Abstract
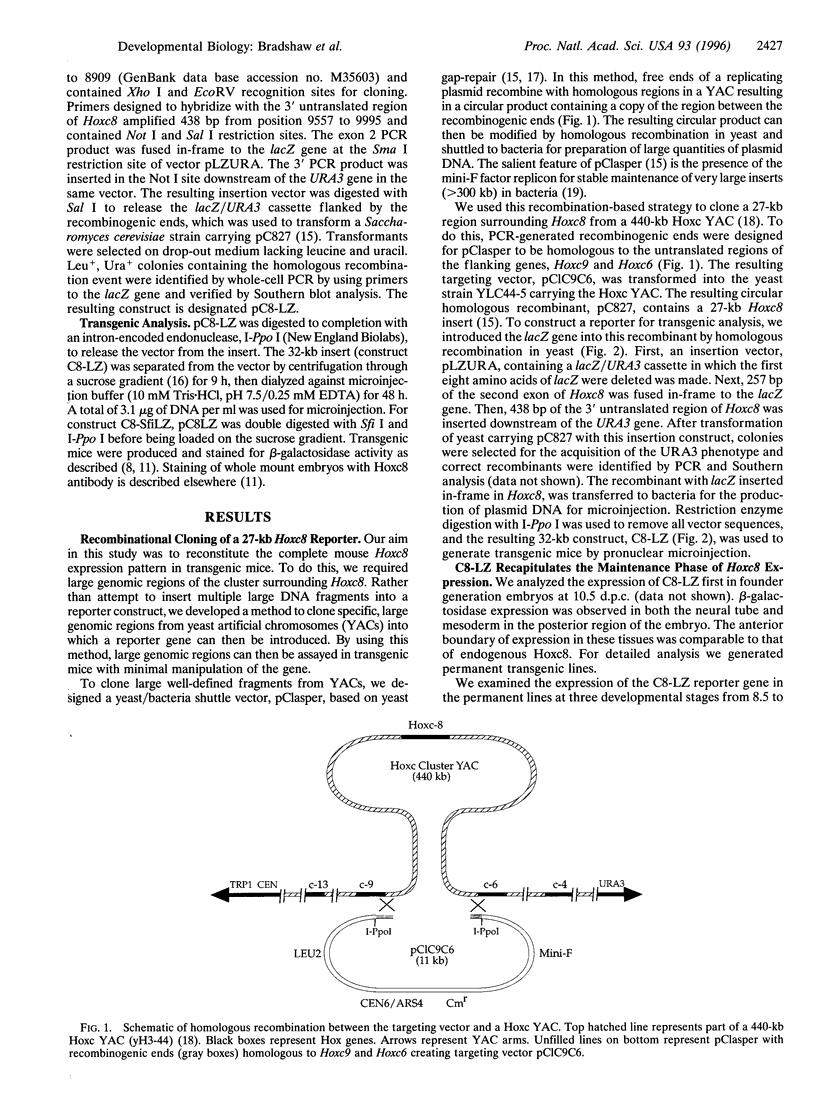
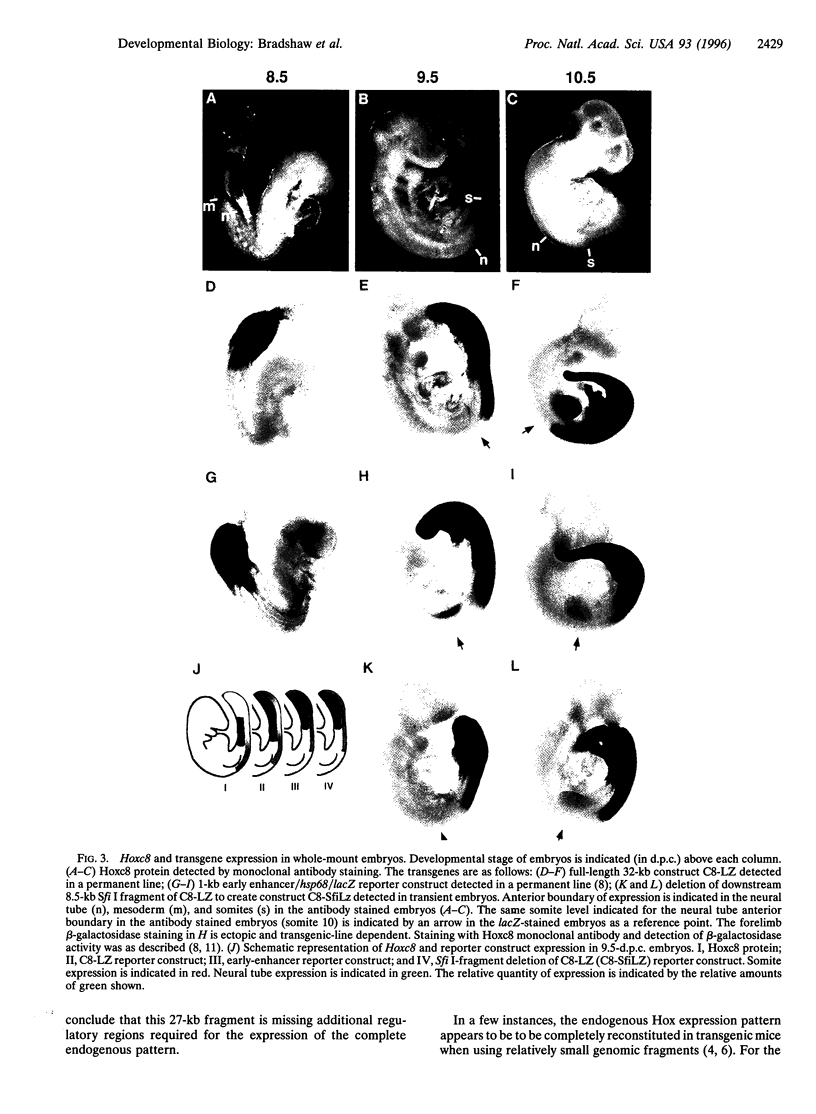
Hox genes are located in highly conserved clusters. The significance of this organization is unclear, but one possibility is that regulatory regions for individual genes are dispersed throughout the cluster and shared with other Hox genes. This hypothesis is supported by studies on several Hox genes in which even large genomic regions immediately surrounding the gene fail to direct the complete expression pattern in transgenic mice. In particular, previous studies have identified proximal regulatory regions that are primarily responsible for early phases of mouse Hoxc8 expression. To locate additional regulatory regions governing expression during the later periods of development, a yeast homologous recombination-based strategy utilizing the pClasper vector was employed. Using homologous recombination into pClasper, we cloned a 27-kb region around the Hoxc8 gene from a yeast artificial chromosome. A reporter gene was introduced into the coding region of the isolated gene by homologous recombination in yeast. This large fragment recapitulates critical aspects of Hoxc8 expression in transgenic mice. We show that the regulatory elements that maintain the anterior boundaries of expression in the neural tube and paraxial mesoderm are located between 11 and 19 kb downstream of the gene.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Awgulewitsch A., Utset M. F., Hart C. P., McGinnis W., Ruddle F. H. Spatial restriction in expression of a mouse homoeo box locus within the central nervous system. 1986 Mar 27-Apr 2Nature. 320(6060):328–335. doi: 10.1038/320328a0. [DOI] [PubMed] [Google Scholar]
- Behringer R. R., Crotty D. A., Tennyson V. M., Brinster R. L., Palmiter R. D., Wolgemuth D. J. Sequences 5' of the homeobox of the Hox-1.4 gene direct tissue-specific expression of lacZ during mouse development. Development. 1993 Mar;117(3):823–833. doi: 10.1242/dev.117.3.823. [DOI] [PubMed] [Google Scholar]
