Summary
Phosphatidylinositolphosphates (PIPs) are phospholipids that contain a phosphorylated inositol head group. PIPs represent a minor fraction of the total phospholipids, yet they are involved in many regulatory processes such as cell signalling and intracellular trafficking. Membrane compartments are enriched or depleted in specific PIPs, which constitute a signature for these compartments and contribute to their identity. The precise subcellular localisation and dynamics of most PIP species is not fully understood in plants. Here, we designed genetically encoded biosensors with distinct relative affinities and expressed them stably in Arabidopsis thaliana. Analysis of this multi-affinity “PIPline” marker set revealed previously unrecognized localisation for various PIPs in root epidermis. Notably, we found that PI(4,5)P2 is able to drive PIP2-interacting protein domains to the plasma membrane in non-stressed root epidermal cells. Our analysis further revealed that there is a gradient of PI4P, with the highest concentration at the plasma membrane, intermediate concentration in post-Golgi/endosomal compartments and lowest concentration in the Golgi. Finally, we also uncovered that there is a similar gradient of PI3P from high in late endosomes to low in the tonoplast. All together our library extends the palette of available PIP biosensors and should promote rapid progress in our understanding of PIP dynamics in plants.
Keywords: sensor, phosphoinositide, lipid binding domain, Arabidopsis thaliana, quantitative co-localisation, membrane trafficking, object-based analysis, lipid signalling, fluorescent protein, endosome
Introduction
Phosphatidylinositolphosphates (PIPs) are minor phospholipids, accounting less than a percent of total membrane lipids, yet they are of disproportionate importance for many membrane-associated signalling events: i) PIPs can be precursors of various second messengers (e.g. inositol 1,4,5-trisphosphate, diacylglycerol), ii) they can activate many ion channels and enzymes, iii) they can be involved in virtually all membrane trafficking events including endocytosis and exocytosis and, iv) they can recruit proteins to the plasma membrane (PM) or intracellular compartments through several structured interaction domains (e.g. Pleckstrin Homology domain (PH), Phox homology domain (PX), Fab1/YOTB/Vac1/EEA1 domain (FYVE)) (De Matteis and Godi, 2004; McLaughlin and Murray, 2005; Lemmon, 2008; Balla et al., 2009). PIPs can be phosphorylated at different positions of the inositol head group, which can generate up to seven different PIP species that include three phosphatidylinositol monophosphates [PI3P, PI4P and PI5P], three phosphatidylinositol biphosphate [PI(3,4)P2, PI(3,5)P2 and PI(4,5)P2] and one phosphatidylinositol triphosphate [PI(3,4,5)P3]. PIP kinases and phosphatases modify the phosphorylation state of the inositol head group, and phospholipases hydrolyse PIPs to release the soluble head group into the cytosol (Lemmon, 2008). The combined action of these enzymes produces the PIP signature of a cell, where certain membrane compartments are enriched or depleted of specific PIPs, contributing to their membrane identity (De Matteis and Godi, 2004; Lemmon, 2008; Balla et al., 2009; Balla, 2013).
The localisation of the various PIP species has been an intense area of research (De Matteis and Godi, 2004; Hammond et al., 2009a; Balla, 2013). Functional studies, together with biochemical and live-cell imaging, have built a relatively clear picture of the precise location of each PIP in cultured mammalian cell lines and in yeast. In animal cells, PI3P mainly resides in early endosomes, where it controls endosome maturation, cargo protein degradation/recycling and cell signalling notably through its interplay with Rab5 GTPases (Simonsen et al., 1998; Christoforidis et al., 1999; Jean and Kiger, 2012). PI4P is located in two different pools in the cell, one at the Golgi apparatus and the other one at the PM (Várnai and Balla, 2006; Hammond et al., 2009a). Each pool of PI4P has separate and diverse functions. The main function of PI4P at the Golgi is to control membrane trafficking events, in particular, the sorting of proteins toward the PM or endosomes (Szentpetery et al., 2010; Daboussi et al., 2012; Jean and Kiger, 2012). PI4P, together with other PIPs, recruits strong cationic proteins to the PM (Hammond et al., 2012). In yeast, the PM pool of PI4P also controls Endoplasmic Reticulum (ER)-to-PM tethering sites that regulate cell signalling and ER morphology (Stefan et al., 2011; Manford et al., 2012). Also, PM-localised PI4P is a source of PI(4,5)P2 (Szentpetery et al., 2010; Ling et al., 2012). PI5P accumulates in the nucleus and at the PM under certain stimuli (Gozani et al., 2003). PI(3,5)P2 is thought to reside in late endosomes, where it regulates lysome/vacuole biogenesis in yeast (Friant et al., 2003; Eugster et al., 2004). PI(4,5)P2 is localised at the PM where it has a large spectra of action such as anchoring signalling and membrane trafficking proteins (De Matteis and Godi, 2004; McLaughlin and Murray, 2005; Zoncu et al., 2007; Hammond et al., 2012; Balla et al., 2009; Balla, 2013). PI(4,5)P2 also controls ion channel activation and is a substrate of Phospholipase C, which triggers synthesis of the second messengers IP3 and DAG (McLaughlin and Murray, 2005; Suh et al., 2006). PI(4,5)P2 is the source of PI(3,4,5)P3, which together with PI(3,4)P2, accumulate at the PM but only when specific signalling pathways are activated (e.g. growth factor signalling) (McLaughlin and Murray, 2005; Balla, 2013). PI(3,4)P2 also controls late-stage clathrin-coated pit formation, independent of PI(3,4,5)P3 (Posor et al., 2013).
Much less is known about the function and localisation of PIPs in plants (Munnik and Vermeer, 2010; Munnik and Nielsen, 2011). The function of PIPs have been clearly established during polarized cell growth (e.g. tip growth of root hairs and pollen tubes), during membrane trafficking and response to stresses (Thole and Nielsen, 2008; Ischebeck et al., 2010; Munnik and Nielsen, 2011). Most of the enzymes involved in PIP metabolism are encoded in plant genomes, with the notable exception of type I- and type II- PI3-kinases (PI3Ks) (Munnik and Nielsen, 2011). These PI3Ks are able to phosphorylate PI4P and PI(4,5)P2 to produce PI(3,4)P2 and PI(3,4,5)P3 and their absence suggest that these two PIPs are not produced in plants. Congruent with these observations, they have never been found in plant extracts (Meijer and Munnik, 2003; Munnik and Nielsen, 2011). The localisations of PI3P, PI4P and PI(4,5)P2 have been studied using genetically encoded biosensors in various plant cell types including transgenic Arabidopsis (Vincent et al., 2005; Vermeer et al., 2006; van Leeuwen et al., 2007; Ischebeck et al., 2008; Thole et al., 2008; Mishkind et al., 2009; Vermeer et al., 2009; Ischebeck et al., 2011; Munnik and Nielsen, 2011). However, when data are available, only one marker per PIP species has been analysed. Here, we built a collection of transgenic Arabidopsis lines expressing various biosensors for each PIP species. This “PIPline” marker set allowed us to quantitatively analyse the localisation of various PIPs with respect to known compartment markers. Our collection provides a new toolbox to study PIP localisation and dynamics in the model plant Arabidopsis. We focused our analysis on the root epidermis but the PIPline collection should provide a new resource for the community to study PIPs in various cell types, developmental contexts or stress conditions.
Results
Generation of a set of transgenic marker lines that highlight PIPs associated with membrane compartments in Arabidopsis
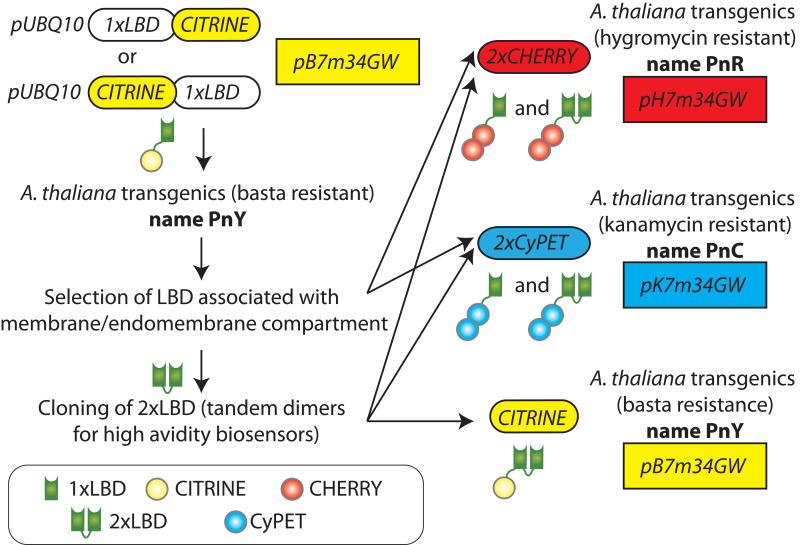
Genetically encoded biosensors have been extensively used to indirectly reveal the localisation and dynamics of PIPs in intact living cells (Várnai and Balla, 2006; Balla, 2013). These markers consist of lipid-binding domains (LBD) that interact specifically with known PIP species in vitro. These domains localise in the compartments of the cell that accumulate the targeted PIPs and can be easily traced when fused with a fluorescent protein. We built a collection of biosensors that include, when available, several independent domains for each of the seven PIPs. By using LBDs from different proteins and from different species, we hope to limit the effects of endogenous cellular proteins on the localisation of the marker. Furthermore, each domain is likely to have a different PIP binding affinity in vivo. For our set of marker lines, we chose only LBDs with extensive evidences of specific interactions with a given lipid in vitro (Table S1). Another potential pitfall of LBD over-expression is that it might titrate the targeted lipid and subsequently compromise the localisation and function of endogenous PIP effectors (Várnai and Balla, 2006; Balla et al., 2009). In order to limit potential over-expression problem, we drove the expression of our markers under the control of the promoter of the UBIQUITIN10 (UBQ10prom) gene (Figure 1). Compared to the strong 35S promoter, UBQ10prom provides a mild uniform expression pattern and this endogenous intron-bearing promoter limits the problems of silencing and mosaic expression often observed with the viral 35S promoter (Geldner et al., 2009). In order to obtain high contrast fluorescence with biosensors expressed at relatively low level, we counterbalanced the use of the mild UBQ10 promoter, by fusing the LBDs with CITRINE, a brighter and monomeric version of the Yellow Fluorescent Protein (YFP) (Heikal et al., 2000; Jaillais et al., 2011) (Figure 1).
