRKY23 is required for defining the apical–basal boundary and for the proper specification of the root pole during early embryonic patterning. Given the strong effect of altered WRKY23 expression on plant development, we hypothesize that the expression of WRKY23 should be under stringent control. Auxin (likely through the MP–BDL module) activates WRKY23 expression but another control mechanism executed by the non-cell autonomous SHR–SCR pathway negatively affects—indirectly—WRKY23 expression (Fig. 4G). As dosage control of WRKY23 is important for root stem cell niche maintenance [10], this is likely also the case in the embryo and it is possible that SHR modulates WRKY23 expression in a constant level instead of just repressing its expression. Recently another WRKY protein, WRKY2, was shown to regulate the expression of WOX8 and WOX9 that are necessary during early embryogenesis for the specification of the basal cell lineage [20]. Interestingly, WRKY23 and WRKY2 have contrasting expression patterns during embryogenesis. As WRKY proteins have been shown to interact with each other and to form transcriptional networks during plant defence responses (also referred to as the ‘WRKY-web’) [21], it will be interesting to investigate the existence of a similar WRKY-web during plant development.
Methods
Plant material and growth conditions. Arabidopsis thaliana (L.) Heynh. seeds were sterilized with chloral gas, plated on half-strength Murashige and Skoog medium (1% sucrose; 0.01% myo-inositol; 0.05% 2-(N-morpholino)ethanesulfonic acid, 0.8% agar; pH 5.7), stored for 2 days at 4 °C, and grown vertically at 21 °C under continuous light. Previously described plant materials were plt1plt2 [4], RPS5A::GAL4 activator line [4], 35S::PLT2-GR (plt1plt2) [5], mpB4149 [6], bdl [7], shr-2 [22], scr-4 [22], DR5::GUS [23], QC184 [9], WRKY23::GUS [17], WRKY23::WRKY23RNAi [10], 35S::WRKY23-SRDX [10]; RPS5A::BDL-GR [3], RPS5A::bdl-GR [3].
Cloning. WRKY23-related constructs are described in the supplementary Information online. All obtained constructs were mobilized to the Agrobacterium tumefaciens strain C58C1RifR and introduced into A. thaliana ecotype Col-0 using the floral dip method [24].
Histological analyses and microscopy. GUS staining of embryos is described in the supplementary Information online. Embryos were transferred to slides and mounted with 10% glycerol and analysed with a DIC fluorescence microscope (Olympus). Whole-mount in situ hybridization of embryos was done as previously described [25] using a full-length WRKY23 RNA probe. Embryos were analysed by clearing the ovules in Hoyer medium (for 50 ml: dissolve 30 g Arabic gum in 30 ml distilled water, add 200 g chloralhydrate and finish with 20 ml glycerol [26]).
RNA extraction, cDNA synthesis and qRT–PCR analysis. Total RNA was isolated with Trizol (Invitrogen) according to the manufacturer's instructions. Poly(dT) cDNA was prepared from 2 μg total RNA with Superscript III reverse transcriptase (Invitrogen) and quantified on an LightCycler 480 apparatus (Roche Diagnostics) with the SYBR Green I Master kit (Roche Diagnostics) according to the manufacturer’s instructions using gene-specific primers (supplementary Table S4 online). Unless stated otherwise, we performed three biological repeats on a pool of seedlings and all individual reactions were done in triplicate. Graphs show one representative analysis. Data were analysed with qBase [27] and normalized to EEF1α4, CDKA, and/or ACT7 (supplementary Table S4 online).
PIN1 immunolocalization. PIN1 immunolocalization was performed as previously described [28].
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Carina Braeckman from the PSB Plant transformation unit for the Arabidopsis transformations, Stijn Dhondt for providing the shr-2 × DR5::GUS seeds, and Ross Sozzani for analysing micro-array data. W.G. is a post-doctoral fellow of the Research Foundation Flanders, B.D.R. is supported by a long-term FEBS fellowship and a Marie Curie long-term FP7 Intra-European fellowship (IEF–2009–252503) and I.D.S. by a Biological Sciences Research Council David Phillips Fellowship (BB_BB/H022457/1) and a Marie Curie European Reintegration Grant (PERG06-GA-2009-256354).
Author contributions: W.G., I.D.S., T.B. and J.F. designed research; W.G., B.D.R., B.V.D.C. H.S.R. and I.D.S. performed research; W.G., I.D.S., B.D.R., V.W., G.G., D.W., T.B. and J.F. analysed the data; and W.G., I.D.S., T.B. and J.F. wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Wendrich JR, Weijers D (2013) The Arabidopsis embryo as a miniature morphogenesis model. New Phytol 199: 14–25 [DOI] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jürgens G (2006) Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10: 265–270 [DOI] [PubMed] [Google Scholar]
- Schlereth A, Moller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D (2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Aida M et al. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Berleth T, Jürgens G (1993) The role of the MONOPTEROS gene in organising the basal body region of the Arabidopsis embryo. Development 118: 575–587 [Google Scholar]
- Hamann T, Benkova E, Baurle I, Kientz M, Jürgens G (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16: 1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W et al. (2012) Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci USA 109: 1554–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller B, Weijers D (2009) Auxin control of embryo patterning. Cold Spring Harb Perspect Biol 1: a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Peer WA, Murphy AS (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Levesque MP et al. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R et al. (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Karimi M, Wieczorek K, Van de Cappelle E, Wischnitzki E, Grundler F, Inzé D, Beeckman T, Gheysen G (2008) A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiology 148: 358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. Embo J 17: 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Zhang Z, Laux T (2011) Transcriptional activation of arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell 20: 264–270 [DOI] [PubMed] [Google Scholar]
- Eulgem T (2006) Dissecting the WRKY web of plant defense regulators. Plos Pathogens 2: 1028–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Di Laurenzio L, Willemsen V, Hauser MT, Janmaat K, Weisbeek P, Benfey PN (1995) Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62 [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Hejatko J, Blilou I, Brewer PB, Friml J, Scheres B, Benkova E (2006) In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat Protoc 1: 1939–1946 [DOI] [PubMed] [Google Scholar]
- Bougourd S, Marrison J, Haseloff J (2000) Technical advance: an aniline blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. Plant J 24: 543–550 [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Paciorek T, Benkova E, Friml J (2006) Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc 1: 98–103 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.