Abstract
ORMDL3 (orosomucoid like 3) has been strongly linked with asthma in genetic association studies. As allergen challenge induces lung ORMDL3 expression in WT mice, we have generated human ORMDL3 Zona Pellucida 3 Cre (hORMDL3zp3-Cre) mice that overexpress human ORMDL3 universally to investigate the role of ORMDL3 in regulating airway inflammation and remodeling. These hORMDL3zp3-Cre mice have significantly increased levels of airway remodeling including increased airway smooth muscle, subepithelial fibrosis, and mucus. hORMDL3zp3-Cre mice had spontaneous increased AHR to methacholine compared to WT mice. This increased airway remodeling was associated with selective activation of the Unfolded Protein Response pathway transcription factor ATF6 (but not Ire1 or PERK). The ATF6 target gene SERCA2b, implicated in airway remodeling in asthma, was strongly induced in the lungs of hORMDL3zp3-Cre mice. In addition, increased levels of expression of genes associated with airway remodeling (TGF-β1, ADAM8) were detected in airway epithelium of these mice. Increased levels of airway remodeling preceded increased levels of airway inflammation in hORMDL3zp3-Cre mice. hORMDL3zp3-Cre mice had increased levels of IgE, with no change in levels of IgG, IgM, and IgA. These studies provide evidence that ORMDL3 plays an important role in vivo in airway remodeling potentially through ATF6 target genes such as SERCA2b, and/or through ATF6 independent genes (TGF-β1, ADAM8).
INTRODUCTION
ORMDL3 (orosomucoid like 3) is a gene localized to chromosome 17q21 which was initially linked to asthma in a genome wide association study (GWAS)(1) with subsequent confirmation in multiple additional GWAS (2–4) and non-GWAS genetic association studies in populations of diverse ethnic backgrounds (5–10). ORMDL3 has been linked to severe asthma (4,9), childhood onset of asthma (1,7,8), exposure of children to environmental tobacco smoke and risk of asthma (2,10), as well as to rhinoviral wheezing illness and genetic risk of childhood onset of asthma (11), underscoring the importance of understanding its function. ORMDL3 is a member of the three member ORMDL gene family (ORMDL1,-2,-3) which encode transmembrane proteins located at the endoplasmic reticulum (ER)(12). ORMDL1 (chromosome 2)(12), and ORMDL2 (chromosome 12)(12) are on different chromosomes from ORMDL3 (chromosome 17q21)(12) and have not been linked to asthma. Both humans and mice express the same three ORMDL family members with ORMDL3 exhibiting 96% identity between these two species (12). ORMDL3 is a 153 amino acid protein with two predicted transmembrane domains (12). We recently demonstrated that in wild type (WT) mice ORMDL3 is an allergen and Th2 cytokine (IL-4, or IL-13) inducible gene localized to the endoplasmic reticulum (ER) and highly expressed in airway epithelial cells (13). Allergen challenge induced a 127 fold increase in ORMDL3 mRNA in bronchial epithelium in WT mice, with lesser 15 fold increases in ORMDL2, and no changes in ORMDL1 (13). We also demonstrated that transfection of ORMDL-3 in human bronchial epithelial cells in vitro induced expression of CC chemokines (CCL-20 also known as MIP-3α)(13), CXC chemokines (IL-8; CXCL-10 also known as IP-10; CXCL-11 also known as ITAC)(13), metalloproteases (MMP-9; ADAM-8)(13), and selectively activated ATF6 (13), one of three ER Unfolded Protein Response (UPR) pathway transcription factors (14) with subsequent regulation of SERCA2b (sarco/endoplasmic reticulum Ca2+ ATPase) which has been implicated in airway remodeling in asthma (15). Thus, these studies with bronchial epithelium in WT mice and in normal human bronchial epithelial cells suggest an important role for a pathway in which initial induction of ORMDL3 with subsequent activation of both ATF6 dependent pathways (i.e. SERCA2b) and/or ATF6 independent pathways (MMP9, ADAM8, CCL20, CXCL10, CXCL11) may contribute to the pathogenesis of asthma.
Although our previous studies demonstrated that Ormdl3 is an allergen and Th2 cytokine inducible gene that is dependent upon Stat6 for expression (13), these prior studies in WT mice did not determine which downstream pathways were regulated by ORMDL3 in vivo. To address this question we have generated ORMDL3 transgenic (TG) mice, and in this study we demonstrate that TG mice overexpressing human ORMDL3 (hORMDL3) spontaneously develop significantly increased levels of airway remodeling (smooth muscle, fibrosis, mucus) that precede the development of airway inflammation. In addition, allergen challenge of ORMDL3 TG mice resulted in enhanced OVA specific IgE responses compared to OVA challenged WT mice and was associated with increased Major Basic Protein (MBP) positive peribronchial eosinophils and lung levels of IL-4. These studies in ORMDL3 TG mice also provide evidence that the ER localized ORMDL3 plays an important role in selective activation of one of the three UPR pathways in vivo (i.e. ATF6), and that expression of ORMDL3 in vivo regulates airway remodeling (smooth muscle, fibrosis, mucus) potentially through ATF6 target genes such as SERCA2b, and/or through ATF6 independent-genes (TGF-β1, ADAM8) which we detected at increased levels in the lungs of ORMDL3 TG mice. ORMDL3 may therefore activate several pathways important to the pathogenesis of airway remodeling and asthma in vivo.
MATERIALS AND METHODS
Zp3-Cre mice
Zp3-Cre mice (embryonic Cre expression) on a C57Bl/6 background were acquired from Jackson labs.
hORMDL3zp3-Cre mouse generation
All the mouse experimental protocols were approved by the UCSD Institutional Animal Care and Use Committee.
Targeting plasmid construction
The hORMDL3 transgenic construct pCAGEN Lox mRFP-H2B STOP Lox hORMDL3 was generated by cloning the 462bp hORMDL3 open reading frame (orf) from pCMV6-AC-ORMDL3 (Origene) with Agel/Notl into a construct previously developed and generously provided by AJ Holland and DW Celeveland (Ludwig Institute for Cancer Research at the University of California, San Diego).
RFP-StopFLhORMDL3-TG mouse generation
Spel/Pvul linearized pCAGEN Lox mRFP-H2B STOP Lox hORMDL3 (Fig 1A, B) was microinjected into the pronuclei of fertilized mouse embryos and implanted into a pseudopregnant mouse (all on a C57Bl/6 background) via an established protocol by the UCSD mouse transgenic core. Progeny were then screened by PCR for the presence of the transgene. Subsequent mouse genotyping was performed using PCR with the following primers: F1-3530: 5′-GCA ACG TGC TGG TTA TTG TG; F2-4009: 5′-CCC CCT GAA CCT GAA ACA TA-3′; R-4644: 5′-TAC AGC ACG ATG GGT GTG AT-3′ (Fig 1C). These RFP-StopFLhORMDL3-TG mice (C57Bl/6 background) were crossed with Zp3-Cre mice (C57Bl/6 background) to generate hORMDL3zp3-Cre mice (C57Bl/6 background).
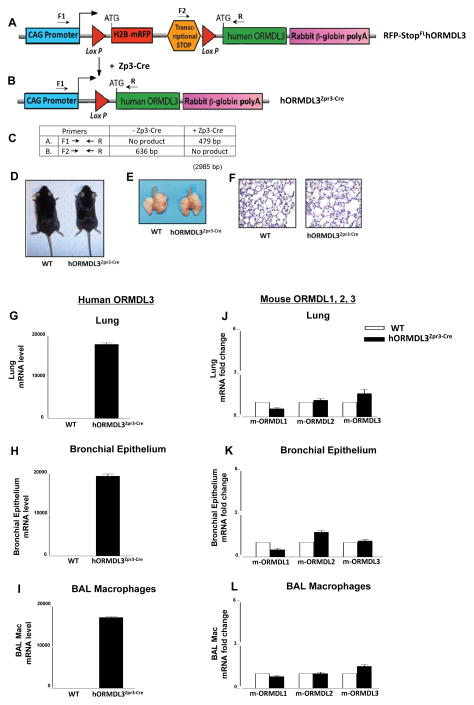
Figure 1. Generation of ORMDL3 TG mice.