Figure 1. Strategy for the generation of the “PIPline” collection.
All PIP biosensor constructs (hereafter referred to as “PIPline”) were cloned into multisite gateway destination vectors (pB7m34GW, pH7m34GW and pK7m34GW). All the PIPlines are expressed under the control of the mild constitutive UBQ10 promoter. Each PIPline has been ascribed a number (n). Yellow PIPlines are named PnY, red PIPline PnR, and cyan PIPline PnC. 1xLBD means that only one LDB is fused to the fluorescent protein; 2xLBD means that two identical LBDs are fused in tandem dimer with the fluorescent protein.
Next, we transformed Arabidopsis (Columbia accession) with each of the created biosensor constructs (see Table S1 for a list of all the 17 LBDs used in this study). None of the selected transgenic lines harboured any visible developmental phenotypes, suggesting that the mild ubiquitous expression of each LBD is not deleterious for the plants and likely does not extensively compete with endogenous proteins. Figure S1 shows the localisation of each marker that was sufficiently stable for observation by confocal microscopy (13 LBDs out of 17). For all our subsequent analyses, we decided to keep only the LBDs that interacted with membranes, either the PM, intracellular compartments, or both (Figure S1), as a soluble localisation is a default localisation in the absence of any targeting.
With the exception of the PH domain of OSBP, each of these LBDs have been extensively studied for their PIP binding properties in vitro by at least 4 distinct techniques (Table 1). The PH domain of OSBP is the domain that has been less characterized in vitro, with only liposome-binding assay and Surface Plasmon Reasonance (SPR) experiments (Levine and Munro, 2002). Therefore, we verified independently the specificity of this PI4P probe by a protein-lipid overlay experiment. For this purpose, we developed an assay to directly test the PIP binding properties of our fluorescently tagged PHOSBP produced in transgenic Arabidopsis. In this assay, we found that CITRINE-PHOSBP is binding preferentially to PI4P as well as PI(3,4)P2 (Figure S2). As discussed above, PI(3,4)P2 has never been found in plants, which is very likely due to the lack of type I and type II PI3Ks (Meijer and Munnik, 2003; Munnik and Nielsen, 2011). Therefore, PHOSBP should be a bona fide PI4P reporter in plants. Similarly, the C-terminal domain of the TUBBY protein (TUBBY-C) has been shown to bind to PI(4,5)P2, PI(3,4)P2 and PI(3,4,5)P3 in vitro (Santagata et al., 2001). Because PI(3,4)P2 and PI(3,4,5)P3 are not synthesized in plants, TUBBY-C, like PHPLC, is a reporter for PI(4,5)P2 in planta.
Table 1. In vitro and in vivo PIP binding specificities of LBDs used in the PIPline collection.
| Domain | In vitro interaction | In vitro techniques | Kd | In vivo localizatio : yeast | In vivo localization animal cells | Evidences for in vivo binding (yeast/animal) | References |
|---|---|---|---|---|---|---|---|
| 1xFYVEHRS | PI3P | liposomes, fat blot, SPR, crystalography, monolayer penetration analysis | ≈0.5μM | endosome / vacuole | mostly cytosolic | Dependent on type III PI3K (VPS34), sensitive to PI3K inhibitor (e.g. Wm) | Burd et al., 1998; Gillooly et al., 2000; Misra and Hurley 2000; Sankaran et al., 2001; Raiborg et al., 2001; Stahelin et al., 2002; He et al., 2009 |
| 1xPXp40 | PI3P | liposomes, fat blot, SPR, ITC, crystalography, monolayer penetration analysis | ≈5μM | endosome / vacuole | early endosome | Dependent on type III PI3K (VPS34), sensitive to PI3K inhibitor (e.g. Wm) | Ago et al., 2001; Kanai et al., 2001; Bravo et al., 2001; Ellson et al., 2001; Zhan et al., 2002, Malkova et al. 2006 |
| 1xPHFAPP1 | PI4P | liposomes, fat blot, SPR, crystalography, monolayer penetration analysis, NMR spectroscopy | ≈18.6 μM | golgi | golgi | Dependent on PI4K (Pik1 in yeast, PI4K-IIIa in human), in vivo protection of PI4P, expression mask binding of anti-PI4P antibody, sensitive to PI4K inhibitor (e.g. PAO), sensitive to PI4-pptases (e.g. SAC1, Inp54) | Dowler et al., 2001; Levine et al., 2002; Godi et al. 2004; Balla et al., 2005; Hammond et al., 2009a, Lenoir et al., 2010; He at al., 2011 |
| 1xPHOSBP | PI4P | fat blot (this study), liposomes, SPR | ≈3.5μM | golgi | golgi | Dependent on PI4K (Pik1 in yeast, PI4K-IIIa in human), sensitive to PI4K inhibitor (e.g. PAO), sensitive to PI4-pptases (e.g. SAC1, Inp54) | Levine et al., 1998; Levine et al., 2002; Balla et al., 2005; Niu et al., 2013 |
| 1xPHPLC | PI(4,5)P2 | fat blot (this study), liposomes, SPR, crystalography, NMR spectroscopy | ≈1μM | mostly cytosolic | PM | localization sensitive to PLC inhibitor and activator sensitive to 5-pptases (e.g. INPP5B, OCRL), dependent on PI4P-5Kinase (e.g. mss4), expression mask binding to anti-PI(4,5)P2 antibody | Ferguson et al., 1995; Lemmon et al., 1995; Levine et al., 2002; Varnai et al., 2002; Tuzi et al., 2003; Yu et al., 2004; Szentpetery et al., 2005; Zoncu et al., 2007 Hammond et al., 2009a; Hammond et al., 2009b |
| 1xTUBBY-C | PI(4,5)P2, PI(3,4)P2, PI(3,4,5)P3 | fat blot (this study), liposomes, SPR, crystalography | nd | PM (this study) | PM | localization sensitive to PLC inhibitor and activator, sensitive to 5-pptases (e.g. INPP5B, OCRL) | Santagata et al., 2001; Quinn et al., 2008; Szentpetery et al., 2009; Hammond et al., 2012 |
Abbreviations: PI3K (PI3-kinase); PI4K (PI4-kinase); pptase (phosphatase); SPR (Surface Plasmon Resonance); ITC (Isothermal Titration Calorimetry), NMR (Nuclear Magnetic Resonance).
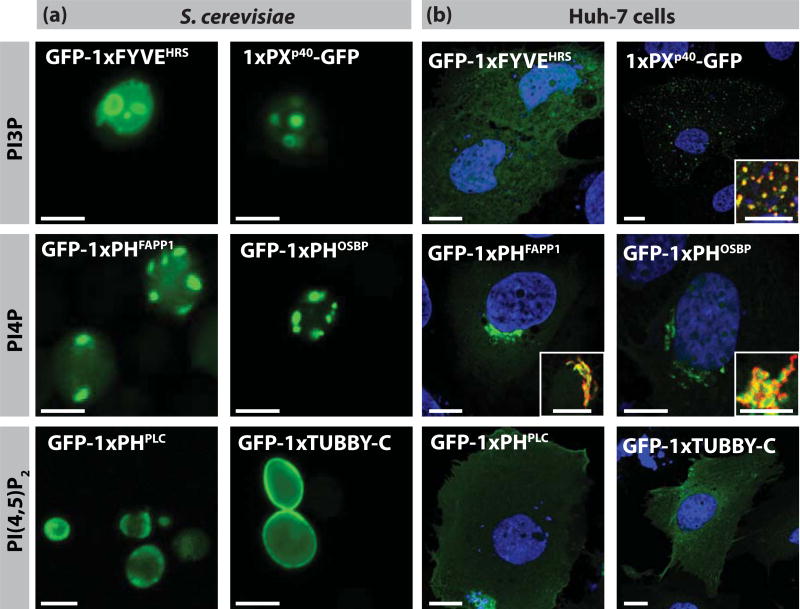
Although we systematically designed our LBD constructs based on previously published data, our sensors might differ by few amino acids at their N- and C-termini. Therefore, we decided to validate our fluorescently-labelled LBDs by expressing them in yeast and human cell lines, two systems in which the localisation of these domains have already been studied and/or the localisation of each PIP is extensively validated (Table 1). These experiments showed that our LBDs behave as previously described constructs both in Saccharomyces cerevisiae (Figure 2a) and the human hepatocarcinoma cell line, Huh-7 (Figure 2b). PI3P probes (1xFYVEHRS and 1xp40PX) were localised to endosomes and vacuole in yeast (Burd and Emr, 1998) and to early endosomes in Huh-7 cells; except for 1xFYVEHRS which was mostly diffuse in the cytoplasm in human cells in agreement with previous report (Gillooly et al., 2000). PI4P markers (1xPHFAPP1 and 1xPHOSBP) were localised in the Golgi apparatus in both systems (Levine and Munro, 2002; A., Balla et al., 2005) and PI(4,5)P2 sensors (1xPHPLC and 1xTUBBY-C) were localised at the PM in both system (Szentpetery et al., 2009); except 1xPHPLC which accumulated mostly in the cytoplasm in yeast (Levine and Munro, 2002; Yu et al., 2004)(Figure 2).
Figure 2. Localisation of the LBD used in this study in yeast and human cells.
Confocal pictures of S. cerevisiae (a) and human hepatocarcinoma cell line Huh-7 (b) expressing GFP-tagged LBD. Inset in (b) are immuno-localisation showing that 1xPXp40 (green) co-localises with the early endosome marker EEA1 (red) and that 1xPHFAPP1/1xPHOSBP (green) co-localise with the Golgi marker GM130 (red). Blue: Hoechst-stained nuclei. Scale bars 5 μm.
