Abstract
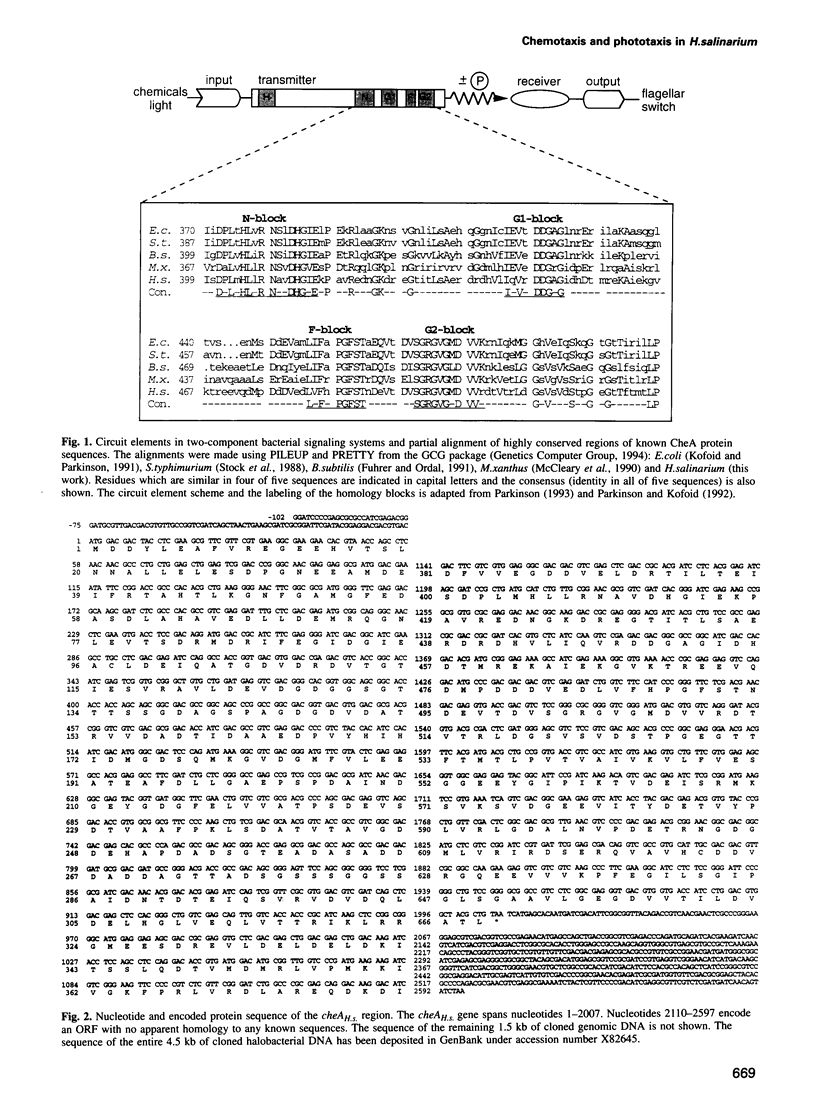
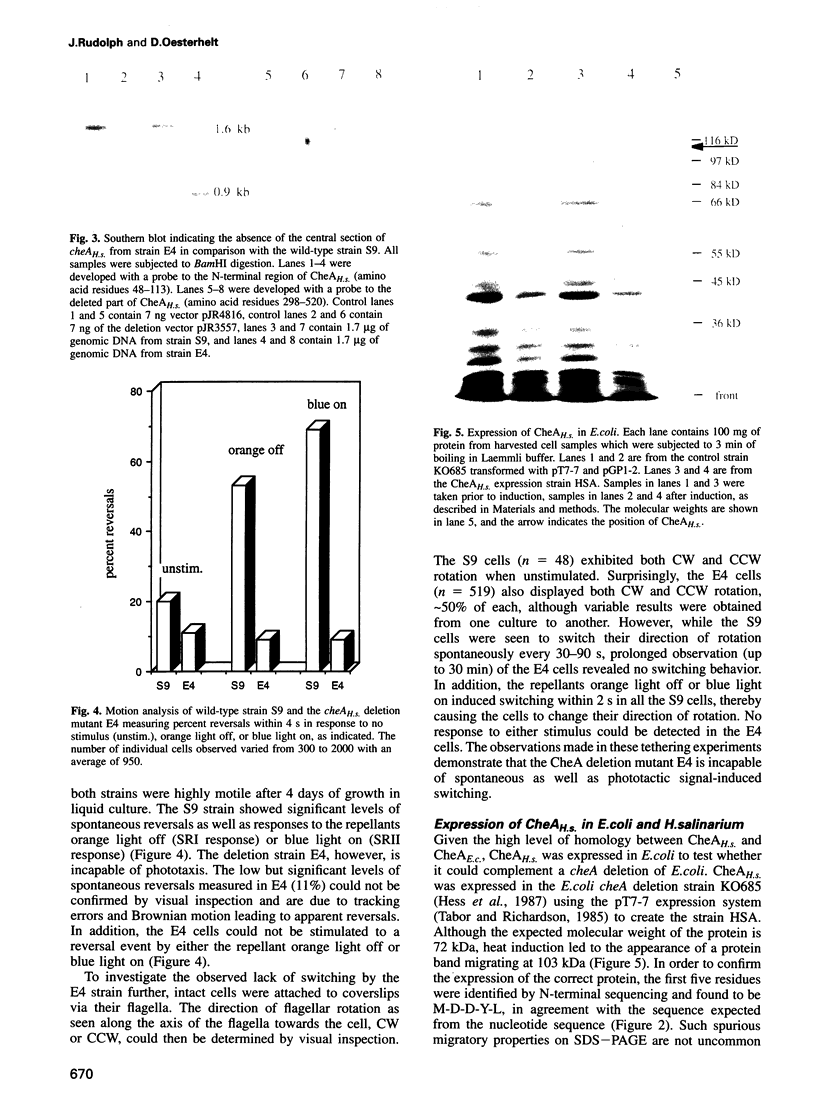
Histidine kinases are part of the two-component signal transduction system responsible for eubacterial responses to diverse environmental signals. They have recently been detected in eukaryotes but their existence in the kingdom Archaea remains uncertain. Here we report the sequence and function of a histidine kinase (CheAH.s.) from Halobacterium salinarium, the first such transmitter in Archaea. The protein CheAH.s. (668 residues) has significant sequence identity with the CheA proteins known from eubacterial signal transduction (e.g. 34% identity with CheA from Bacillus subtilis). Antibodies were raised against CheAH.s. as expressed in Escherichia coli and were used in Western blotting to demonstrate the expression of cheAH.s. in H. salinarium. As has been observed for other halophilic proteins, CheAH.s. has a deviant electrophoretic migration, with an apparent molecular weight of 103 kDa on SDS-PAGE compared with a calculated molecular weight of 72 kDa. Deletion of a part of the cheAH.s. gene leads to loss of both chemotactic and phototactic responses in H. salinarium as measured by swarm plate assays, motion analysis and tethering experiments. This indicates that CheAH.s. plays a crucial role in chemical and light signal integration, presumably interacting with at least two phototransducers and a number of chemoreceptors.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alam M., Hazelbauer G. L. Structural features of methyl-accepting taxis proteins conserved between archaebacteria and eubacteria revealed by antigenic cross-reaction. J Bacteriol. 1991 Sep;173(18):5837–5842. doi: 10.1128/jb.173.18.5837-5842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M., Lebert M., Oesterhelt D., Hazelbauer G. L. Methyl-accepting taxis proteins in Halobacterium halobium. EMBO J. 1989 Feb;8(2):631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M., Oesterhelt D. Morphology, function and isolation of halobacterial flagella. J Mol Biol. 1984 Jul 15;176(4):459–475. doi: 10.1016/0022-2836(84)90172-4. [DOI] [PubMed] [Google Scholar]
- Barak R., Eisenbach M. Fumarate or a fumarate metabolite restores switching ability to rotating flagella of bacterial envelopes. J Bacteriol. 1992 Jan;174(2):643–645. doi: 10.1128/jb.174.2.643-645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikov S. I., Grishanin R. N., Kaulen A. D., Marwan W., Oesterhelt D., Skulachev V. P. Bacteriorhodopsin is involved in halobacterial photoreception. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9446–9450. doi: 10.1073/pnas.90.20.9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanck A., Oesterhelt D., Ferrando E., Schegk E. S., Lottspeich F. Primary structure of sensory rhodopsin I, a prokaryotic photoreceptor. EMBO J. 1989 Dec 20;8(13):3963–3971. doi: 10.1002/j.1460-2075.1989.tb08579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Kwok S. F., Bleecker A. B., Meyerowitz E. M. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993 Oct 22;262(5133):539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Cline S. W., Lam W. L., Charlebois R. L., Schalkwyk L. C., Doolittle W. F. Transformation methods for halophilic archaebacteria. Can J Microbiol. 1989 Jan;35(1):148–152. doi: 10.1139/m89-022. [DOI] [PubMed] [Google Scholar]
- Ferrando-May E., Krah M., Marwan W., Oesterhelt D. The methyl-accepting transducer protein HtrI is functionally associated with the photoreceptor sensory rhodopsin I in the archaeon Halobacterium salinarium. EMBO J. 1993 Aug;12(8):2999–3005. doi: 10.1002/j.1460-2075.1993.tb05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer D. K., Ordal G. W. Bacillus subtilis CheN, a homolog of CheA, the central regulator of chemotaxis in Escherichia coli. J Bacteriol. 1991 Dec;173(23):7443–7448. doi: 10.1128/jb.173.23.7443-7448.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegner J. A., Graham D. R., Roth A. F., Dahlquist F. W. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992 Sep 18;70(6):975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- Hargrave P. A., McDowell J. H. Rhodopsin and phototransduction: a model system for G protein-linked receptors. FASEB J. 1992 Mar;6(6):2323–2331. doi: 10.1096/fasebj.6.6.1544542. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Matsumura P., Simon M. I. Protein phosphorylation is involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7609–7613. doi: 10.1073/pnas.84.21.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand E., Dencher N. Two photosystems controlling behavioural responses of Halobacterium halobium. Nature. 1975 Sep 4;257(5521):46–48. doi: 10.1038/257046a0. [DOI] [PubMed] [Google Scholar]
- Hildebrand E., Schimz A. Integration of photosensory signals in Halobacterium halobium. J Bacteriol. 1986 Jul;167(1):305–311. doi: 10.1128/jb.167.1.305-311.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagramanova V. K., Mankin A. S., Baratova L. A., Bogdanov A. A. The 3'-terminal nucleotide sequence of the Halobacterium halobium 16 S rRNA. FEBS Lett. 1982 Jul 19;144(1):177–180. doi: 10.1016/0014-5793(82)80595-4. [DOI] [PubMed] [Google Scholar]