(A). The human ORMDL3 transgenic construct contains a CAG promoter for universal over-expression (light blue), H2B-mRFP (red fluorescent protein)(red), followed by a transcriptional stop site (orange), the human ORMDL3 open reading frame (green), and a rabbit B-globin poly-adenylation sequence (pink). The H2B-mRFP (red fluorescent protein) and transcriptional stop site are flanked by LoxP sites (triangles). Cells containing this pCAGEN Lox mRFP-H2B STOP Lox hORMDL3 construct will express RFP (red fluorescent protein) and not express hORMDL3 as the transcriptional stop codon (orange) prevents transcription of hORMDL-3. (B). Expression of the hORMDL3 gene is Cre recombinase dependent since the transcriptional stop codon preventing hORMDL-3 expression (and RFP) are excised by Cre recombinase via the Lox P sites resulting in over-expression of hORMDL3 only in those cells expressing Cre recombinase. (C) Primer sets (F1, R; F2, R) used to detect progeny of RFP-StopFLhORMDL3-TG mice crossed with Zp3-cre mouse and predicted sizes of transgene mRNA assessed by PCR. (D) WT mice (left) and hORMDL3zp3-Cre mice (right) aged 26 weeks appeared morphologically similar. (E) The gross appearance of lungs of WT mice (left) and hORMDL3zp3-Cre mice (right) aged 26 weeks appeared morphologically similar. (F). Other than airway remodeling the appearance of lungs of WT mice (left) and hORMDL3zp3-Cre mice (right) aged 26 weeks appeared morphologically similar. (G) Expression of the human ORMDL3 (hORMDL3) transgene was detected by qRT/PCR in mouse lung, (H) mouse bronchial epithelium, and (I) mouse BAL macrophages in hORMDL3zp3-Cre mice, but not in WT mice. The human ORMDL3 transgene did not increase levels of mouse ORMDL1 (m-ORMDL1), mouse ORMDL2 (m-ORMDL2), or mouse ORMDL3 (m-ORMDL3) as assessed by qRT/PCR in mouse lung (J), mouse bronchial epithelium (K), and mouse BAL macrophages (L) in either hORMDL3zp3-Cre or WT mice.
Processing of lungs, BAL, and blood
hORMDL3zp3-Cre mice and littermate controls were euthanized at different ages (4 weeks, 8 weeks, 26 weeks) to quantitate levels of airway inflammation, airway remodeling, as well as expression of cytokines, chemokines, and remodeling genes. In addition to examining whole lung, purified populations of selected lung cell types (bronchial epithelium, BAL macrophages) were also studied. Levels of IgE and other immunoglobulins were quantitated in peripheral blood.
Lung processing
Lungs were processed for protein and RNA extraction, as well as for immunohistology (paraffin-embedded lung sections) as previously described in this laboratory (13, 16). For protein and RNA extractions, lungs were initially snap-frozen in liquid nitrogen and stored at −80°C. For paraffin-embedded sections, lungs were equivalently inflated with an intratracheal injection of the same volume of 4% paraformaldehyde solution (Sigma Chemicals, St. Louis, MO) to preserve the pulmonary architecture. Lung sections were processed for immunohistochemistry to detect major basic protein (MBP)(anti-mouse MBP Ab kindly provided by James Lee PhD, Mayo Clinic, Scottsdale, Arizona), neutrophil elastase (anti-mouse neutrophil elastase Ab; Santa Cruz Biotech), F4/80 (anti-mouse F4/80 Ab; Santa Cruz Biotech), and CD4 (anti-mouse CD4 Ab; GeneTex). The number of individual cells staining positive for different cell types in the peribronchial space was counted using a light microscope. Results are expressed as the number of peribronchial cells staining positive per bronchiole with 150–200 μm of internal diameter. At least ten bronchioles were counted in each slide.
BAL macrophages
In selected experiments, purified populations of BAL macrophages (>98% purity) were obtained by adhesion by placing BAL cells in a 10-cm Petri dish in complete media for 4 h at 37°C as previously described in this laboratory (13). Pooled BAL macrophages from 4 mice/group were used for RNA and protein extraction.
Isolation of bronchial epithelial cells
The isolation of bronchial epithelial cells was performed as previously described in this laboratory (13, 17). Briefly, the epithelial brushing was performed using a sterile plastic feeding tube (Solomon Scientific) inserted into the right main and left main bronchus with gentle brushing and immediately placed in RNA-STAT-60 (Tel-Test) for RNA extraction. Bronchial epithelial cells were of >95% purity as assessed by E-cadherin expression on FACS, and histologic detection of ciliated epithelial cells (13, 17). Four mice/group were used for each data point.
BAL Fluid collection
BAL fluid was collected by lavaging the lung with 1 ml PBS via a tracheal catheter as previously described (16). BAL fluid was centrifuged, and the supernatant frozen at 80 °C for subsequent cytokine analysis.
Peripheral Blood
Peripheral blood was obtained from mice by cardiac puncture into tubes without anticoagulant added for quantitation of serum immunoglobulin levels.
Detection of ORMDL3 and ORMDL3 regulated genes by qRT/PCR
qRT/PCR was performed as previously described in this laboratory (13). In brief, total RNA was extracted with RNA-STAT-60 (Tel-Test) and reverse transcribed with Oligo-dT and SuperScript II kit (Life Technologies). qPCR was performed with TaqMan PCR Master Mix and ORMDL1, ORMDL2, ORMDL3 (human and mouse), SERCA2b, TGF-β1, ADAM8, MMP9, ITAC, and IP-10 primers (all from Applied Biosystems). The relative amounts of transcripts were normalized to those of housekeeping gene (GAPDH) mRNA and compared between the different genes by the ΔΔCt method as previously described in this laboratory (13).
Detection of airway remodeling
Peribronchial smooth muscle layer
The thickness of the airway smooth muscle layer was measured by α-smooth muscle actin immunohistochemistry as previously described (16). Lung sections were immunostained with an anti-α-smooth muscle actin primary antibody (Sigma-Aldrich) to detect peribronchial smooth muscle. Species- and isotype-matched Abs were used as controls in place of the primary Ab. The area of peribronchial α-smooth muscle actin staining in paraffin-embedded lungs was outlined and quantified under a light microscope (Leica DMLS) attached to an image analysis system (Image-Pro plus) as previously described (16). Results are expressed as the area of peribronchial α-smooth muscle actin staining per μm length of basement membrane of bronchioles 150–200 μm of internal diameter.
Peribronchial trichrome staining
The area of peribronchial trichrome staining in paraffin-embedded lungs was outlined and quantified under a light microscope (Leica DMLS, Leica Microsystems) attached to an image analysis system (Image-Pro plus, Media Cybernetics) as previously described (16). Results are expressed as the area of trichrome staining per μm length of basement membrane of bronchioles 150–200 μm of internal diameter.
Lung collagen
The amount of lung collagen was measured as previously described in this laboratory (16) with a collagen assay kit that uses a dye reagent that selectively binds to the [Gly-X-Y]n tripeptide sequence of mammalian collagens (Biocolor; Newtonabbey). In all experiments, a collagen standard was used to calibrate the assay.
Airway mucus expression
To quantitate the level of mucus expression in the airway, the number of periodic acid Schiff (PAS)-positive and PAS-negative epithelial cells in individual bronchioles was counted as previously described in this laboratory (16). At least ten bronchioles were counted in each slide. Results are expressed as the percentage of PAS-positive cells per bronchiole, which is calculated from the number of PAS-positive epithelial cells per bronchus divided by the total number of epithelial cells of each bronchiole.
Lung protein for Elisa and western blot
Lung tissue was homogenized in 500 μl of lysis buffer and 100 mg of stainless steel beads (Next Advance, Inc) for 5 min using the Bullet Blender™ homogenizer (Next Advance, Inc). The lysate was centrifuged at 13,000rpm for 10 min at 4°C. The supernatant was used for Elisa and western blots.
Quantitation of lung cytokines
BAL fluid and lung levels of selected Th2 cytokine (IL-4, IL-5, IL-13), and chemokines (eotaxin-1) was performed by ELISA according to the manufacturer’s instructions (R & D Systems). Levels of lung IL-4, IL-5, IL-13, and eotaxin-1 are expressed as the amount of cytokine/chemokine in pg per mg lung protein. Lung protein levels were quantitated by the BCA method (Thermo Scientific).
Detection of SERCA2b by western blot
Lung protein was separated on a SDS/PAGE gel and transferred to a PVDF membrane. Membranes were blocked in 5% (wt/vol) milk in 1× Tris-buffered saline with Tween for 1 h and then incubated with the primary antibody overnight at 4 °C. The primary antibodies used in this study were mouse monoclonal anti-SERCA2b (Abcam), and rabbit monoclonal anti-GAPDH (Genetex).
Quantitation of bronchial epithelial TGF-β1, ADAM8, Serca2b, MMP9 by image analysis
Lung sections from hORMDL3zp3-Cre and WT mice were immunostained with either anti-TGFβ1, anti-ADAM8, anti-Serca2b, or anti-MMP9 Abs. Levels of expression of either TGFβ1, ADAM8, Serca2B, or MMP-9 by bronchial epithelial cells (outlined and visualized with a light microscope attached to an image-analysis system) was quantitated by image analysis as described (18). The images were saved and analyzed with Image-Pro Plus 3 software (Media Cybernetics, Bethesda, MD). The mean value of bronchial epithelial immuostain intensity was normalized for the total area of bronchial epithelium.
Quantitation of serum IgE, IgG, IgM, and IgA
Serum total IgE was quantitated with an IgE ELISA kit (BD Biosciences). Serum IgG (IgG1 and IgG2a), IgM, and IgA were quantitated using a mouse immunoglobulin isotyping ELISA kit (BD Pharmingen) with results reported as OD at 450nm per manufacturer’s instructions. Serum OVA-specific IgE was quantitated with a mouse OVA-specific IgE kit (Biolegend) with results reported as ng/ml. All ELISA plates were read with a BioRad Model 680 microplate reader.