- Bieberich C. J., Utset M. F., Awgulewitsch A., Ruddle F. H. Evidence for positive and negative regulation of the Hox-3.1 gene. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8462–8466. doi: 10.1073/pnas.87.21.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw M. S., Bollekens J. A., Ruddle F. H. A new vector for recombination-based cloning of large DNA fragments from yeast artificial chromosomes. Nucleic Acids Res. 1995 Dec 11;23(23):4850–4856. doi: 10.1093/nar/23.23.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw M. S., Ruddle F. H. Identification of the murine Hox-c12 and Hox-c13 homeoboxes on yeast artificial chromosomes. Genomics. 1994 Jul 1;22(1):234–236. doi: 10.1006/geno.1994.1371. [DOI] [PubMed] [Google Scholar]
- Charité J., de Graaff W., Vogels R., Meijlink F., Deschamps J. Regulation of the Hoxb-8 gene: synergism between multimerized cis-acting elements increases responsiveness to positional information. Dev Biol. 1995 Oct;171(2):294–305. doi: 10.1006/dbio.1995.1282. [DOI] [PubMed] [Google Scholar]
- Duboule D., Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994 Oct;10(10):358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Eid R., Koseki H., Schughart K. Analysis of LacZ reporter genes in transgenic embryos suggests the presence of several cis-acting regulatory elements in the murine Hoxb-6 gene. Dev Dyn. 1993 Mar;196(3):205–216. doi: 10.1002/aja.1001960307. [DOI] [PubMed] [Google Scholar]
- Erselius J. R., Goulding M. D., Gruss P. Structure and expression pattern of the murine Hox-3.2 gene. Development. 1990 Oct;110(2):629–642. doi: 10.1242/dev.110.2.629. [DOI] [PubMed] [Google Scholar]
- Frasch M., Chen X., Lufkin T. Evolutionary-conserved enhancers direct region-specific expression of the murine Hoxa-1 and Hoxa-2 loci in both mice and Drosophila. Development. 1995 Apr;121(4):957–974. doi: 10.1242/dev.121.4.957. [DOI] [PubMed] [Google Scholar]
- Gérard M., Duboule D., Zákány J. Structure and activity of regulatory elements involved in the activation of the Hoxd-11 gene during late gastrulation. EMBO J. 1993 Sep;12(9):3539–3550. doi: 10.1002/j.1460-2075.1993.tb06028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegalian B. G., Miller R. W., Wright C. V., Blum M., De Robertis E. M. A Hox 3.3-lacZ transgene expressed in developing limbs. Mech Dev. 1992 Dec;39(3):171–180. doi: 10.1016/0925-4773(92)90044-k. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994 Jul 29;78(2):191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Le Mouellic H., Condamine H., Brûlet P. Pattern of transcription of the homeo gene Hox-3.1 in the mouse embryo. Genes Dev. 1988 Jan;2(1):125–135. doi: 10.1101/gad.2.1.125. [DOI] [PubMed] [Google Scholar]