- Kofoid E. C., Parkinson J. S. Tandem translation starts in the cheA locus of Escherichia coli. J Bacteriol. 1991 Mar;173(6):2116–2119. doi: 10.1128/jb.173.6.2116-2119.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krah M., Marwan W., Verméglio A., Oesterhelt D. Phototaxis of Halobacterium salinarium requires a signalling complex of sensory rhodopsin I and its methyl-accepting transducer HtrI. EMBO J. 1994 May 1;13(9):2150–2155. doi: 10.1002/j.1460-2075.1994.tb06491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S. C., Koshland D. E., Jr Multiple kinetic states for the flagellar motor switch. J Bacteriol. 1989 Nov;171(11):6279–6287. doi: 10.1128/jb.171.11.6279-6287.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam W. L., Doolittle W. F. Mevinolin-resistant mutations identify a promoter and the gene for a eukaryote-like 3-hydroxy-3-methylglutaryl-coenzyme A reductase in the archaebacterium Haloferax volcanii. J Biol Chem. 1992 Mar 25;267(9):5829–5834. [PubMed] [Google Scholar]
- Maeda T., Wurgler-Murphy S. M., Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994 May 19;369(6477):242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Marwan W., Oesterhelt D. Signal formation in the halobacterial photophobic response mediated by a fourth retinal protein (P480). J Mol Biol. 1987 May 20;195(2):333–342. doi: 10.1016/0022-2836(87)90654-1. [DOI] [PubMed] [Google Scholar]
- Marwan W., Schäfer W., Oesterhelt D. Signal transduction in Halobacterium depends on fumarate. EMBO J. 1990 Feb;9(2):355–362. doi: 10.1002/j.1460-2075.1990.tb08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrone M., Marwan W., Grünberg H., Musseleck S., Starostzik C., Oesterhelt D. Sensory rhodopsin-controlled release of the switch factor fumarate in Halobacterium salinarium. Mol Microbiol. 1993 Dec;10(5):1077–1085. doi: 10.1111/j.1365-2958.1993.tb00978.x. [DOI] [PubMed] [Google Scholar]
- Nordmann B., Lebert M. R., Alam M., Nitz S., Kollmannsberger H., Oesterhelt D., Hazelbauer G. L. Identification of volatile forms of methyl groups released by Halobacterium salinarium. J Biol Chem. 1994 Jun 10;269(23):16449–16454. [PubMed] [Google Scholar]
- Olson K. D., Spudich J. L. Removal of the transducer protein from sensory rhodopsin I exposes sites of proton release and uptake during the receptor photocycle. Biophys J. 1993 Dec;65(6):2578–2585. doi: 10.1016/S0006-3495(93)81295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa K., Mutoh N., Simon M. I. Cloning of the C-terminal cytoplasmic fragment of the tar protein and effects of the fragment on chemotaxis of Escherichia coli. J Bacteriol. 1988 Jun;170(6):2521–2526. doi: 10.1128/jb.170.6.2521-2526.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota I. M., Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993 Oct 22;262(5133):566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S., Kofoid E. C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S. Signal transduction schemes of bacteria. Cell. 1993 Jun 4;73(5):857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J Bacteriol. 