Activation of ATF6, IRE1, and PERK
Purified populations of macrophages (> 95% pure) were generated from hORMDL3zp3-Cre and WT mouse bone marrow cells cultured in complete DMEM media (Gibco) supplemented with stem cell growth factors (L-cells media, ATCC) for 10 days as described (19). Activation of ATF6 was detected with immunofluorescence microscopy to detect nuclear localization of ATF6 using an ATF6 Ab (Imgenex) as previously described (13, 20). Activation of Ire1 was detected by PCR as it removes the UPR intron from the un-spliced form of XBP1 (XBP1u) to generate the spliced form of XBP1 (XBP1s) mRNA (13, 20). Activation of PERK was assessed by increased levels of phospho-eIF2α by western blot using an antibody specific to the phosphorylated form of eIF2α (13, 20). In all UPR experiments, thapsigargin a known activator of the UPR was incubated with cells for one hour before collecting cells for either RNA or protein analyses. As insufficient numbers of bronchial epithelial cells were available for UPR western blot studies, the UPR studies were performed on macrophages a cell type that like epithelial cells express high levels of ORMDL3 (13).
Acute OVA Challenge Model
hORMDL3zp3-Cre and littermate control mice aged 12 weeks were sensitized and challenged with OVA (Worthington, Lakewood, NJ) as previously described (13). In brief, mice were sensitized i.p. with 100 μg OVA and 2 mg aluminum hydroxide (Imject Alum; Thermo Fisher Scientific, Waltham, MA) in a total volume of 200 μl PBS on days 0 and 10 followed by intranasal administration of 200 μg OVA in 20 μl PBS on days 21, 23, and 25. Non-OVA challenged ORMDL3 TG and littermate groups of mice were sensitized and challenged with PBS only. Twenty-four hours after the last challenge, bronchoalveolar lavage (BAL) fluid, lungs, and blood, were collected as described above.
Airway hyperreactivity to methacholine
Airway responsiveness to methacholine (MCh) was assessed in intubated and ventilated mice aged 12 weeks (n=8 mice/group)(flexiVent ventilator; Scireq) anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg) intraperitoneally as previously described (16). The dynamic airway resistance and elastance were determined using Scireq software in mice exposed to nebulized PBS and MCh (0, 3, 24, 48 mg/ml). The following ventilator settings were used: tidal volume (10 ml/kg), frequency (150/min), and positive end-expiratory pressure (3 cmH2O). Increased elastance values signal an increased stiffness of the lungs, and elastance is the inverse of compliance (Scireq).
Statistical Analysis
All results are presented as mean ± SEM. A statistical software package (Graph Pad Prism, San Diego, CA) was used for the analysis. P values of < 0.05 were considered statistically significant.
RESULTS
Generation of hORMDL3 TG mice
To perform studies in hORMDL3 TG mice, we generated conditional hORMDL3 TG floxed mice (RFP-StopFLhORMDL3-TG mice) with the pCAGEN lox mRFP-H2B STOP lox hORMDL3 transgene construct (Fig 1 A, B), and crossed these with Zona Pellucida 3 (Zp3) Cre mice resulting in offspring with expression of human ORMDL3 in all cells (hORMDL3zp3-Cre mice)(21). The transgene construct we developed has a loxP flanked red fluorescent protein (RFP) and transcriptional stop codon positioned at the transcriptional initiation site of the hORMDL3 transgene which prevents transcription of hORMDL3 (Fig 1A). Thus, all cells in this RFP-StopFLhORMDL3-TG mouse will not express hORMDL3 until crossed with a Cre expressing mouse which excises the transcriptional stop codon and RFP (Fig 1B). We used PCR (Fig 1C) to confirm successful expression of the hORMDL3 transgene in hORMDL3zp3-Cre mice. The presence of the non-expressed floxed transgene construct in RFP-StopFLhORMDL3-TG mice tissues/cells was assessed by red fluorescence prior to crossing to Zp3 Cre mice. Crossing RFP-StopFLhORMDL3-TG mice to Zp3 Cre mice results in the loss of RFP expression in cells that was designed to provide a rapid initial screen for successful expression of hORMDL3 (Fig 1A, 1B). However, as levels of the red fluorescent protein as detected by immunofluorescence microscopy varied widely in RFP-StopFLhORMDL3-TG mice, we utilized hORMDL3 PCR, rather than loss of red fluoresecence, to detect hORMDL3 expression. hORMDL3zp3-Cre mice overexpressing hORMDL3 constitutively in all cells were viable with no obvious developmental or morphologic defects (Fig 1 D) and their lung size and weights and were similar to that of WT mice (Fig 1E–F).
Levels of the human ORMDL3 transgene were highly expressed in hORMDL3zp3-Cre mouse lung (Fig 1G), bronchial epithelium (Fig 1H), and BAL macrophages (Fig 1I) as assessed by qRT/PCR. In contrast, no expression of the human ORMDL3 transgene was detected in wild type littermate mice (referred to as WT subsequently) (Fig 1G–I). Levels of mouse ORMDL1, mouse ORMDL2, and mouse ORMDL3 in hORMDL3zp3-Cre mice were not altered in mouse lung (Fig 1J), bronchial epithelium (Fig 1K), and BAL macrophages (Fig 1L) as assessed by qRT/PCR.
Increased airway remodeling in hORMDL3zp3-Cre mice
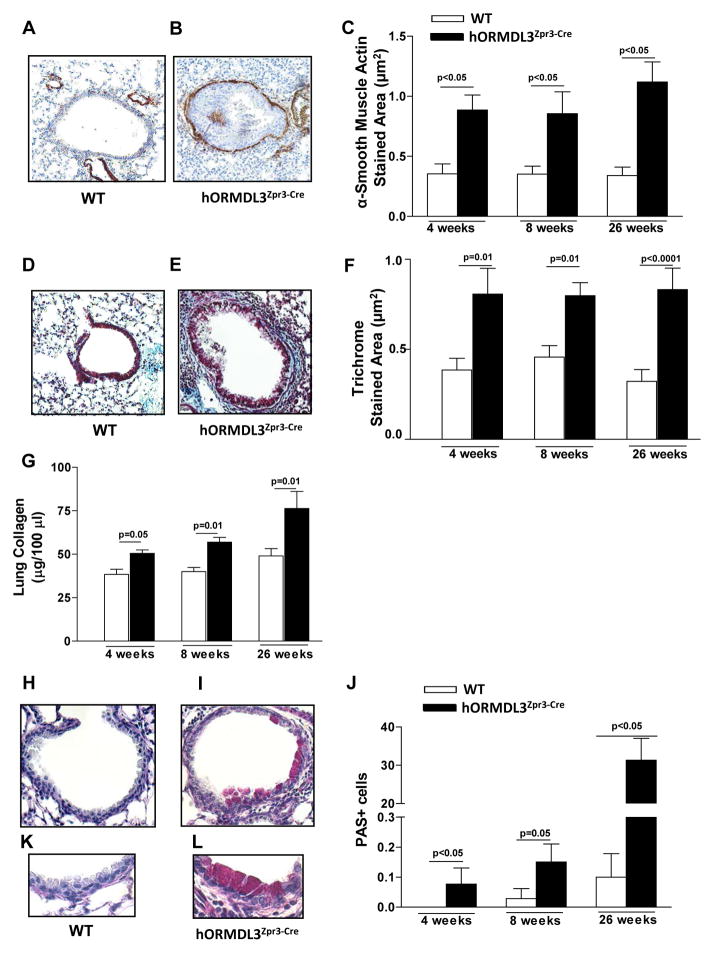
hORMDL3zp3-Cre mice have evidence of airway remodeling characteristic of asthma at 4 weeks of age which persisted through 26 weeks of life (Fig 2). Features of airway remodeling that are evident in hORMDL3zp3-Cre mice include an increase in the area of peribronchial smooth muscle as assessed by immunostaining with an anti-α-smooth muscle actin Ab (Fig 2A–C), an increase in peribronchial fibrosis as assessed by the area of peribronchial trichrome staining to detect lung collagen (Fig 2D–F), and an increase in total lung collagen (Fig 2G). In addition, there was a significant increase in mucus expression detected by PAS staining (Fig 2H–L). While the levels of peribronchial smooth muscle (Fig 2C), and peribronchial fibrosis as assessed by trichrome staining (Fig 2F) remained stably increased from week 4 to week 26, levels of mucus continued to increase from week 4 to week 26 (Fig 2J).
Figure 2. Airway remodeling in hORMDL3zp3-Cre mice.
A.–C. Levels of peribronchial smooth muscle were quantitated by immunohistochemistry using an anti-α-smooth muscle actin antibody and image analysis. Results are expressed as the α-smooth muscle actin stained area (μm2) per μm length of basement membrane of bronchioles 150–200 μm of internal diameter in (A) WT and (B,C) hORMDL3zp3-Cre mice aged 4, 8, and 26 weeks of age (n=12 mice/group). D–F. Levels of peribronchial trichrome staining were quantitated by image analysis. Results are expressed as the trichrome stained area (μm2) per μm length of basement membrane of bronchioles 150–200 μm of internal diameter in (D) WT and (E,F) hORMDL3zp3-Cre mice 4, 8, and 26 weeks of age (n=12 mice/group). G. Levels of lung collagen were quantitated by a Sircol assay in WT and hORMDL3zp3-Cre mice aged 4, 8, and 26 weeks of age (n=12 mice/group). H–L. Levels of mucus were quantitated by PAS staining in (H) WT and (I, J) hORMDL3zp3-Cre mice aged 4, 8, and 26 weeks of age (n=12 mice/group). Higher magnification view of PAS staining in (K) WT and (L) hORMDL3zp3-Cre mice.