- Le Mouellic H., Lallemand Y., Brûlet P. Homeosis in the mouse induced by a null mutation in the Hox-3.1 gene. Cell. 1992 Apr 17;69(2):251–264. doi: 10.1016/0092-8674(92)90406-3. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Püschel A. W., Balling R., Gruss P. Position-specific activity of the Hox1.1 promoter in transgenic mice. Development. 1990 Mar;108(3):435–442. doi: 10.1242/dev.108.3.435. [DOI] [PubMed] [Google Scholar]
- Püschel A. W., Balling R., Gruss P. Separate elements cause lineage restriction and specify boundaries of Hox-1.1 expression. Development. 1991 May;112(1):279–287. doi: 10.1242/dev.112.1.279. [DOI] [PubMed] [Google Scholar]
- Sham M. H., Hunt P., Nonchev S., Papalopulu N., Graham A., Boncinelli E., Krumlauf R. Analysis of the murine Hox-2.7 gene: conserved alternative transcripts with differential distributions in the nervous system and the potential for shared regulatory regions. EMBO J. 1992 May;11(5):1825–1836. doi: 10.1002/j.1460-2075.1992.tb05234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashikant C. S., Bieberich C. J., Belting H. G., Wang J. C., Borbély M. A., Ruddle F. H. Regulation of Hoxc-8 during mouse embryonic development: identification and characterization of critical elements involved in early neural tube expression. Development. 1995 Dec;121(12):4339–4347. doi: 10.1242/dev.121.12.4339. [DOI] [PubMed] [Google Scholar]
- Shizuya H., Birren B., Kim U. J., Mancino V., Slepak T., Tachiiri Y., Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels R., Charité J., de Graaff W., Deschamps J. Proximal cis-acting elements cooperate to set Hoxb-7 (Hox-2.3) expression boundaries in transgenic mice. Development. 1993 May;118(1):71–82. doi: 10.1242/dev.118.1.71. [DOI] [PubMed] [Google Scholar]
- Whiting J., Marshall H., Cook M., Krumlauf R., Rigby P. W., Stott D., Allemann R. K. Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev. 1991 Nov;5(11):2048–2059. doi: 10.1101/gad.5.11.2048. [DOI] [PubMed] [Google Scholar]