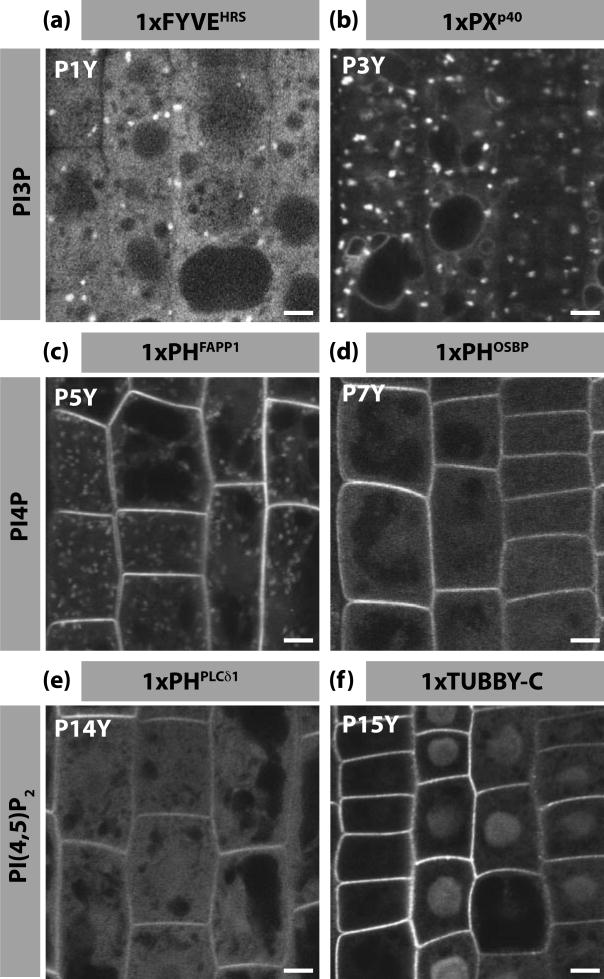
In A. thaliana root, LBDs that bind to the same lipid did not always exhibit exactly overlapping localisation domains. For example, the PI3P sensor 1xFYVEHRS was mainly cytosolic and weakly associated with intracellular compartments (Figure 3a), while 1xPXp40 was localised in intracellular compartments as well as weakly in the tonoplast (Figure 3b). Furthermore, the PI4P biosensor 1xPHFAPP1 was localised at the PM and intracellular compartments (Figure 3c), while 1xPHOSBP was more restricted to the PM (Figure 3d). Finally, both PI(4,5)P2 sensors (1xPHPLC and 1xTUBBY-C) were localised at the PM, although 1xPHPLC was also localised in the cytosol and 1xTUBBY-C in the nucleus (Figure 3e and f). These slight differences in localisation of LBDs that bind the same lipid might be due to various parameters such as differences in binding affinities (Table 1), expression level, protein stability, local pH, local electrostatic potential of the membrane, the protein affinity for a given membrane curvature or the need to bind to other cellular co-factors. Overall, these results highlight the need to use multiple independent biosensors for each PIP in order to have a more complete and dynamic view of PIP cellular localisation.
Figure 3. Localisation of PI3P, PI4P and PI(4,5)P2 in Arabidopsis root epidermis.
(a-f) Confocal pictures of Arabidopsis root epidermal cells expressing various CITRINE-tagged LBDs: (a) CITRINE-1xFYVEHRS, (b) 1xPXp40-CITRINE, (c) CITRINE-1xPHFAPP1, (d) CITRINE-1xPHOSBP, (e) CITRINE-1xPHPLC, (f) CITRINE-1xTUBBY-C. The respective PIPline name is indicated in the top left corner. Scale bars 5 μm.
Engineering of biosensors with different affinities for their cognate lipids
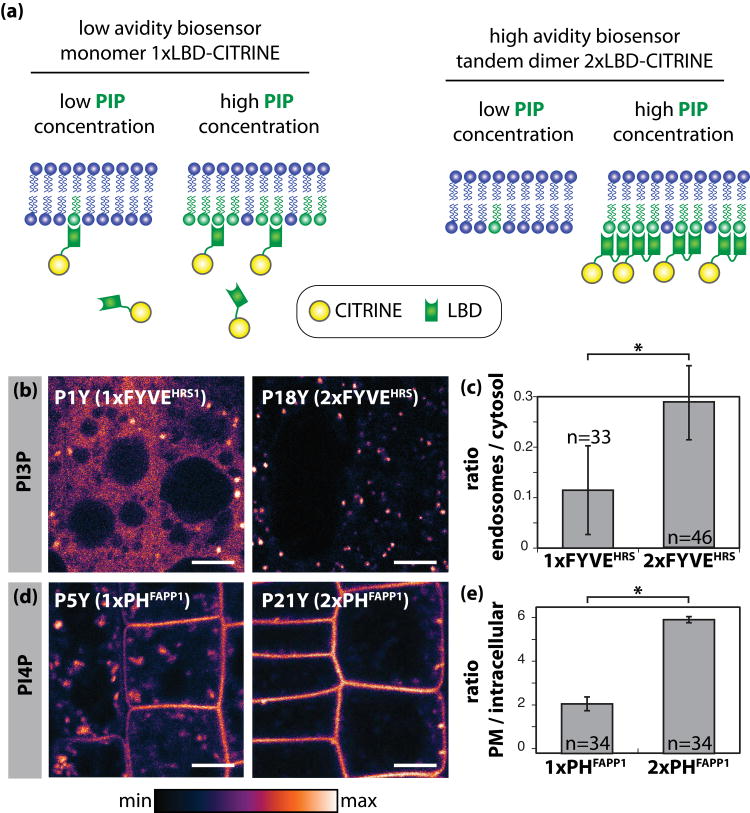
Most cellular proteins are not localised only by one membrane interacting domain (Lemmon, 2008). It is often the combination of several LBDs or the joint action of a LBD with a lipid anchor or transmembrane segment that drive the protein to its final location (Lemmon, 2008). It is well established that this bipartite lipid recognition is key in generating membrane specificity and/or extended residence time at the target membrane (Schultz, 2010). Therefore, in order to create high avidity biosensors (avidity being the combined strength of multiple bond interactions), we fused in tandem dimers the LBDs previously identified as interacting with membranes (Figure 1 and 4a). This strategy has previously been used to increase the binding avidity of several lipid biosensors (Gillooly et al., 2000; Roy and Levine, 2004; Godi et al., 2004). Increasing the relative avidity of a lipid sensor might have two effects on its localisation: i) it increases the proportion of membrane-bound sensor and ii) it preferentially targets the high avidity sensors toward membranes that are the most enriched with their lipid partners, because it increases its residence time at that particular membrane (Lemmon, 2008; Schultz, 2010). Low avidity sensors are less efficient in discriminating between two membranes with two different concentrations of their targeted PIP and they might highlight several pools of this PIP within the cell. By contrast, high avidity sensors will have increased residence time at the membrane that is the most enriched in the targeted PIP and they might therefore reveal variation in PIP concentration within the cell. In other word, high avidity sensors work like Velcro: they will grab more strongly to a surface with more spikes (in this case the spikes being PIPs) (Figure 4a).
Figure 4. Engineering and analysis of low and high avidity PI3P and PI4P biosensors in Arabidopsis root epidermal cells.
(a) Schematic representation of the strategy used to obtain low and high avidity PIP biosensors. (b) Confocal pictures of Arabidopsis root epidermal cells expressing CITRINE-tagged 1xFYVEHRS and 2xFYVEHRS. (c) Graph representation of the ratio of 1xFYVEHRS (P1Y) and 2xFYVEHRS (P18Y) endosomal signal relative to the levels of cytosolic signal. (d) Confocal pictures of Arabidopsis root epidermal cells expressing CITRINE-tagged 1xPHFAPP1 and 2xPHFAPP1. (e) Graph representation of the ratio of 1xPHFAPP1 (P5Y) and 2xPHFAPP1 (P21Y) at the PM relative to the intracellular levels. Confocal pictures are colour-coded in pixel intensity following the LUT scale shown at the bottom. Scale bars 5 μm. Error bars represent standard deviation (s.d.). Asterisk mark: statistical difference (p<0.05) according to Student's t-test. n is the number of cells used in each quantitative analysis.
Using SPR experiments, Gillooly et al. reported that 1xFYVE showed dissociation kinetics characteristic of a 1:1 binding, while 2xFYVE showed complex association / dissociation kinetics that could be fit into a bivalent model (Gillooly et al., 2000). Therefore, the ability of one 2xFYVE molecule to interact with two molecules of PI3P likely explain its superior PI3P binding compared with 1xFYVE. This observation was further verified in vivo in human cells as the 2xFYVE probe strongly localises to early endosomes while 1xFYVE localisation is mostly diffuse in the cytosol (Gillooly et al., 2000). In Arabidopsis root, the low avidity sensor (1xFYVE) was largely cytosolic and only weakly associated with intracellular compartments (Figure 4b and c). By contrast, the high avidity sensor (2xFYVE) was more strongly associated with intracellular compartments (Figure 4b and c) and from time to time also localised to the tonoplast. This result was confirmed independently by the localisation of the PXp40 PI3P sensor that also localised in intracellular compartments (Figure 3b).
The comparison of 1x and 2xPHFAPP1 previously suggested that 2xPHFAPP1 has a stronger PI4P binding in vitro that 1xPHFAPP1 (Godi et al., 2004). In Arabidopsis roots, we found that the high avidity PI4P sensor 2xPHFAPP1 was more strongly localised to the PM and less to endomembrane compartments than the low avidity sensor 1xPHFAPP1 (Figure 4d and e). As explained above and illustrated in Figure 4a, these results suggest that the concentration of PI4P is greater at the PM than in intracellular compartments. Together with the observation that 1xPHOSBP localises almost exclusively to the PM (Figure 3d), our results establish that PI4P accumulates primarily at the PM and, to a lesser extent, to one or various intracellular compartments.