Activation of ATF6 but not Ire1 or PERK in hORMDL3zp3-Cre mice
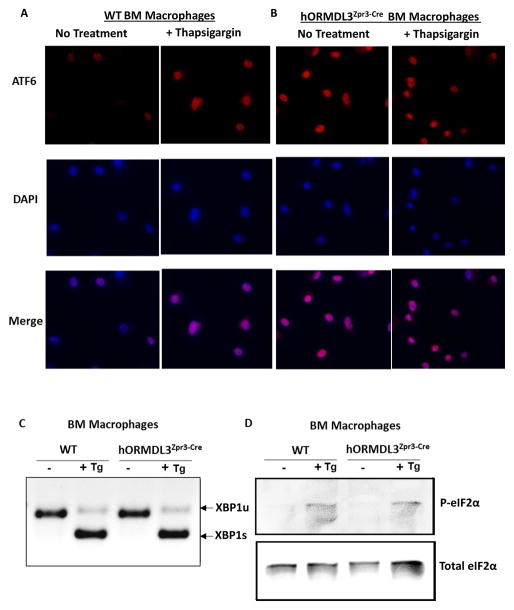
We have previously demonstrated that in vitro transfection of ORMDL3 activates only one of the three pathways of the UPR (i.e. activates the ATF6 pathway and not the Ire1 or PERK pathway) (13). To determine whether in vivo hORMDL3 activated the UPR, we cultured bone marrow derived macrophages from WT and hORMDL3zp3-Cre mice to have sufficient numbers of cells to perform a western blot. hORMDL3zp3-Cre mouse macrophages, but not WT macrophages, spontaneously activated the ATF6 pathway as assessed by translocation of ATF6 from being dispersed in the ER to the nucleus using immunofluorescence microscopy (Fig 3A–B). In contrast, both WT and hORMDL3zp3-Cre mouse macrophages did not activate either Ire1 (Fig 3C), or PERK (Fig 3D) pathways. Incubation of WT and hORMDL3zp3-Cre mouse macrophages with thapsigargin a known activator of the UPR induced activation of the ATF6 (Fig 3A–B), Ire1 (Fig 3C), and PERK (Fig 3D) pathways in both WT and hORMDL3zp3-Cre mouse macrophages. Thus, bone marrow derived macrophages from hORMDL3zp3-Cre mice, like cells transfected with ORMDL3 in vitro (13), exhibit selective activation of the ATF6 UPR pathway.
Figure 3. ORMDL3 AND UPR.
Bone marrow derived macrophages from WT or hORMDL3zp3-Cre mice were treated with Thapsigargin (Tg), a known activator of the UPR, for 1 hour. (A–B) ATF6. Immunofluorescence against ATF6 (red) was performed. Active ATF6 is shown by nuclear localization (merged purple color) as depicted by co-localization with DAPI (blue). (C). Ire1. Activation of Ire1 was detected by RT/PCR as it removes the UPR intron from the un-spliced form of XBP1 (XBP1u) to generate the spliced form of XBP1 (XBP1s) mRNA. (D) PERK. Activation of PERK is assessed by phosphorylation of elF2α (P-elF2α) blotted by anti-phospho specific elF2α antibody on SDS-PAGE by western blot. Total elF2α was blotted by anti-elF2α antibody.
Increased remodeling genes in lungs of hORMDL3zp3-Cre mice
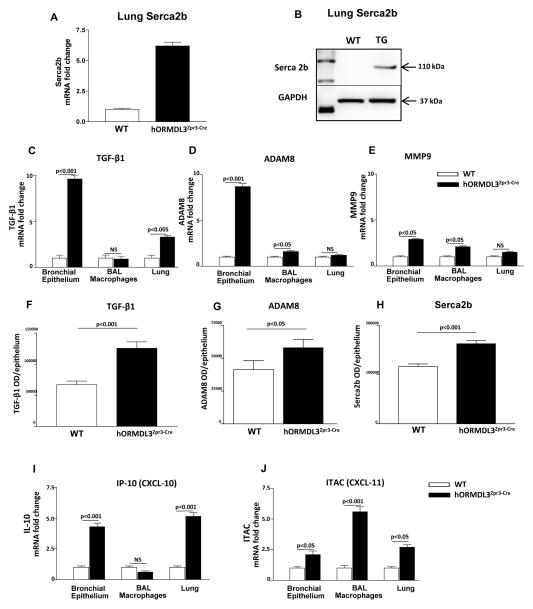
As Serca2b has been implicated in airway remodeling in asthma (15), we examined whether levels of Serca2b were modulated in hORMDL3zp3-Cre mice. Our studies demonstrate that hORMDL3zp3-Cre mice have increased lung levels of Serca2b as assessed by qRT/PCR (Fig 4A), and western blot (Fig 4B). In addition, hORMDL3zp3-Cre mice have increased levels of bronchial epithelial expression of TGF-β1 (Fig 4C), and ADAM8 (Fig 4D), with a smaller increase in MMP-9 (Fig 4E) as assessed by qRT/PCR. Levels of TGF-β1 (Fig 4F), ADAM8 (Fig 4G), Serca2b (Fig 4H), but not MMP-9 (data not shown), were increased in bronchial epithelium in hORMDL3zp3-Cre mice as assessed by immunohistochemistry with quantitation by image analysis.
Figure 4. hORMDL3zp3-Cre mice express SERCA2b, TGFβ1, ADAM8, MMP9, CXCL10, and CXCL11.
Levels of SERCA2b were quantitated in the lungs of hORMDL3zp3-Cre and WT mice by (A) qPCR, and (B), western blot. Levels of (C) TGFβ1, (D) ADAM8, and (E) MMP9, were quantitated by qPCR in the lungs, bronchial epithelium, and BAL macrophages of hORMDL3zp3-Cre and WT mice. Levels of (F) TGFβ1, (G) ADAM8, and (H) Serca2b were quantitated by immunohistochemistry and image analysis in bronchial epithelium of hORMDL3zp3-Cre and WT mice. Levels of (I) CXCL10 (IP-10), and (J) CXCL11 (ITAC), were quantitated by qPCR in the lungs, bronchial epithelium, and BAL macrophages of hORMDL3zp3-Cre and WT mice. (n=12 mice/group).
Levels of selected CXC and CC chemokines in hORMDL3zp3-Cre mice
As we had previously demonstrated that transfection of ORMDL-3 in vitro induced high levels of expression of CXC chemokines (IL-8; CXCL-10 also known as IP-10; CXCL-11 also known as ITAC)(13), and lower levels of CC chemokines (CCL-20 also known as MIP-3α)(13), we measured levels of these chemokines by qPCR in the hORMDL3zp3-Cre mice. hORMDL3zp3-Cre mice had significantly increased levels of lung CXC chemokine mRNA including CXCL10 in bronchial epithelial cells (Fig 4I), as well as increased levels of CXCL11 mRNA in BAL macrophages (Fig 4J), while levels of lung KC mRNA the murine equivalent of IL-8 was not increased (data not shown). Levels of selected CC chemokine mRNA (CCL-20 and eotaxin-1) were also not different in lung, bronchial epithelium or BAL macrophages in hORMDL3zp3-Cre compared to WT mice (data not shown).
Airway remodeling preceded airway inflammation in hORMDL3zp3-Cre mice
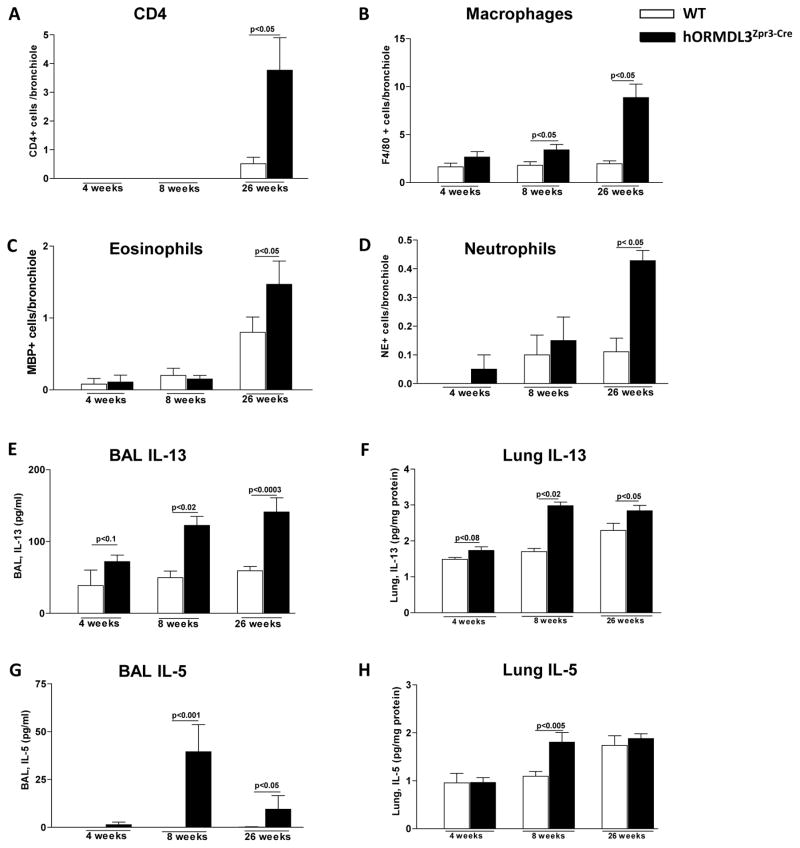
hORMDL3zp3-Cre mice exhibit significant airway remodeling at 4 weeks of age (Fig 2), a time point at which there is no evidence of any increase in peribronchial CD4+ cells (Fig 5A), F4/80+ macrophages (Fig 5B), MBP+ eosinophils (Fig 5C), or neutrophil elastase+ neutrophils (Fig 5D) compared to WT mice. At 8 weeks of age there is a small increase in F4/80+ peribronchial macrophages (Fig 5B) in the lungs of hORMDL3zp3-Cre mice, with no change in CD4+ cells, eosinophils, or neutrophils (Fig 5A, C, D). At 26 weeks of age there is a significant increase in peribronchial CD4+ cells (Fig 5A), F4/80+ macrophages (Fig 5B), eosinophils (Fig 5C), and neutrophils (Fig 5D) in hORMDL3zp3-Cre compared to WT mice.