1976 May;126(2):758–770. doi: 10.1128/jb.126.2.758-770.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schegk E. S., Oesterhelt D. Isolation of a prokaryotic photoreceptor: sensory rhodopsin from halobacteria. EMBO J. 1988 Sep;7(9):2925–2933. doi: 10.1002/j.1460-2075.1988.tb03151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimz A., Hildebrand E. Chemosensory responses of Halobacterium halobium. J Bacteriol. 1979 Dec;140(3):749–753. doi: 10.1128/jb.140.3.749-753.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S. C., Swanson R. V., Alex L. A., Bourret R. B., Simon M. I. Assembly and function of a quaternary signal transduction complex monitored by surface plasmon resonance. Nature. 1993 Sep 23;365(6444):343–347. doi: 10.1038/365343a0. [DOI] [PubMed] [Google Scholar]
- Soppa J. Two hypotheses--one answer. Sequence comparison does not support an evolutionary link between halobacterial retinal proteins including bacteriorhodopsin and eukaryotic G-protein-coupled receptors. FEBS Lett. 1994 Mar 28;342(1):7–11. doi: 10.1016/0014-5793(94)80573-3. [DOI] [PubMed] [Google Scholar]
- Spudich E. N., Hasselbacher C. A., Spudich J. L. Methyl-accepting protein associated with bacterial sensory rhodopsin I. J Bacteriol. 1988 Sep;170(9):4280–4285. doi: 10.1128/jb.170.9.4280-4285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Takahashi T., Spudich J. L. Sensory rhodopsins I and II modulate a methylation/demethylation system in Halobacterium halobium phototaxis. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7746–7750. doi: 10.1073/pnas.86.20.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984 Dec 6;312(5994):509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A., Chen T., Welsh D., Stock J. CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1403–1407. doi: 10.1073/pnas.85.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R. V., Simon M. I. Signal transduction. Bringing the eukaryotes up to speed. Curr Biol. 1994 Mar 1;4(3):234–237. doi: 10.1016/s0960-9822(00)00052-x. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M., Oosawa K., Aizawa S. I., Eisenbach M. Effects of phosphorylation, Mg2+, and conformation of the chemotaxis protein CheY on its binding to the flagellar switch protein FliM. Biochemistry. 1994 Aug 30;33(34):10470–10476. doi: 10.1021/bi00200a031. [DOI] [PubMed] [Google Scholar]
- Wolfe A. J., Stewart R. C. The short form of the CheA protein restores kinase activity and chemotactic ability to kinase-deficient mutants. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1518–1522. doi: 10.1073/pnas.90.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao V. J., Spudich J. L. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]