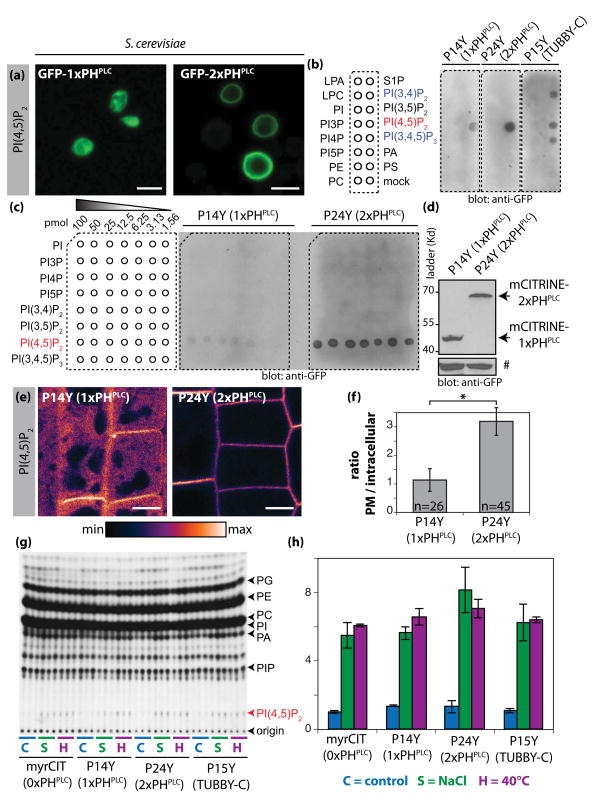
Next, we investigated the properties and localisation of the PI(4,5)P2 probes 1x and 2xPHPLC. In both yeast and human cell lines, the 2xPHPLC fusion protein is extensively targeted to the PM, while 1xPHPLC is more cytosolic (Levine and Munro, 2002; Hammond et al., 2009b). We could not find in the literature any in vitro characterisation that compared 1x and 2xPHPLC binding to PI(4,5)P2. Therefore, in order to validate our constructs, we first verified whether they behaved as expected when expressed in yeast (Table 1). In agreement with previous report (Levine and Munro, 2002), we found that 1xPHPLC is mainly cytosolic, while 2xPHPLC is specifically targeted to the PM when expressed in S. cerevisiae (Figure 5a). Next, we validated that both probes are exquisitely specific for PI(4,5)P2 in protein-lipid overlay assay, as they do not interact with any other lipids (Figure 5b). Using a similar assay, we verified that the TUBBY-C domain interacted in vitro with PI(4,5)P2, PI(3,4)P2 and PI(3,4,5)P3 as previously reported (Santagata et al., 2001) (Figure 5b).
Figure 5. PI(4,5)P2 is able to drive PIP2-interacting domains to the PM in non-stressed root epidermal cells.
(a) Confocal pictures of S. cerevisiae expressing GFP-1xPHPLC (left) and GFP-2xPHPLC (right). Scale bars 5 μm. (b) Protein-lipid overlay assay with CITRINE-1xPHPLC (left), CITRINE-2xPHPLC (middle) and CITRINE-TUBBY-C (right) proteins extracted from P14Y, P15Y and P24Y transgenic plants. The position of each lipid is indicated on the map on the left panel. (c) Protein-lipid overlay assay with the same quantities of CITRINE-1xPHPLC (left), CITRINE-2xPHPLC (right) extracted from P14Y and P24Y transgenic lines. The position and quantity of each lipid is indicated on the map on the left panel. (d) Western blot showing similar expression level of transgenic proteins. The non-specific band indicated by a sharp sign serves as a loading control. (e) Confocal pictures of Arabidopsis root epidermal cells expressing CITRINE-tagged 1xPHPLC and 2xPHPLC. Confocal pictures are colour-coded in pixel intensity following the LUT scale shown at the bottom. Scale bars 5 μm. (f) Graph representing the ratio of 1xPHPLC (P14Y) and 2xPHPLC (P24Y) at the PM relative to the intracellular signal. Error bars represent standard deviation (s.d.). Asterisk mark indicates statistical difference (p<0.05) according to Student's t-test. n is the number of cells used in each quantitative analysis. (g) Alkaline TLC profile of Arabidopsis seedlings labelled for 16H with 32Pi and then incubated for 30 min at: 22°C in control buffer (C = control, blue), 22°C in control buffer supplemented with 250mM NaCl (S = Salt, green) or 40°C in control buffer (H = Heat, purple). Each lane is a pool of 3 seedlings and each condition was analysed in triplicate using the following genotypes: myristoylated 2xCITRINE (myrCIT) as a non-PIP2 binding control (0xPHPLC), P14Y (CITRINE-1xPHPLC), P24Y (CITRINE-2xPHPLC) and P15Y (CITRINE-TUBBY-C). An autoradiograph of a typical experiment is shown. (h) Quantification of PIP2 levels by densitometry of the autoradiograph shown in (g). The fold change was calculated relative to levels of PI(4,5)P2 present in myrCIT (0xPHPLC) in the control condition from two independent experiments.
Furthermore, we found that when using protein extract containing comparable quantity of sensors, CITRINE-2xPHPLC binds more tightly to PI(4,5)P2 than CITRINE-1xPHPLC in protein-lipid overlay assay (Figure 5c and d). Next, we compared the subcellular localisation of the low (1xPHPLC) and high (2xPHPLC) avidity PI(4,5)P2 sensors in Arabidopsis roots. Although 1xPHPLC was mainly cytoplasmic, it also showed a clear PM localisation in non-dividing/non-stressed root epidermal cells (Figure 5e and f). In contrast, the 2xPHPLC reporter was almost exclusively localised to the PM (Figure 5e and f). This observation was independently confirmed by the extensive PM localisation of the 1xTUBBY-C domain (Figure 3f). These results were surprising because in absence of stresses, PI(4,5)P2 levels are known to be very low in plants (Munnik and Nielsen, 2011). This suggested either that i) PI(4,5)P2 is able to drive proteins to the PM in non-stressed epidermal cells or ii) that expression of our biosensors up-regulated PI(4,5)P2 metabolism to maintain the amount of free PI(4,5)P2. Such feedback mechanism has been previously observed in tobacco BY-2 cells stably expressing YFP-2xFYVE (Vermeer et al., 2006) and might explain why we did not observed any phenotypes in our transgenic lines. In order to discriminate between these two possibilities, we measured the quantities of the various phospholipid species in transgenic lines expressing CITRINE-1xPHPLC, CITRINE-2xPHPLC, CITRINE-TUBBY-C and a myristoylated CITRINE (myrCIT) as a non-PIP binding control (Jaillais et al., 2011). As shown in Figure 5g and 5h, no significant differences between the four genotypes in term of lipid species quantity was found, nor in their response to salt or heat stress, which both trigger a rapid PIP2 response (Figure 5g and h) ( van Leeuwen et al., 2007; Mishkind et al., 2009). Altogether, our results suggest that the local concentration of PI(4,5)P2 at the PM is sufficient to drive PIP2-interacting proteins to this compartment in non-stressed root epidermal cells.
A multi-colour marker set for rapid co-localisation with other PIP biosensors and known membrane compartment markers
Next, all the LBDs that were associated with membranes (including both 1x and 2x versions, 9 LBDs in total, highlighted in red in Table S1) were further engineered as fusion proteins with additional fluorescent proteins, allowing their rapid combinatorial analysis in Arabidopsis. Cyan Wave lines used the CERULEAN fluorescent protein and had extremely weak fluorescence (Geldner et al., 2009). We therefore decided to use a brighter cyan fluorescent protein, CyPet (Nguyen and Daugherty, 2005), fused in tandem dimer in order to further increase its brightness. Unfortunately, this strategy resulted in finding only four marker lines that exhibited sufficient level of fluorescence for confocal microscopy detection, although they were still very weak (Figure S3). In parallel, we fused the nine LBDs with a tandem dimer of the monomeric red fluorescent protein CHERRY (2xCHERRY) (Shaner et al., 2004). Each CHERRY marker exhibited good fluorescence and had a similar cellular localisation to those of CITRINE lines (Figure S3). In reference to the name of the Wave line collection (Geldner et al., 2009), we named our PIP biosensor set, the “PIPlines” (PnY for the CITRINE lines, PnR for the CHERRY lines and PnC for the CyPET lines, Figure 1 and Table S4). All DNA constructs and transgenic lines will be deposited in the stock centre for fast distribution of the PIPline collection.
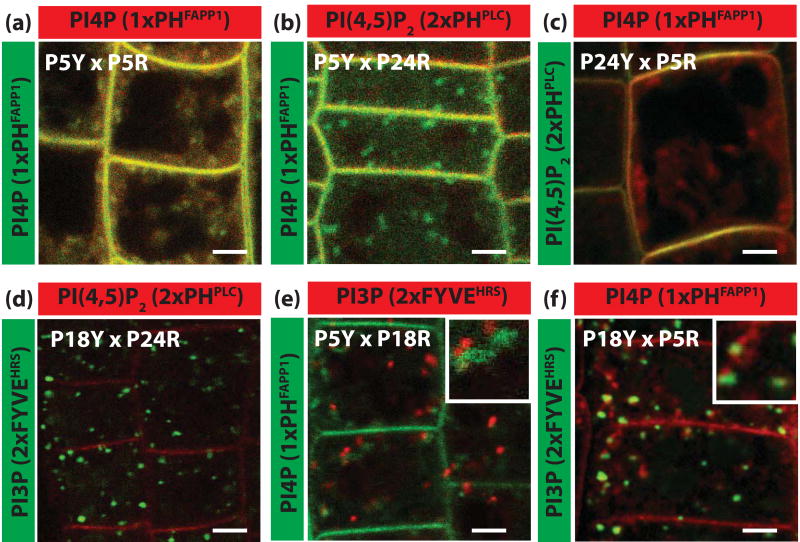
As a proof of concept that our PIPlines are suitable for co-localisation analyses, we crossed yellow and red biosensors for PI3P (2xFYVEHRS), PI4P (1xPHFAPP1) and PI(4,5)P2 (2xPHPLC) (Figure 6). These crosses provide an additional resource to visualize the localisation of two different PIP species simultaneously in planta. As expected, we found extensive co-localisation when the same PIP was highlighted with both yellow and red biosensors (Figure 6a). Since the sequence and structure of CITRINE and CHERRY are distinct, these results rule out the potential non-specific targeting of the sensors by the fluorescent proteins. Moreover, we detected co-localisation at the PM between PI4P and PI(4,5)P2 biosensors (Figure 6b and c) and no co-localisation between PI3P and PI(4,5)P2 biosensors, that localise in intracellular compartments and the PM respectively (Figure 4d). We also detected very limited co-localisation between PI3P and PI4P biosensors both of which are found in intracellular compartments (Figure 6e and f). These results suggest, similarly to tobacco BY2 cells (Vermeer et al., 2009), that these PIP species largely accumulate in different compartments in Arabidopsis epidermal cells. These results are in accordance with the notion that the PIP composition of a given compartment represents a unique signature marking the identity for this organelle (De Matteis and Godi, 2004; Munro, 2004; Lemmon, 2008) and raise the question of the identity of these compartments in the Arabidopsis root epidermis.