Figure 5. Levels of lung immune and inflammatory cells, and Th2 cytokines in hORMDL3zp3-Cre mice.
The number of (A) CD4+, (B) F4/80+, (C) MBP+, and (D) neutrophil elastase+ (NE+) cells per bronchiole of 150–200 μm of internal diameter was quantitated by immunohistochemistry and image analysis in WT and hORMDL3zp3-Cremice at 4, 8, and 26 weeks of age (n=12 mice/group). Levels of IL-13 in (E) BAL and (F) lung, as well as levels of IL-5 in (G) BAL and (H) lung were quantitated by ELISA (n=12 mice/group).
Airway remodeling precedes increase in Th2 cytokines in hORMDL3zp3-Cre mice
Levels of Th2 cytokines IL-5, IL-13 were not increased in either BAL or lung of hORMDL3zp3-Cre mice at 4 weeks of age (Fig 5E–H), a time point at which hORMDL3zp3-Cre mice exhibit significant airway remodeling. Levels of BAL IL-13 (Fig 5E) and lung IL-13 (Fig 5F) were significantly increased in hORMDL3zp3-Cre mice at 8 weeks and 26 weeks of age as assessed by ELISA. Levels of BAL IL-5 (Fig 5G) were significantly increased in hORMDL3zp3-Cre mice at 8 weeks and 26 weeks of age, while levels of lung IL-5 were only increased at 8 weeks (Fig 5H). In contrast, there was no increase in levels of lung IL-4 in hORMDL3zp3-Cre mice at 4, 8, or 26 weeks of age (data not shown).
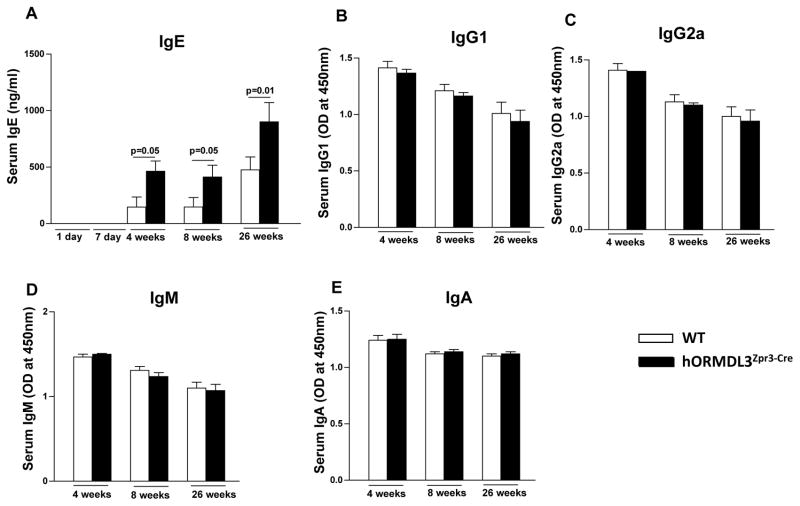
Increased IgE but not IgG, IgM, or IgA in hORMDL3zp3-Cre mice
Levels of IgE were significantly increased in hORMDL3zp3-Cre compared to WT mice at 4 weeks of age, and this increase in IgE persisted at 8 weeks and 26 weeks (Fig 6A). The increase in IgE was selective, as there was no increase in IgG (IgG1 or IgG2)(Fig 6B, 6C), IgM (Fig 6D), or IgA (Fig 6E) in hORMDL3zp3-Cre mice.
Figure 6. Levels of IgE, IgG, IgM, and IgA in hORMDL3zp3-Cre mice.
Levels of (A) IgE, (B) IgG1, (C) IgG2, (D) IgM, and (E) IgA were quantitated by ELISA in hORMDL3zp3-Cre (black bar) and WT (white bar) mice at 4, 8, and 26 weeks of age (n=12 mice/group).
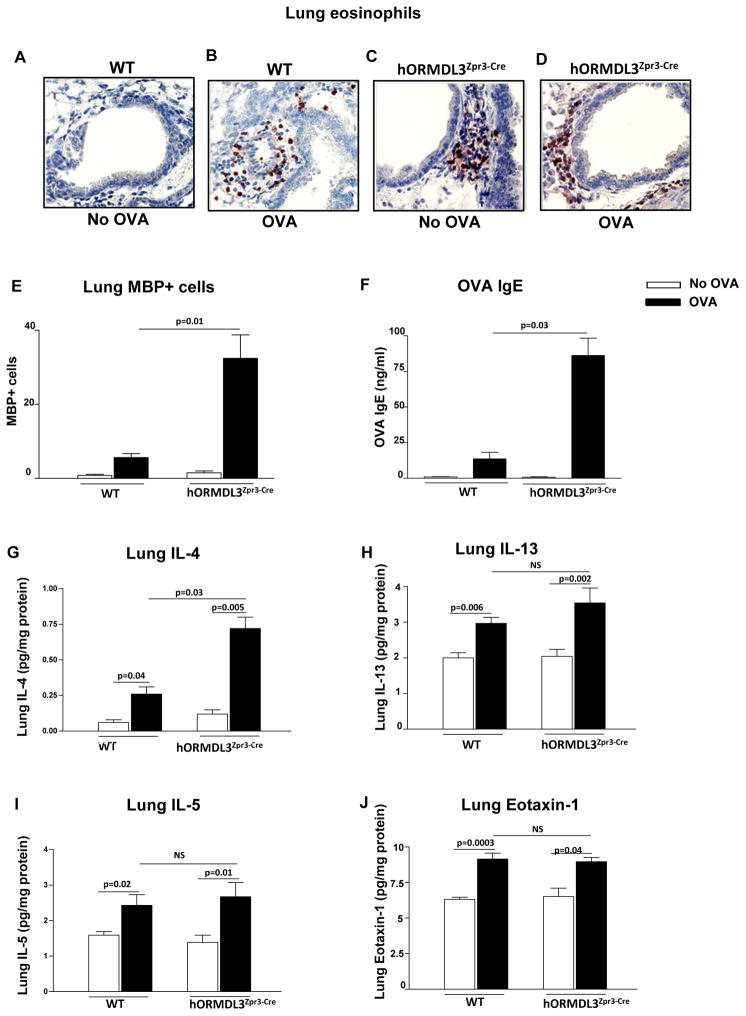
Acute OVA allergen challenge enhances peribronchial eosinophilic inflammation, OVA specific IgE, and IL-4 in hORMDL3zp3-Cre mice
Acute OVA challenge induced a greater increase in peribronchial eosinophils (Figure 7A–E) and OVA specific IgE (Fig 7F) in hORMDL3zp3-Cre mice compared to WT mice. This was associated with increased lung IL-4 levels in hORMDL3zp3-Cre mice compared to WT mice (Fig 7G). While acute OVA challenge induced increased levels of lung IL-5 (Fig 7I), IL-13 (Fig 7H), and eotaxin-1 (Fig 7J) in hORMDL3zp3-Cre mice (OVA vs no OVA), these levels were not different from that detected in OVA challenged WT mice.
Figure 7. Effect of acute OVA allergen challenge on airway inflammation, IgE, and Th2 cytokines in hORMDL3zp3-Cre mice.
hORMDL3zp3-Cre and WT mice were sensitized to OVA and challenged with OVA allergen by inhalation. Compared to OVA challenged WT mice, OVA challenged hORMDL3zp3-Cre mice had significantly increased levels of (A–E) lung MBP+ peribronchial eosinophils as assessed by immunohistochemistry, (F) OVA specific IgE, and (G) lung IL-4. Levels of lung (H) IL-13, (I) IL-5, and (J) eotaxin-1 were similar in OVA challenged WT and OVA challenged hORMDL3zp3-Cre mice (n=12 mice/group).