Figure 6. Simultaneous labelling of two PIP species in Arabidopsis root epidermis.
(a-f) Confocal pictures of root epidermal cells co-expressing one CITRINE- and one CHERRY-tagged PIPline. Each image is an overlay of the green channel (CITRINE) and red channel (CHERRY), co-localisation being visualised by the yellow colour. (a) CITRINE-1xPHFAPP1 x 2xCHERRY-1xPHFAPP1, (b) CITRINE-1xPHFAPP1 x 2xCHERRY-2xPHPLC, (c) CITRINE-2xPHPLC x 2xCHERRY-1xPHFAPP1, (d) CITRINE-2xFYVEHRS x 2xCHERRY-2xPHPLC, (e) CITRINE-1xPHFAPP1 x 2xCHERRY-2xFYVEHRS, (f) CITRINE-2xFYVEHRS x 2xCHERRY-1xPHFAPP1. The names of the PIPlines used in each cross are indicated at the top and left of each panel. Scale bars 5 μm.
PI3P localises to late endosomes/PVC in Arabidopsis root epidermis
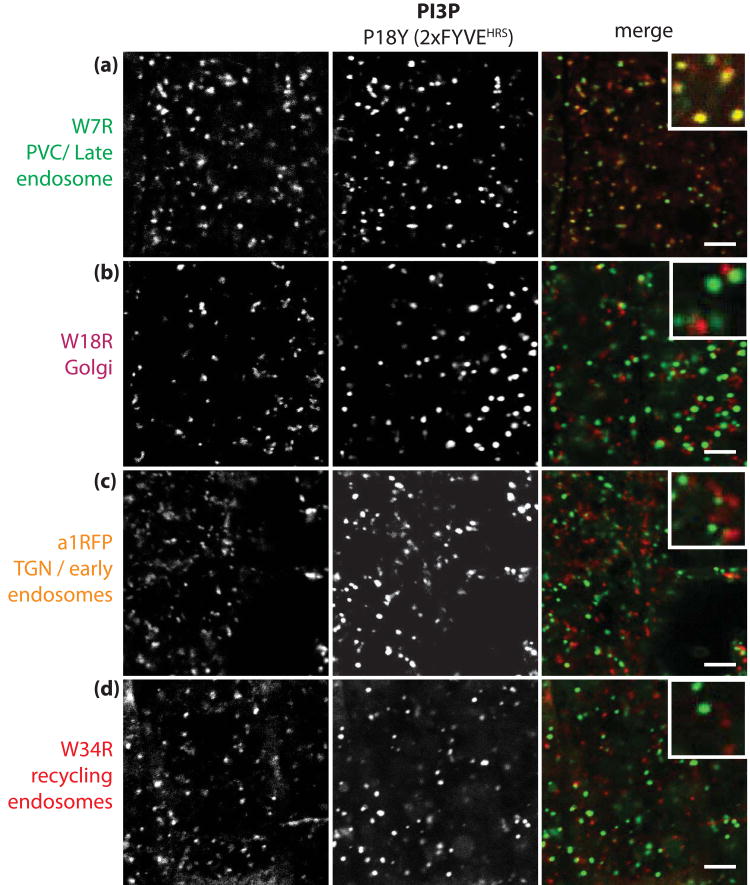
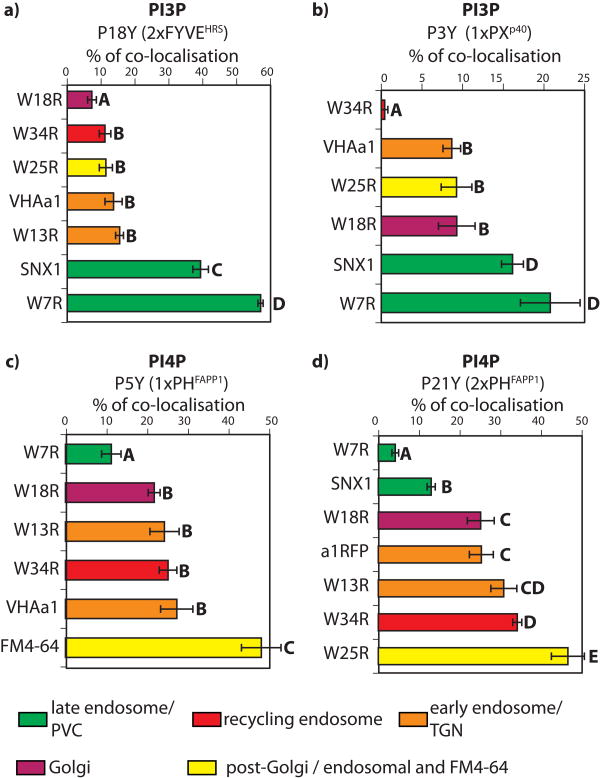
In animal cells, PI3P mainly resides in early endosomes (Table 1, Figure 2b) (De Matteis and Godi, 2004; Lemmon, 2008). The subcellular localisation of PI3P has been previously analysed in tobacco BY2 cells using a 2xFYVE reporter (Vermeer et al., 2006). In this system, PI3P was found to accumulate in late endosomes/PVC rather than Golgi bodies (Vermeer et al., 2006). The fact that our PXp40 sensor localises to the tonoplast suggests that PI3P might also accumulate in a late endosomal compartment in Arabidopsis roots. To verify this hypothesis, we crossed the P18Y (CITRINE-2xFYVEHRS) and P3Y (PXp40-CITRINE) lines with transgenic lines expressing various red fluorescent organelle markers (Dettmer et al., 2006; Jaillais et al., 2006; Geldner et al., 2009). In the F2 generation, we analysed the co-localisation between our PI3P biosensors and these compartment markers, qualitatively (Figure 7 and Figure S4) and quantitatively (Figure 8), using an object-based analysis. We determined that 2xFYVEHRS co-localises extensively with markers of the late endosomes/PVC (Figure 7a and 8a). Similarly, 1xPXp40 also co-localises preferentially with late endosomal markers (Figure 8b and S4); although to a lesser extent, due to its additional localisation to the tonoplast (Figure S4). Altogether, our result showed that PI3P, like in tobacco BY-2 cells (Vermeer et al., 2006), mainly accumulate in late endosomes/PVC and to a lesser degree at the tonoplast.
Figure 7. CITRINE-2xFYVEHRS localises to late endosomes in Arabidopsis root epidermis.
(a-d) Confocal pictures of root epidermal cells co-expressing CITRINE-2xFYVEHRS with intracellular compartment markers fused with a red fluorescent protein: (a) W7R (late endosomes/PVC), (b) W18R (Golgi apparatus), (c) VHAa1-RFP (early endosomes/TGN) and (d) W34R (recycling endosomes). Left pictures correspond to the compartment markers, middle pictures correspond to CITRINE-2xFYVEHRS (both depicted in grey scale for increased contrast), while the right pictures correspond to the overlay of both channels with the compartment markers in red and the 2xFYVEHRS sensor in green. Scale bars 5 μm.
Figure 8. Quantitative analysis of intra-cellular co-localisations.
Quantitative co-localisation data obtained by object-based analysis between various compartment markers and 2xFYVEHRS (a), 1xPXp40 (b), 1xPHFAPP1 (c), 2xPHFAPP1 (d). Error bars represent standard deviation. Bold capital letters indicate statistical difference (p<0.05) according to Steel-Dwass-Critchlow-Fligner bilateral test. Co-localisations were quantified in 30 cells per conditions only on intra-cellular signals (i.e. excluding the PM).
PI4P accumulates in endosomal compartments in Arabidopsis root epidermis
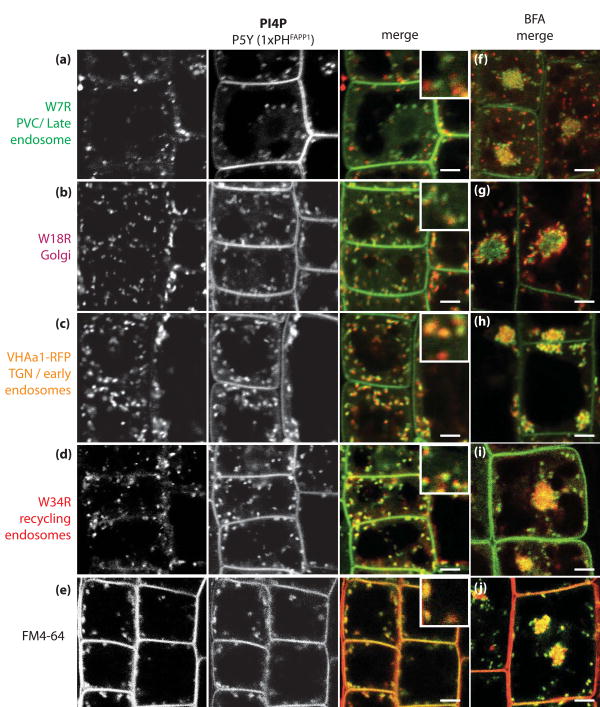
Next, we conducted a similar qualitative (Figure 9 and Fig S5) and quantitative (Figure 8) co-localisation analysis for our PI4P sensors in order to determine in which intracellular compartments they localise. Our quantitative analyses revealed that both 1xPHFAPP1 and 2xPHFAPP1 localised to early endosomes/TGN and recycling endosomes (collectively referred to as post-Golgi/endosomal compartments by Geldner et al., (2009), about 45% of co-localisation) as well as to the Golgi apparatus (Figure 8). In order to confirm that PI4P resides in a post-Golgi/endosomal compartment in Arabidopsis epidermal cells, we performed a co-localisation experiment with the endocytic tracer FM4-64 (Figure 9e and Figure 8c). FM4-64 is a vital dye that fluoresces in a lipophilic environment and cannot pass through membranes. It can therefore enter inside the cell only by endocytosis, where it labels endosomes. We found a good co-localisation between FM4-64 and the CITRINE-1xPHFAPP1 marker (about 48% of co-localisation) (Figure 8), further confirming that PI4P accumulates significantly in a post-Golgi/endosomal compartment (Figure 9e). As a third approach, we also performed treatment with the fungal toxin Brefeldin A (BFA) (Figure 9f-j). BFA allows for a good discrimination between intracellular compartments of root epidermal cells, notably between Golgi and post-Golgi compartments that segregate around and inside the “BFA compartment”, respectively (Grebe et al., 2003; Jaillais et al., 2008; Geldner et al., 2009). In accordance with our quantitative co-localisation analysis, a significant proportion of CITRINE-1xPHFAPP1-labelled compartments were found to reside at the heart of the BFA compartment, together with FM4-64 as well as markers of early endosomes/TGN and recycling endosomes (Figure 9h-j). On the other hand, BFA largely dissociated the PI4P sensor from the Golgi that was surrounding the CITRINE-1xPHFAPP1-labelled BFA compartment (Figure 9g). Furthermore, we noticed that a significant proportion of CITRINE-1xPHFAPP1-labelled compartments were resistant to BFA (Figure 9j). These BFA-insensitive PI4P-containing compartments mainly co-localised with early endosomes/TGN marker that is also partially insensitive to BFA (Figure 9h and (Geldner et al., 2009)). Altogether, our results showed that in Arabidopsis root epidermis, PI4P localises at the PM and to one or possibly several post-Golgi/endosomal compartments (early endosomes/TGN and recycling endosomes) and to a lesser extent, the Golgi apparatus.
Figure 9. Intra-cellular CITRINE-1xPHFAPP1 localises to post-golgi/endosomal compartments in Arabidopsis root epidermis.
(a-d) Confocal pictures of root epidermal cells co-expressing CITRINE-1xPHFAPP1 with intracellular compartment markers fused with a red fluorescent protein: (a) W7R (late endosomes/PVC), (b) W18R (Golgi apparatus), (c) VHAa1-RFP (early endosomes/TGN) and (d) W34R (recycling endosomes). (e) Co-localisation with red endocytic tracer FM4-64. Left pictures correspond to the compartment markers (a-d) or FM6-64 (e), middle picture correspond to CITRINE-1xPHFAPP1 (both depicted in grey scale for increased contrast), while the two right columns of pictures correspond to the overlay of both channels with the compartment markers in red and the 1xPHFAPP1 sensor in green. (f-j) Co-localisation between CITRINE-1xPHFAPP1 and the corresponding compartment markers in the presence of BFA at 25 μM for 1 hour. Scale bars 5 μm.
Discussion
The PIPline collection as a tool to dissect PIP function in Arabidopsis
Our set of PIP marker lines provides a comprehensive collection to study the localisation and dynamics of distinct PIP species in Arabidopsis thaliana. The PIPline marker set implements several new features over the already existing PIP reporters in Arabidopsis. First, we systematically engineered reporters with various avidities for each PIP species. The comparison of their respective localisation is indicative of the relative concentration of PIP in different cellular compartments. Second, our PIPline collection is multi-coloured, which allows for the co-labelling of several PIP species at the same time as well as their fast co-localisation with already established markers independent of their colour. Third, although we restricted our co-localisation analysis to root epidermal tissue, we used a broadly expressed promoter that will enable the study of plant PIPs in a variety of tissues and developmental contexts. Thus, it will be possible to analyse the impact of both abiotic and biotic stresses in the relevant tissue. Finally, we used several independent LBDs to report on the localisation of the same lipids, which revealed previously unrecognized localisation for various PIPs in root epidermis.
A map of PIP localisation in Arabidopsis root epidermis
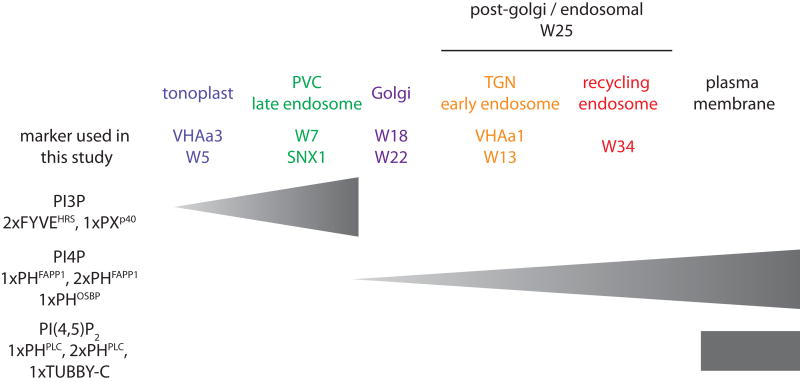
Although genetically encoded PIP sensors have known limitations, our work, together with previous studies (Vermeer et al., 2006; van Leeuwen et al., 2007; Mishkind et al., 2009; Vermeer et al., 2009), suggests the model presented in Figure 10 for the localisation of PI3P, PI4P and PI(4,5)P2 in non-stressed Arabidopsis root epidermis. Interestingly, we found that in this situation PI(4,5)P2 is already present at the PM in sufficient quantity to localise PIP2 binding proteins such as PHPLC and 1xTUBBY-C. Previous analyses using the 1xPHPLC sensor found that it is not localised to the PM in Arabidopsis root cell with the exception of the root hair tips and stressed cells (e.g. osmotic stress) (van Leeuwen et al., 2007). In our growth conditions, we found a significant proportion of 1xPHPLC at the PM of epidermal cells in the absence of any specific stresses. This might be due to differences of growth conditions (e.g. slightly different media). Another possible explanation is that we used the PH domain of PLCδ1 of Rattus norvegicus (Levine and Munro, 2002) while van Leeuwen et al., used the PH domain of human PLCδ1 (van Leeuwen et al., 2007). It is possible the rat PLCδ1 has a slightly different (i.e. higher) affinity for PI(4,5)P2 when expressed in Arabidopsis than its human counterpart, which could account for the differences observed in vivo. The differences of localisation might also be due to differences in the constructs design (e.g. linkers, promoters, fluorescent proteins, domain size). In any case, the YFP-1xPHHsPLC previously described (van Leeuwen et al., 2007) and our CITRINE-1xPHRnPLC should be complementary tools, which together with the CITRINE-2xPHRnPLC and CITRINE-1xTUBBY-C provide biosensors with four different apparent affinities for PI(4,5)P2.
Figure 10. Summary of PI3P, PI4P and PI(4,5)P2 localisation in Arabidopsis epidermal cells.
The gradient of intensity of localisation in intracellular compartments is represented by the broadness of the triangle.
PI3P is localised in late endosomes/PVC and to a less extent to the tonoplast. The late endosomes/PVC localisation is consistent with previous reports in tobacco BY-2 cells (Vermeer et al., 2006) as well as the localisation of PI3P effectors in this compartment (Jaillais et al., 2006; Pourcher et al., 2010). In animals, PI3P accumulates in early endosomes rather than late endosomes (Balla, 2013; Lemmon, 2008; De Matteis and Godi, 2004). However it is not surprising since plant late endosomes share many similarities with animal early endosomes (Jaillais et al., 2008), such as the presence of the small GTPases of the Rab5 family (Jaillais et al., 2008; Geldner et al., 2009). The coordinate action of Rab5 GTPases and PI3P at the surface of the plant late endosomes and animal early endosomes will attract their effector proteins (Jean and Kiger, 2012), many of which are conserved between the two kingdoms (e.g. SORTING NEXIN family (Jaillais et al., 2006; Lemmon, 2008; Pourcher et al., 2010; Cullen and Korswagen, 2012)).
PI4P was reported to accumulate in the Golgi apparatus in cowpea mesophyll protoplasts as well as in tobacco BY2 cells (Vermeer et al., 2009), but its co-localisation with post-Golgi markers was not investigated. We accumulated several lines of evidence suggesting that our PI4P biosensors localise in a post-Golgi compartment in Arabidopsis epidermal cells, including i) co-localisation with markers of the early endosomes/TGN and the recycling endosomes, ii) co-localisation with the endocytic tracer FM4-64 and, iii) sensitivity to BFA. Our results are in accordance with the localisation of the Arabidopsis PI4-kinase β1 and β2 in post-Golgi compartments in Arabidopsis root (Kang et al., 2011). Furthermore, loss of PI4-kinase β1 and β2 and of PI4-phosphatase (RHD4) activity induces TGN morphology defects in Arabidopsis roots (Preuss et al., 2006; Thole et al., 2008; Kang et al., 2011). Because of the dual nature of the plant TGN as the early endosome, it is likely that PI4P will have major functions in both protein exocytosis and endocytosis. This key cellular position is highlighted by the function of PI4P in polarized cell expansion (Preuss et al., 2006; Thole et al., 2008; Thole and Nielsen, 2008; Vermeer et al., 2009). However, it is clear that many more studies are required to understand how the various cellular pools of PI4P control specific cellular pathways. We believe that our PIPline marker set will catalyse future research on the various functions of PIPs in Arabidopsis on diverse topics including but not limited to membrane trafficking, cell signalling, cell morphogenesis, reproduction, development, response to abiotic and biotic stresses and adaptation to the environment.
Experimental procedures
Material and growth conditions
Plants were grown in soil with long daylight at 21°C and 70% humidity. For root analysis, seedlings were grown vertically on MS medium [(pH 5.7, 0.8% plant agar (Duchefa)] in the absence of sucrose, with continuous daylight condition for 6 to 9 days. Plants from the Columbia 0 accession and yeast from the BY4743 strain were used for transformation. The wave-lines, VHAa1/VHAa3 compartment markers and myrCIT lines were described before (Dettmer et al., 2006; Geldner et al., 2009; Jaillais et al., 2011).