Airway hyperreactivity to methacholine
hORMDL3zp3-Cre mice had spontaneous increased AHR to methacholine compared to WT mice at 12 weeks of age (Fig 8A). In addition, hORMDL3zp3-Cre mice had increased lung elastance compared to WT mice (Fig 8B).
Figure 8. Airway Responsiveness in hORMDL3zp3-Cre mice.
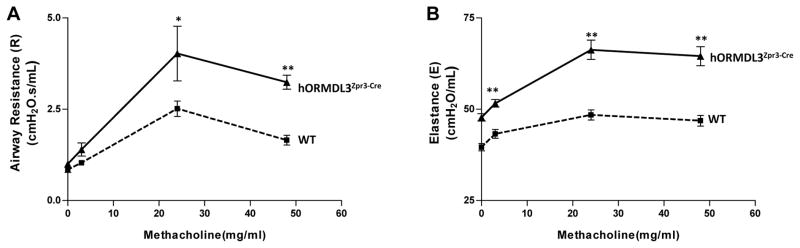
Airway responsiveness to methacholine (MCh) was assessed in intubated and ventilated hORMDL3zp3-Cre and WT mice aged 12 weeks (flexiVent ventilator; Scireq)(n=8 mice/group)(A). Lung elastance (the inverse of compliance) was also recorded in hORMDL3zp3-Cre and WT mice (n=8mice/group) (B). * p< 0.01; ** P<0.002
DISCUSSION
Although multiple genetic association studies have demonstrated that ORMDL3 is highly linked to asthma (1–10), the mechanism by which ORMDL3 may contribute to the pathogenesis of asthma in vivo is at present unknown. In this study using a mouse model in which the human ORMDL3 gene is overexpressed, we demonstrate the novel findings that expression of the human ORMDL3 transgene in vivo is associated with significantly increased airway remodeling including increased airway smooth muscle, subepithelial fibrosis, and mucus. These airway remodeling changes in hORMDL3zp3-Cre mice were associated with the spontaneous development of increased airway responsiveness. In addition, the remodeling changes were associated with an increased lung elastance (the inverse of lung compliance) which indicates an increased stiffness of the remodeled lungs. The mechanism of the increased airway remodeling did not appear to be dependent upon increased airway inflammation, as significant airway remodeling was evident at 4 weeks of age in the hORMDL3zp3-Cre mice, a time point not associated with an increased number of peribronchial eosinophils, neutrophils, macrophages, or CD4 cells. Increased levels of expression of genes associated with airway remodeling were detected in the lung (SERCA2b) and airway epithelium (TGF-β1, ADAM8, MMP-9) of hORMDL3zp3-Cre mice suggesting that these pathways may contribute to airway remodeling detected in these mice. The importance of these genes that are highly expressed in hORMDL3zp3-Cre mice to airway remodeling and asthma is suggested from studies demonstrating expression of these pathways in the airways of human asthmatics (15, 22–27), induction of these mediators by allergen inhalation in asthmatics (TGF-β1, MMP-9)(28–30), and inhibition of asthma outcomes in mice deficient in these genes (ADAM-8, Smad3, MMP-9)(31–35), or in mice treated with neutralizing antibodies (TGF-β1) (36).
We have previously performed in vitro studies and demonstrated that transfection of ORMDL3 induces activation of one of the three pathways of the endoplasmic reticulum UPR, namely the ATF6 pathway (13). Using hORMDL3zp3-Cre mice we have made the novel observation that the ATF6 pathway of the UPR, is selectively activated by the human ORMDL-3 transgene in vivo. ATF6 (consisting of the closely related ATF6α and ATF6β in mammals)(37) is a transcription factor known to regulate genes involved in ER protein folding (14), as well as expression of SERCA2b which has been implicated in airway remodeling in asthma (15). In this study we demonstrate that hORMDL3zp3-Cre mice exhibit both activation of ATF6 and increased levels of lung SERCA2b, suggesting that the ATF6 and SERCA2b pathways are downstream of ORMDL3 in vivo as previously demonstrated in vitro (13). In prior in vitro studies we have demonstrated that transfection of ORMDL3 induced activation of ATF6 and expression of SERCA2b, while knockdown of ATF6α inhibited SERCA2b expression (13). Activation of ATF6 was maximum as further induction of the UPR upon treatment of the cells with thapsigargin, a well characterized ER stress inducer, did not increase the nuclear localization of ATF6. Overall, these studies of hORMDL3zp3-Cre mice provide in vivo evidence of ATF-6α dependent pathways (SERCA2b) and ATF-6α independent pathways (TGF-β1, ADAM-8, MMP-9) through which the ER localized ORMDL3 may be linked to airway remodeling and asthma. However, only future studies in which the ATF6 pathway is selectively inhibited will be able to determine the role played by the ATF6 pathway in any remodeling changes we have noted in hORMDL3zp3-Cre mice.
hORMDL3zp3-Cre mice also had an age related increase in levels of airway inflammation, as well as increase in lung cytokines and chemokines. The increased levels of airway inflammation were not evident in hORMDL3zp3-Cre mice aged 4 weeks, and only started to be evident at 8 weeks (increase in peribronchial macrophages). At 26 weeks hORMDL3zp3-Cre mice had significantly increased levels of peribronchial CD4+ cells, eosinophils, macrophages, and neutrophils, suggesting that the airway inflammatory response in hORMDL3zp3-Cre mice increased during the period from 8 weeks to 26 weeks of age. As the inflammatory response in hORMDL3zp3-Cre mice had evidence of both Th2 mediated inflammation (increased CD4+ cells and eosinophils), as well as increased neutrophils and macrophages we investigated whether their lungs expressed cytokines and chemokines associated with Th2 mediated inflammation (IL-4, IL-5, IL-13, eotaxin-1), or chemokines known to be regulated by ORMDL3 in vitro (CXCL10, CXCL11, IL-8, CCL20)(13). Levels of lung Th2 cytokines (IL-5 and IL-13) were increased in hORMDL3zp3-Cre mice at 8 and 26 weeks of age. In contrast, there was no increase in lung IL-4 or eotaxin-1 at any time point. Allergen challenged hORMDL3zp3-Cre mice had significantly increased levels of peribronchial eosinophils and lung IL-4 compared to allergen challenged WT mice. IL-4 can contribute to increased lung eosinophilic inflammation as evident in studies of IL-4 transgenic mice (38). Our studies of hORMDL3zp3-Cre mice confirmed that chemokines (CXCL10, CXCL11) we had observed to be highly expressed in vitro in ORMDL3 transfected cells (13), were highly expressed in hORMDL3zp3-Cre mice bronchial epithelium (CXCL10) and alveolar macrophages (CXCL11). Although asthma has predominantly been associated with expression of CC chemokines, CXC chemokines have also been linked to asthma in studies in humans with asthma (39–42), as well as in studies in animal models (43, 44)(i.e. CXCL-10 KO have significant reduction in Th2-type allergic airway inflammation and AHR)(43).
hORMDL3zp3-Cre mice also had increased levels of IgE without increases in IgG, IgM, or IgA. The level of OVA specific IgE increased significantly more in hORMDL3zp3-Cre mice challenged with OVA compared to WT mice challenged with OVA. As IL-4, and IL-13 switch B cells to IgE production (45), we examined whether either cytokine was increased in association with increases in OVA specific IgE or increases in total IgE. The increase in OVA specific IgE in hORMDL3zp3-Cre mice following acute OVA challenge was associated with increased levels of IL-4. In contrast, the increase in total IgE detected in hORMDL3zp3-Cre mice preceded the increase in IL-13. Preliminary studies did not demonstrate a difference in the number of splenic B cells expressing IgE in hORMDL3zp3-Cre compared to WT mice (data not shown). Further studies are needed to determine whether the increased total IgE, and allergen specific IgE, production we have noted in hORMDL3zp3-Cre mice is due to either cytokines (IL-4, IL-13), or T cell and/or B cell pathways that are regulated by ORMDL3. In epidemiologic studies of asthma chromosome 17q21 has been linked to IgE in some (5, 8) but not all studies (3, 7). Studies of ORMDL3 in Puerto-Rican asthmatics have demonstrated a significant association between SNP rs12603332 and log10 IgE levels (5), while, subgroup analysis showed important associations between SNPs rs4378650 and rs12603332 in patients with allergic asthma (IgE > 100 IU/ml). The association between these SNPs and asthma became stronger in patients with IgE levels exceeding 100 IU/ml (5).