Imaging
All imaging were performed on an inverted Zeiss LSM710 confocal microscope using a 40x Plan-apochromatic objective (numerical aperture 1.2). Dual-colour images were acquired by sequential line switching, allowing the separation of channels by both excitation and emission. In the case of co-localisation, we also controlled for a complete absence of channel crosstalk. Hoechst was excited with a 405nm laser, CyPET was excited with a 445nm laser, GFP was excited with a 488nm laser, CITRINE was excited with a 515nm laser and mCHERRY/Alexa555 were excited with a 561nm laser. FM4-64 (Invitrogen) was applied at a concentration of 3 μM; BFA (Sigma-Aldrich) was applied at a concentration of 25 μM for 1 hour in liquid medium. Stock solutions were prepared in DMSO at 3 μM and 50 μM, respectively. Imaging was performed in the root epidermis in cells that are at the onset of elongation. For quantitative imaging, pictures of epidermal root cells were taken with detector settings optimised for low background and at the limit of pixel saturation in order to obtain the best dynamic range possible.
Statistical analyses
Sample size was determined by variance analysis value (p<0.05) using Excel software (Microsoft). For the quantitative analysis of membrane localisation of low and high affinity biosensors, Student's t-tests were performed using Excel (Microsoft) (p<0.05). Quantitative co-localisation results were statistically compared using a bilateral test (Steel-Dwass-Critchlow-Fligner) using XLstat software (http://www.xlstat.com/). This non-parametric test is used to make all possible pairwise comparisons between groups with a probability of detecting localisation differences of 0.05%. Graphs were drawn using the Deltagraph5 software (http://www.rockware.com/).
See supplementary methods for: cloning of the PIP-line constructs (the protein IDs and primers used are presented in Table S2 and S3 and the sequences of all the constructs can be downloaded at http://www.ens-lyon.fr/RDP/SiCE/PIPline.html), 32P-phospholipid labelling and lipid analysis, plant transformation and selection, quantitative image analysis, protein extraction and protein-lipid overlay assay and, Huh-7 transfection and immunofluorescence analysis.
Supplementary Material
Figure S1. Identification of PIP biosensors targeted to the endomembrane system and/or plasma membrane in transgenic Arabidopsis roots.
Figure S2. CITIRNE-1xPHOSBP interacts with PI4P and PI(3,4)P2 in protein-lipid overlay assay
Figure S3. Multi-colour imaging of the PIPline collection.
Figure S4. 1xPXp40-CITRINE localises to late endosomes and tonoplast in Arabidopsis root epidermis.
Figure S5. Intracellular CITRINE-2xPHFAPP1 mainly accumulates to post-Golgi/endosomal compartments in Arabidopsis root epidermis.
Table S1. Overview of the LBDs used in this study.
Table S2. Information on the proteins used for the PIPline constructs (accession number, domain size, linker, primers used for cloning of the PIPline constructs)
Table S3. Primers used for cloning of the fluorescent protein reporters.
Table S4. Table recapitulating all the transgenic lines included in the PIPline marker set.
Acknowledgments
We thank all the members of the SiCE group for discussions, N. Geldner, K. Schumacher, J. Long, H. Stenmark, M. Yaffe, T. Levine, J.L. Farré, A. Marshall, S. Friant, L. Shapiro, C. Montell, W. Hahn, D. Root and R. Tsien for sharing material. This work was initially supported by grants from the U.S. National Institutes of Health (GM094428), and NSF (IOS-1045256) and The Howard Hughes Medical Institute (to J.C.), and then by grants from the Agence National pour la Recherche (ANR-12-JSV2-0002-01), and the Marie Curie Action (PCIG-GA-2011-303601) (to Y.J.) M.S. and S.A. are supported by a Ph.D. fellowship (MENRT) from the French ministry of higher education and research.
References
- Ago T, Takeya R, Hiroaki H, Kuribayashi F, Ito T, Kohda D, Sumimoto H. The PX Domain as a Novel Phosphoinositide- Binding Module. Biochem Biophys Res Commun. 2001;287:733–738. doi: 10.1006/bbrc.2001.5629. [DOI] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Tsiomenko A, Várnai P, Balla T. A Plasma Membrane Pool of Phosphatidylinositol 4- Phosphate Is Generated by Phosphatidylinositol 4-Kinase Type-III Alpha: Studies with the PH Domains of the Oxysterol Binding Protein and FAPP1. Molecular biology of the cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiological Reviews. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T, Szentpetery Z, Kim YJ. Phosphoinositide signaling: new tools and insights. Physiology (Bethesda) 2009;24:231–244. doi: 10.1152/physiol.00014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJ, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Korswagen HC. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol. 2012;14:29–37. doi: 10.1038/ncb2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi L, Costaguta G, Payne GS. Phosphoinositide-mediated clathrin adaptor progression at the trans-Golgi network. Nat Cell Biol. 2012;14:239–250. doi: 10.1038/ncb2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis MA, Godi A. PI-loting membrane traffic. Nat Cell Biol. 2004;6:487–492. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellson CD, Gobert-Gosse S, Anderson KE, Davidson K, Erdjument-Bromage H, Tempst P, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Gaffney PR, Coadwell J, Chilvers ER, Hawkins PT, Stephens LR. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox) Nat Cell Biol. 2001;3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- Eugster A, Pécheur EI, Michel F, Winsor B, Letourneur F, Friant S. Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Molecular biology of the cell. 2004;15:3031–3041. doi: 10.1091/mbc.E03-11-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré JC, Vidal J, Subramani S. A cytoplasm to vacuole targeting pathway in P. pastoris. Autophagy. 2007;3:230–234. doi: 10.4161/auto.3905. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- Franke TF, Kaplan DR, Cantley LC, Toker A. Direct Regulation of the Akt Proto-Oncogene Product by Phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- Friant S, Pécheur EI, Eugster A, Michel F, Lefkir Y, Nourrisson D, Letourneur F. Ent3p Is a PtdIns(3,5)P2 effector required for protein sorting to the multivesicular body. Dev Cell. 2003;5:499–511. doi: 10.1016/s1534-5807(03)00238-7. [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 2009;59:169–178. doi: 10.1111/j.1365-313X.2009.03851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A, Campli AD, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Grebe M, Xu J, Möbius W, Ueda T, Nakano A, Geuze HJ, Rook MB, Scheres B. Arabidopsis Sterol Endocytosis Involves Actin-Mediated Trafficking via ARA6-Positive Early Endosomes. Curr Biol. 2003;13:1378–1387. doi: 10.1016/s0960-9822(03)00538-4. [DOI] [PubMed] [Google Scholar]
- Hammond GRV, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GRV, Schiavo G, Irvine R. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns (4, 5) P2. Biochem J. 2009a;422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GRV, Sim Y, Lagnado L, Irvine RF. Reversible binding and rapid diffusion of proteins in complex with inositol lipids serves to coordinate free movement with spatial information. J Cell Biol. 2009b;184:297–308. doi: 10.1083/jcb.200809073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Scott JL, Heroux A, Roy S, Lenoir M, Overduin M, Stahelin RV, Kutateladze TG. Molecular basis of phosphatidylinositol 4-phosphate and ARF1 GTPase recognition by the FAPP1 pleckstrin homology (PH) domain. J Biol Chem. 2011;286:18650–18657. doi: 10.1074/jbc.M111.233015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Vora M, Haney RM, Filonov GS, Musselman CA, Burd CG, Kutateladze AG, Verkhusha VV, Stahelin RV, Kutateladze TG. Membrane insertion of the FYVE domain is modulated by pH. Proteins. 2009;76:852–860. doi: 10.1002/prot.22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikal AA, Hess ST, Baird GS, Tsien RY, Webb WW. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine) Proc Natl Acad Sci USA. 2000;97:11996–12001. doi: 10.1073/pnas.97.22.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T, Seiler S, Heilmann I. At the poles across kingdoms: phosphoinositides and polar tip growth. Protoplasma. 2010;240:13–31. doi: 10.1007/s00709-009-0093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T, Stenzel I, Heilmann I. Type B phosphatidylinositol-4-phosphate 5-kinases mediate Arabidopsis and Nicotiana tabacum pollen tube growth by regulating apical pectin secretion. Plant Cell. 2008;20:3312–3330. doi: 10.1105/tpc.108.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T, Stenzel I, Hempel F, Jin X, Mosblech A, Heilmann I. Phosphatidylinositol-4,5-bisphosphate influences Nt-Rac5-mediated cell expansion in pollen tubes of Nicotiana tabacum. Plant J. 2011;65:453–468. doi: 10.1111/j.1365-313X.2010.04435.x. [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miège C, Gaude T. Evidence for a sorting endosome in Arabidopsis root cells. Plant J. 2008;53:237–247. doi: 10.1111/j.1365-313X.2007.03338.x. [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miège C, Rollin C, Gaude T. AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature. 2006;443:106–109. doi: 10.1038/nature05046. [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Hothorn M, Belkhadir Y, Dabi T, Nimchuk ZL, Meyerowitz EM, Chory J. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25:232–237. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean S, Kiger AA. Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat Rev Mol Cell Biol. 2012;13:463–470. doi: 10.1038/nrm3379. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, Caponigro G, Hieronymus H, Murray RR, Salehi-Ashtiani K, Hill DE, Vidal M, Zhao JJ, Yang X, Alkan O, Kim S, Harris JL, Wilson CJ, Myer VE, Finan PM, Root DE, Roberts TM, Golub T, Flaherty KT, Dummer R, Weber BL, Sellers WR, Schlegel R, Wargo JA, Hahn WC, Garraway LA. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, Brown GE, Cantley LC, Yaffe MB. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA. Electron Tomography of RabA4b- and PI-4Kβ1-Labeled Trans Golgi Network Compartments in Arabidopsis. Traffic. 2011;12:313–329. doi: 10.1111/j.1600-0854.2010.01146.x. [DOI] [PubMed] [Google Scholar]