In summary, these studies in hORMDL3zp3-Cre mice provide evidence that ORMDL3 plays an important role in activation of the ATF6 UPR pathway in vivo, and that expression of ORMDL3 in vivo regulates airway remodeling (smooth muscle, fibrosis, mucus) potentially through ATF6 target genes such as SERCA2b, and/or through ATF6 independent genes (TGF-β1, ADAM8, MMP-9) detected in airway epithelium, as well as through as yet unidentified and/or uninvestigated pathways. In this regard sphingolipids are known to be regulated by ORMDL3 and sphingolipid pathways have been associated with asthma but not airway remodeling in mouse models (46–48). ORMDL3 also regulates ER mediated calcium signaling (49) and lymphocyte activation in vitro (50). As ORMDL3 is expressed in multiple cell types important to the pathogenesis of asthma (i.e. epithelial cells, macrophages, eosinophils, T cells)(13, 51), in this study we have examined how increased expression of ORMDL3 in multiple cell types contribute to the pathogenesis of asthma. Future studies with selective over-expression of ORMDL3 in particular cell types will provide insight into the role of ORMDL3 in these individual cell types to the pathogenesis of asthma. Interestingly, increased levels of airway remodeling preceded increased levels of airway inflammation (CD4+ cells, macrophages, eosinophils, neutrophils) in the lungs of hORMDL3zp3-Cre mice, suggesting that airway remodeling can be dissociated from these pathways by activation of ORMDL3. As airway remodeling can be detected not only in adults with asthma, but also in childhood asthma (52), increased expression of ORMDL3 in childhood asthmatics could contribute to the early development of airway remodeling. Overall, these studies provide preliminary evidence for an in vivo mechanism to link an ER localized protein such as ORMDL-3 to the pathogenesis of airway remodeling, airway hyperreactivity, and asthma.
Acknowledgments
Grant support: NIH grants AI 107779, AI 38425, AI 70535, AI 72115 to D.H.B, GM087415 and American Cancer Society (RSG10-027-01) to M.N., and K08 AI080938, and AAAAI/ALA to T.D.
Abbreviation used in this article
- ADAM8
a disintegrin and metalloproteinase domain-containing protein 8
- ATF6
activating transcription factor 6
- AHR
airway responsiveness
- BAL
bronchoalveolar lavage
- ER
endoplasmic reticulum
- IP-10
interferon gamma-induced protein 10
- Ire1
inositol-requiring-enzyme 1
- ITAC
interferon-inducible T-cell alpha
- MBP
major basic protein
- ORMDL
orosomucoid like
- PAS
periodic acid Schiff
- PERK
protein-kinase-regulated-by-RNA-like-ER-associated-kinase
- Serca2B
sarco/endoplasmic reticulum Ca2+ ATPase
- TGF-β1
transforming growth factor beta 1
- UPR
unfolded protein response
- WB
western blot
- WT
wild-type
- XBP1
x-box-binding protein 1
References
- 1.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 2.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, Chateigner N, Gormand F, Just J, Le Moual N, Scheinmann P, Siroux V, Vervlot D, Zelenika D, Pin I, Kauffmann F, Lathrop M, Demenais F. Effect of 17q21variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–1994. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 3.Moffatt MF, I, Gut G, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO. GABRIEL Consortium. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan YI, Shrine NR, Soler Artigas M, Wain LV, Blakey JD, Moffatt MF, Bush A, Chung KF, Cookson WO, Strachan DP, Heaney L, Al-Momani BA, Mansur AH, Manney S, Thomson NC, Chaudhuri R, Brightling CE, Bafadhel M, Singapuri A, Niven R, Simpson A, Holloway JW, Howarth PH, Hui J, Musk AW, James AL, Brown MA, Baltic S, Ferreira MA, Thompson PJ, Tobin MD, Sayers I, Hall IP Australian Asthma Genetics Consortium. Genome-wide association study to identify genetic determinants of severe asthma. Thorax. 2012;67:762–768. doi: 10.1136/thoraxjnl-2011-201262. [DOI] [PubMed] [Google Scholar]
- 5.Galanter J, Choudhry S, Eng C, Nazario S, Rodríguez-Santana JR, Casal J, Torres-Palacios A, Salas J, Chapela R, Watson HG, Meade K, LeNoir M, Rodríguez-Cintrón W, Avila PC, Burchard EG. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sleiman PM, Annaiah K, Imielinski M, Bradfield JP, Kim CE, Frackelton EC, Glessner JT, Eckert AW, Otieno FG, Santa E, Thomas K, Smith RM, Glaberson W, Garris M, Gunnlaugsson S, Chiavacci RM, Allen J, Spergel J, Grundmeier R, Grunstein MM, Magnusson M, Bisgaard H, Grant SF, Hakonarson H. ORMDL3 variants associated with asthma susceptibility in North Americans of European ancestry. J Allergy Clin Immunol. 2008;122:1225–1227. doi: 10.1016/j.jaci.2008.06.041. [DOI] [PubMed] [Google Scholar]
- 7.Hirota T, Harada M, Sakashita M, Doi S, Miyatake A, Fujita K, Enomoto T, Ebisawa M, Yoshihara S, Noguchi E, Saito H, Nakamura Y, Tamari M. Genetic polymorphism regulating ORM1-like 3 (Saccharomyces cerevisiae) expression is associated with childhood atopic asthma in a Japanese population. J Allergy Clin Immunol. 2008;121:769–770. doi: 10.1016/j.jaci.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 8.Leung TF, Sy HY, Ng MC, Chan IH, Wong GW, Tang NL, Waye MM, Lam CW. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy. 2009;64:621–628. doi: 10.1111/j.1398-9995.2008.01873.x. [DOI] [PubMed] [Google Scholar]
- 9.Binia A, Khorasani N, Bhavsar PK, Adcock I, Brightling CE, Chung KF, Cookson WO, Moffatt MF. Chromosome 17q21 SNP and severe asthma. J Hum Genet. 2011;56:97–98. doi: 10.1038/jhg.2010.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marinho S, Custovic A, Marsden P, Smith JA, Simpson A. 17q12-21 variants are associated with asthma and interact with active smoking in an adult population from the United Kingdom. Ann Allergy Asthma Immunol. 2012;108:402–411.e9. doi: 10.1016/j.anai.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Calikan M, Bochkov YA, Kreiner-Møller E, Bønnelykke K, Stein MM, Du G, Bisgaard H, Jackson DJ, Gern JE, Lemanske RF, Jr, Nicolae DL, Ober C. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hjemqvist L, Tuson M, Marfany G, Herrero E, Balcells S, Gonzalez-Durate R. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 2002;3:RESEARCH0027. doi: 10.1186/gb-2002-3-6-research0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller M, Tam AB, Cho JY, Doherty TA, Pham A, Khorram N, Rosenthal P, Mueller JL, Hoffman HM, Suzukawa M, Niwa M, Broide DH. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci U S A. 2012;109:16648–16653. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 15.Mahn K, Hirst SJ, Ying S, Holt MR, Lavender P, Ojo OO, Siew L, Simcock DE, McVicker CG, Kanabar V, Snetkov VA, O’Connor BJ, Karner C, Cousins DJ, Macedo P, Chung KF, Corrigan CJ, Ward JP, Lee TH. Diminished sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) expression contributes to airway remodelling in bronchial asthma. Proc Natl Acad Sci U S A. 2009;106:10775–10780. doi: 10.1073/pnas.0902295106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho JY, Miller M, Baek KJ, Han JW, Nayar J, Rodriguez M, Lee SY, McElwain K, McElwain S, Friedman S, Broide DH. Inhibition of airway remodeling in IL-5 deficient mice. J Clin Invest. 2004;113:551–560. doi: 10.1172/JCI19133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doherty TA, Khorram N, Sugimoto K, Sheppard D, Rosenthal P, Cho JY, Pham A, Miller M, Croft M, Broide D. Alternaria induces STAT-6 dependent acute airway eosinophilia and epithelial FIZZ1 expression that promotes airway fibrosis and epithelial thickness. J Immunol. 2012;188:2622–2629. doi: 10.4049/jimmunol.1101632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasad K, Prabhu GK. Image Analysis Tools for Evaluation of Microscopic Views of Immunohistochemically Stained Specimen in Medical Research. J Med Syst. 2012;36:2621–2631. doi: 10.1007/s10916-011-9737-7. [DOI] [PubMed] [Google Scholar]
- 19.Manzanero S. Generation of mouse bone marrow-derived macrophages. Methods Mol Biol. 2012;844:177–181. doi: 10.1007/978-1-61779-527-5_12. [DOI] [PubMed] [Google Scholar]
- 20.Babour A, Bicknell AA, Tourtellotte J, Niwa M. A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance. Cell. 2010;142:256–269. doi: 10.1016/j.cell.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brydges SD, Mueller JL, McGeough MD, Pena CA, Misaghi A, Gandhi C, Putnam CD, Boyle DL, Firestein GS, Horner AA, Soroosh P, Watford WT, O’Shea JJ, Kastner DL, Hoffman HM. Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity. 2009;30:875–887. doi: 10.1016/j.immuni.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vignola AM, Chanez P, Chiappara G, Merendino A, Pace E, Rizzo A, la Rocca AM, Bellia V, Bonsignore G, Bousquet J. Transforming growth factor-beta expression in mucosal biopsies in asthma and chronic bronchitis. Am J Respir Crit Care Med. 1997;156:591–599. doi: 10.1164/ajrccm.156.2.9609066. [DOI] [PubMed] [Google Scholar]
- 23.Redington AE, Madden J, Frew AJ, Djukanovic R, Roche WR, Holgate ST, Howarth PH. Transforming growth factor-beta 1 in asthma. Measurement in bronchoalveolar lavage fluid. Am J Respir Crit Care Med. 1997;156:642–647. doi: 10.1164/ajrccm.156.2.9605065. [DOI] [PubMed] [Google Scholar]
- 24.Wenzel SE, Schwartz LB, Langmack EL, Halliday JL, Trudeau JB, Gibbs RL, Chu HW. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am J Respir Crit Care Med. 1999;160:1001–1008. doi: 10.1164/ajrccm.160.3.9812110. [DOI] [PubMed] [Google Scholar]
- 25.Lemjabbar H, Gosset P, Lamblin C, Tillie I, Hartmann D, Wallaert B, Tonnel AB, Lafuma C. Contribution of 92 kDa gelatinase/type IV collagenase in bronchial inflammation during status asthmaticus. Am J Respir Crit Care Med. 1999;159:1298–1307. doi: 10.1164/ajrccm.159.4.9708080. [DOI] [PubMed] [Google Scholar]
- 26.Hoshino M, Nakamura Y, Sim J, Shimojo J, Isogai S. Bronchial subepithelial fibrosis and expression of matrix metalloproteinase-9 in asthmatic airway inflammation. J Allergy Clin Immunol. 1998;102:783–788. doi: 10.1016/s0091-6749(98)70018-1. [DOI] [PubMed] [Google Scholar]
- 27.Foley SC, Mogas AK, Olivenstein R, Fiset PO, Chakir J, Bourbeau J, Ernst P, Lemière C, Martin JG, Hamid Q. Increased expression of ADAM33 and ADAM8 with disease progression in asthma. J Allergy Clin Immunol. 2007;119:863–871. doi: 10.1016/j.jaci.2006.12.665. [DOI] [PubMed] [Google Scholar]
- 28.Phipps S, Benyahia F, Ou TT, Barkans J, Robinson DS, Kay AB. Acute allergen-induced airway remodeling in atopic asthma. Am J Respir Cell Mol Biol. 2004;31:626–632. doi: 10.1165/rcmb.2004-0193OC. [DOI] [PubMed] [Google Scholar]
- 29.Kariyawasam HH, Pegorier S, Barkans J, Xanthou G, Aizen M, Ying S, Kay AB, Lloyd CM, Robinson DS. Activin and transforming growth factor-beta signaling pathways are activated after allergen challenge in mild asthma. J Allergy Clin Immunol. 2009;124:454–462. doi: 10.1016/j.jaci.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly EA, Busse WW, Jarjour NN. Increased matrix metalloproteinase-9 in the airway after allergen challenge. Am J Respir Crit Care Med. 2000;162:1157–1161. doi: 10.1164/ajrccm.162.3.9908016. [DOI] [PubMed] [Google Scholar]
- 31.Cataldo DD, Tournoy KG, Vermaelen K, Munaut C, Foidart JM, Louis R, Noël A, Pauwels RA. Matrix metalloproteinase-9 deficiency impairs cellular infiltration and bronchial hyperresponsiveness during allergen-induced airway inflammation. Am J Pathol. 2002;161:491–498. doi: 10.1016/S0002-9440(10)64205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim DH, Cho JY, Miller M, McElwain K, McElwain S, Broide DH. Reduced peribronchial fibrosis in allergen-challenged MMP-9-deficient mice. Am J Physiol Lung Cell Mol Physiol. 2006;291:L265–L271. doi: 10.1152/ajplung.00305.2005. [DOI] [PubMed] [Google Scholar]
- 33.King NE, Zimmermann N, Pope SM, Fulkerson PC, Nikolaidis NM, Mishra A, Witte DP, Rothenberg ME. Expression and regulation of a disintegrin and metalloproteinase (ADAM) 8 in experimental asthma. Am J Respir Cell Mol Biol. 2004;31:257–265. doi: 10.1165/rcmb.2004-0026OC. [DOI] [PubMed] [Google Scholar]
- 34.Naus S, Blanchet MR, Gossens K, Zaph C, Bartsch JW, McNagny KM, Ziltener HJ. The metalloprotease-disintegrin ADAM8 is essential for the development of experimental asthma. Am J Respir Crit Care Med. 2010;181:1318–1328. doi: 10.1164/rccm.200909-1396OC. [DOI] [PubMed] [Google Scholar]
- 35.Le AV, Cho JY, Miller M, McElwain S, Golgotiu K, Broide DH. Inhibition of allergen-induced airway remodeling in Smad 3-deficient mice. J Immunol. 2007;178:7310–7316. doi: 10.4049/jimmunol.178.11.7310. [DOI] [PubMed] [Google Scholar]
- 36.McMillan SJ, Xanthou G, Lloyd CM. Manipulation of allergen-induced airway remodeling by treatment with anti-TGF-beta antibody: effect on the Smad signaling pathway. J Immunol. 2005;174:5774–5780. doi: 10.4049/jimmunol.174.9.5774. [DOI] [PubMed] [Google Scholar]
- 37.Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 38.Rankin JA, Picarella DE, Geba GP, Temann UA, Prasad B, DiCosmo B, Tarallo A, Stripp B, Whitsett J, Flavell RA. Phenotypic and physiologic characterization of transgenic mice expressing interleukin 4 in the lung: lymphocytic and eosinophilic inflammation without airway hyperreactivity. Proc Natl Acad Sci U S A. 1996;93:7821–7825. doi: 10.1073/pnas.93.15.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Norzila MZ, Fakes K, Henry RL, Simpson J, Gibson PG. Interleukin-8 secretion and neutrophil recruitment accompanies induced sputum eosinophil activation in children with acute asthma. Am J Respir Crit Care Med. 2000;161:769–774. doi: 10.1164/ajrccm.161.3.9809071. [DOI] [PubMed] [Google Scholar]
- 40.Jatakanon A, Uasuf C, Maziak W, Lim S, Chung KF, Barnes PJ. Neutrophilic inflammation in severe persistent asthma. Am J Respir Crit Care Med. 1999;160:1532–1539. doi: 10.1164/ajrccm.160.5.9806170. [DOI] [PubMed] [Google Scholar]
- 41.Ying S, O’Connor B, Ratoff J, Meng Q, Fang C, Cousins D, Zhang G, Gu S, Gao Z, Shamji B, Edwards MJ, Lee TH, Corrigan CJ. Expression and cellular provenance of thymic stromal lymphopoietin and chemokines in patients with severe asthma and chronic obstructive pulmonary disease. J Immunol. 2008;181:2790–2798. doi: 10.4049/jimmunol.181.4.2790. [DOI] [PubMed] [Google Scholar]
- 42.Bochner BS, Hudson SA, Xiao HQ, Liu MC. Release of both CCR4-active and CXCR3-active chemokines during human allergic pulmonary late-phase reactions. J Allergy Clin Immunol. 2003;112:930–934. doi: 10.1016/j.jaci.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 43.Medoff BD, Sauty A, Tager AM, Maclean JA, Smith RN, Mathew A, Dufour JH, Luster AD. IFN-gamma-inducible protein 10 (CXCL10) contributes to airway hyperreactivity and airway inflammation in a mouse model of asthma. J Immunol. 2002;168:5278–5286. doi: 10.4049/jimmunol.168.10.5278. [DOI] [PubMed] [Google Scholar]
- 44.Lin Y, Yan H, Xiao Y, Piao H, Xiang R, Jiang L, Chen H, Huang K, Guo Z, Zhou W, Lu B, Gao J. Attenuation of antigen-induced airway hyperresponsiveness and inflammation in CXCR3 knockout mice. Respir Res. 2011;12:123. doi: 10.1186/1465-9921-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burton OT, Oettgen HC. Beyond immediate hypersensitivity: evolving roles for IgE antibodies in immune homeostasis and allergic diseases. Immunol Rev. 2011;242:128–143. doi: 10.1111/j.1600-065X.2011.01024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Worgall TS, Veerappan A, Sung B, Kim BI, Weiner E, Bholah R, Silver RB, Jiang XC, Worgall S. Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity. Sci Transl Med. 2013;5:186ra67. doi: 10.1126/scitranslmed.3005765. Erratum in Sci Transl Med 5 192er7. [DOI] [PubMed] [Google Scholar]
- 48.Levy BD. Sphingolipids and susceptibility to asthma. N Engl J Med. 2013;369:976–978. doi: 10.1056/NEJMcibr1306864. [DOI] [PubMed] [Google Scholar]
- 49.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19:111–121. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 50.Carreras-Sureda A, Cantero-Recasens G, Rubio-Moscardo F, Kiefer K, Peinelt C, Niemeyer BA, Valverde MA, Vicente R. ORMDL3 modulates store-operated calcium entry and lymphocyte activation. Hum Mol Genet. 2013;22:519–530. doi: 10.1093/hmg/dds450. [DOI] [PubMed] [Google Scholar]
- 51.Ha SG, Ge XN, Bahaie NS, Kang BN, Rao A, Rao SP, Sriramarao P. ORMDL3 promotes eosinophil trafficking and activation via regulation of integrins and CD48. Nat Commun. 2013;4:2479. doi: 10.1038/ncomms3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malmström K, Pelkonen AS, Mäkelä MJ. Remodeling, inflammation and airway responsiveness in early childhood asthma. Curr Opin Allergy Clin Immunol. 2013;13:203–210. doi: 10.1097/ACI.0b013e32835e122c. [DOI] [PubMed] [Google Scholar]