- Kwon Y, Hofmann T, Montell C. Integration of phosphoinositide- and calmodulin-mediated regulation of TRPC6. Mol Cell. 2007;25:491–503. doi: 10.1016/j.molcel.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, O'Brien R, Sigler PB, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci USA. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir M, Coskun UN, Grzybek M, Cao X, Buschhorn SB, James J, Simons K, Overduin M. Structural basis of wedging the Golgi membrane by FAPP pleckstrin homology domains. EMBO report. 2010;11:279–284. doi: 10.1038/embor.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Munro S. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Current Biology. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Current Biology. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Ling Y, Stefan CJ, MacGurn JA, Audhya A, Emr SD. The dual PH domain protein Opy1 functions as a sensor and modulator of PtdIns(4,5)P2 synthesis. EMBO J. 2012;31:2882–2894. doi: 10.1038/emboj.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Málková Š, Stahelin RV, Pingali SV, Cho W, Schlossman ML. Orientation and Penetration Depth of Monolayer-Bound p40 phox-PX. Biochemistry. 2006;45:13566–13575. doi: 10.1021/bi061133l. [DOI] [PubMed] [Google Scholar]
- Manford AG, Stefan CJ, Yuan HL, MacGurn JA, Emr SD. ER-to-Plasma Membrane Tethering Proteins Regulate Cell Signaling and ER Morphology. Dev Cell. 2012;23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Marshall AJ, Krahn AK, Ma K, Duronio V, Hou S. TAPP1 and TAPP2 are targets of phosphatidylinositol 3-kinase signaling in B cells: sustained plasma membrane recruitment triggered by the B-cell antigen receptor. Mol Cell Biol. 2002;22:5479–5491. doi: 10.1128/MCB.22.15.5479-5491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- Meijer HJG, Munnik T. Phospholipid-based signaling in plants. Annu Rev Plant Biol. 2003;54:265–306. doi: 10.1146/annurev.arplant.54.031902.134748. [DOI] [PubMed] [Google Scholar]
- Mishkind M, Vermeer JEM, Darwish E, Munnik T. Heat stress activates phospholipase D and triggers PIP2 accumulation at the plasma membrane and nucleus. Plant J. 2009;60:10–21. doi: 10.1111/j.1365-313X.2009.03933.x. [DOI] [PubMed] [Google Scholar]
- Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- Munnik T, Nielsen E. Green light for polyphosphoinositide signals in plants. Curr Opin Plant Biol. 2011;14:489–497. doi: 10.1016/j.pbi.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Munnik T, Vermeer JEM. Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ. 2010;33:655–669. doi: 10.1111/j.1365-3040.2009.02097.x. [DOI] [PubMed] [Google Scholar]
- Munro S. Organelle identity and the organization of membrane traffic. Nat Cell Biol. 2004;6:469–472. doi: 10.1038/ncb0604-469. [DOI] [PubMed] [Google Scholar]
- Niu Y, Zhang C, Sun Z, Hong Z, Li K, Sun D, Yang Y, Tian C, Gong W, Liu JJ. PtdIns(4)P regulates retromer-motor interaction to facilitate dynein-cargo dissociation at the trans-Golgi network. Nat Cell Biol. 2013;15:417–429. doi: 10.1038/ncb2710. [DOI] [PubMed] [Google Scholar]
- Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- Posor Y, Eichhorn-Gruenig M, Puchkov D, Schöneberg J, Ullrich A, Lampe A, Müller R, Zarbakhsh S, Gulluni F, Hirsch E, Krauss M, Schultz C, Schmoranzer J, Noé F, Haucke V. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499:233–237. doi: 10.1038/nature12360. [DOI] [PubMed] [Google Scholar]
- Pourcher M, Santambrogio M, Thazar N, Thierry AM, Fobis-Loisy I, Miège C, Jaillais Y, Gaude T. Analyses of sorting nexins reveal distinct retromer-subcomplex functions in development and protein sorting in Arabidopsis thaliana. Plant Cell. 2010;22:3980–3991. doi: 10.1105/tpc.110.078451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Schmitz AJ, Thole JulieM, Bonner HKS, Otegui MS, Nielsen Erik. A role for the RabA4b effector protein PI-4K 1 in polarized expansion of root hair cells in Arabidopsis thaliana. J Cell Biol. 2006;172:991–998. doi: 10.1083/jcb.200508116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn KV, Behe P, Tinker A. Monitoring changes in membrane phosphatidylinositol 4,5-bisphosphate in living cells using a domain from the transcription factor tubby. J Physiol (Lond) 2008;586 doi: 10.1113/jphysiol.2008.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Bremnes B, Mehlum A, Gillooly DJ, D'Arrigo A, Stang E, Stenmark H. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. Journal of Cell Science. 2001;114:2255–2263. doi: 10.1242/jcs.114.12.2255. [DOI] [PubMed] [Google Scholar]
- Rameh LE, Arvidsson AK, Carraway KL, Couvillon AD, Rathbun G, Crompton A, VanRenterghem B, Czech MP, Ravichandran KS, Burakoff SJ. A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J Biol Chem. 1997;272:22059–22066. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- Roy A, Levine TP. Multiple Pools of Phosphatidylinositol 4-Phosphate Detected Using the Pleckstrin Homology Domain of Osh2p. J Biol Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- Salim K, Bottomley MJ, Querfurth E, Zvelebil MJ, Gout I, Scaife R, Margolis RL, Gigg R, Smith CI, Driscoll PC. Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton's tyrosine kinase. EMBO J. 1996;15:6241. [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Klein DE, Sachdeva MM, Lemmon MA. High-Affinity Binding of a FYVE Domain to Phosphatidylinositol 3-Phosphate Requires Intact Phospholipid but Not FYVE Domain Oligomerization. Biochemstry. 2001;40:8581–8587. doi: 10.1021/bi010425d. [DOI] [PubMed] [Google Scholar]
- Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- Schultz C. Challenges in studying phospholipid signaling. Nat Chemical Biology. 2010;6:473–475. doi: 10.1038/nchembio.389. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Lippé R, Christoforidis S, Gaullier JM, Brech A, Callaghank J, Tohk BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J Biol Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh Proteins Regulate Phosphoinositide Metabolism at ER-Plasma Membrane Contact Sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- Suh BC, Inoue T, Meyer T, Hille B. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentpetery Z, Balla A, Kim YJ, Lemmon MA, Balla T. Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate; a comparative study. BMC Cell Biol. 2009;10:67. doi: 10.1186/1471-2121-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentpetery Z, Várnai P, Balla T. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc Natl Acad Sci USA. 2010;107:8225–8230. doi: 10.1073/pnas.1000157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole JM, Nielsen E. Phosphoinositides in plants: novel functions in membrane trafficking. Curr Opin Plant Biol. 2008;11:620–631. doi: 10.1016/j.pbi.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Thole JM, Vermeer JEM, Zhang Y, Gadella TWJ, Nielsen E. ROOT HAIR DEFECTIVE4 Encodes a Phosphatidylinositol-4-Phosphate Phosphatase Required for Proper Root Hair Development in Arabidopsis thaliana. Plant Cell. 2008;20:381–395. doi: 10.1105/tpc.107.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzi S, Uekama N, Okada M, Yamaguchi S, Saitô H, Yagisawa H. Structure and dynamics of the phospholipase C-δ1 pleckstrin homology domain located at the lipid bilayer surface. J Biol Chem. 2003;278:28019–28025. doi: 10.1074/jbc.M300101200. [DOI] [PubMed] [Google Scholar]
- van Leeuwen W, Vermeer JEM, Gadella TWJ, Munnik T. Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J. 2007;52:1014–1026. doi: 10.1111/j.1365-313X.2007.03292.x. [DOI] [PubMed] [Google Scholar]
- Várnai P, Balla T. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim Biophys Acta. 2006;1761:957–967. doi: 10.1016/j.bbalip.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Várnai P, Lin X, Lee SB, Tuymetova G, Bondeva T, Spät A, Rhee SG, Hajnóczky G, Balla T. Inositol lipid binding and membrane localization of isolated pleckstrin homology (PH) domains. Studies on the PH domains of phospholipase C delta 1 and p130. J Biol Chem. 2002;277:27412–27422. doi: 10.1074/jbc.M109672200. [DOI] [PubMed] [Google Scholar]
- Vermeer JEM, Thole JM, Goedhart J, Nielsen E, Munnik T, Gadella TWJ. Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J. 2009;57:356–372. doi: 10.1111/j.1365-313X.2008.03679.x. [DOI] [PubMed] [Google Scholar]
- Vermeer JEM, van Leeuwen W, Tobeña-Santamaria R, Laxalt AM, Jones DR, Divecha N, Gadella TWJ, Munnik T. Visualization of PtdIns3P dynamics in living plant cells. Plant J. 2006;47:687–700. doi: 10.1111/j.1365-313X.2006.02830.x. [DOI] [PubMed] [Google Scholar]
- Vincent P, Chua M, Nogue F, Fairbrother A, Mekeel H, Xu Y, Allen N, Bibikova TN, Gilroy S, Bankaitis VA. A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J Cell Biol. 2005;168:801–812. doi: 10.1083/jcb.200412074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita SI, Oku M, Wasada Y, Ano Y, Sakai Y. PI4P-signaling pathway for the synthesis of a nascent membrane structure in selective autophagy. J Cell Biol. 2006;173:709–717. doi: 10.1083/jcb.200512142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, Dewald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- Zhan Y, Virbasius JV, Song X, Pomerleau DP, Zhou GW. The p40phox and p47phox PX domains of NADPH oxidase target cell membranes via direct and indirect recruitment by phosphoinositides. J Biol Chem. 2002;277:4512–4518. doi: 10.1074/jbc.M109520200. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Perera RM, Sebastian R, Nakatsu F, Chen H, Balla T, Ayala G, Toomre D, De Camilli PV. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc Natl Acad Sci USA. 2007;104:3793–3798. doi: 10.1073/pnas.0611733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Identification of PIP biosensors targeted to the endomembrane system and/or plasma membrane in transgenic Arabidopsis roots.
Figure S2. CITIRNE-1xPHOSBP interacts with PI4P and PI(3,4)P2 in protein-lipid overlay assay
Figure S3. Multi-colour imaging of the PIPline collection.
Figure S4. 1xPXp40-CITRINE localises to late endosomes and tonoplast in Arabidopsis root epidermis.
Figure S5. Intracellular CITRINE-2xPHFAPP1 mainly accumulates to post-Golgi/endosomal compartments in Arabidopsis root epidermis.
Table S1. Overview of the LBDs used in this study.
Table S2. Information on the proteins used for the PIPline constructs (accession number, domain size, linker, primers used for cloning of the PIPline constructs)
Table S3. Primers used for cloning of the fluorescent protein reporters.
Table S4. Table recapitulating all the transgenic lines included in the PIPline marker set.