Abstract
The effect of activation and over-expression of the nuclear receptor PPARβ/δ in human MDA-MB-231 (ER−) and MCF7 (ER+) breast cancer cell lines was examined. Target gene induction by ligand activation of PPARβ/δ was increased by over-expression of PPARβ/δ compared to controls. Over-expression of PPARβ/δ caused a decrease in cell proliferation in MCF7 and MDA-MB-231 cells compared to controls while ligand activation of PPARβ/δ further inhibited proliferation of MCF7 but not MDA-MB-231 cells. Over-expression and/or ligand activation of PPARβ/δ in MDA-MB-231 or MCF7 cells had no effect on experimental apoptosis. Decreased clonogenicity was observed in both MDA-MB-231 and MCF7 over-expressing PPARβ/δ in response to ligand activation of PPARβ/δ as compared to controls. Ectopic xenografts developed from MDA-MB-231 and MCF7 cells over-expressing PPARβ/δ were significantly smaller and ligand activation of PPARβ/δ caused an even greater reduction in tumor volume as compared to controls. Interestingly, the decrease in MDA-MB-231 tumor size after over-expressing PPARβ/δ and ligand activation of PPARβ/δ correlated with increased necrosis. These data show that ligand activation and/or over-expression of PPARβ/δ in two human breast cancer cell lines inhibits relative breast cancer tumorigenicity and provide further support for the development of ligands for PPARβ/δ to specifically inhibit breast carcinogenesis. These new cell-based models will be invaluable tools for delineating the role of PPARβ/δ in breast cancer and evaluating the effects of PPARβ/δ agonists.
Keywords: PPARβ/δ, breast cancer, cell proliferation, xenografts
Introduction
Peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) belongs to the PPAR family of nuclear hormone receptors, which regulate a variety of biological processes, including cell differentiation, cell proliferation, lipid accumulation, and glucose and fatty acid metabolism (1–4). Thus, PPARs also influence the development of human diseases, such as diabetes and cancers. PPARβ/δ primarily acts as a transcription factor through dynamic interactions with chromatin with multiple levels of regulation including, the expression level of the receptor, the expression level of co-effectors proteins, the presence of endogenous/exogenous ligands, and the availability of binding sites on chromatin containing target genes and regulatory regions that are modulated through these interactions (5). However, PPARβ/δ can also directly interact with other transcription factors such as the p65 subunit of NF-kB and repress gene expression; an event associated with its known anti-inflammatory functions (reviewed in (1, 4, 6)).
Despite the large body of evidence that PPARβ/δ is a protein expressed at very high levels in the skin, intestine and liver (7), has known potent anti-inflammatory activities (8), and is required for many important physiological processes (9), the role of PPARβ/δ in carcinogenesis remains controversial. For example, some studies indicate that activating PPARβ/δ promotes tumorigenesis whereas antagonizing PPARβ/δ inhibits tumorigenesis, for multiple types of cancer (reviewed in (1, 4, 6)). In contrast, other studies show that activating PPARβ/δ inhibits tumorigenesis, while antagonizing PPARβ/δ promotes or has no effect on tumorigenesis, for multiple types of cancer (reviewed in (1, 4, 6)).
Thus, controversy exists in the literature indicating that expression and/or activating PPARβ/δ either promotes or inhibits breast cancer based on in vitro and in vivo models. For example, the estrogen receptor-positive (ER+), MCF7 cell line cultured in vitro with a PPARβ/δ ligand exhibited enhanced cell proliferation and when PPARβ/δ was over-expressed in the same cell line, ligand activation of PPARβ/δ caused an even greater increase in average cell proliferation (10). Interestingly, the estrogen receptor-negative (ER−), MDA-MB-231 cell line did not exhibit any change in cell proliferation following treatment with PPARβ/δ ligands. This may suggest that this effect was in part dependent on the presence of a functional ER (10). In contrast, another study showed that increased expression of PPARβ/δ was associated with terminal differentiation of MCF7 cells (11), an effect that is known to coincide with their withdrawal from the cell cycle (12). The latter study is also consistent with another study that found inhibition of cell proliferation following ligand activation of PPARβ/δ in MCF7 cells (13). Interestingly, the difference between results from the above studies and those observed by Stephen and colleagues (10) was not due to differences in the ligands used or the presence or lack of serum. In addition, antagonizing PPARβ/δ with GSK3787 had no effect on MCF7 cell proliferation (14). In vivo analysis examining the effect of PPARβ/δ activation has also led to results that are difficult to interpret. For example, cyclooxygenase-2 (COX2)-null crossed with Pparβ/δ-null mice exhibit reduced tumorigenicity as compared to controls, but tumor multiplicity was not measured (15). This study suggests that PPARβ/δ is required for mammary tumorigenesis in the Cox2-null transgenic mouse line. This observation is consistent with results observed after activating PPARβ/δ by low dose GW501516 in FVB mice co-treated with medroxyprogesterone acetate and 7,12-dimethylbenz[a]anthracene (DMBA) where increased mammary tumorigenesis was observed (16). Unfortunately, tumor multiplicity was again not reported. Most recently, analysis of a single clonal line of transgenic mice over-expressing PPARβ/δ suggested that this receptor can promote mammary tumorigenesis in FVB mice (17). In contrast to these in vivo results, ex vivo analysis showed dose-dependent decrease in clonogenicity in C20 mouse mammary gland cancer cells in response to ligand activation of PPARβ/δ (18). Collectively, these studies demonstrate the large variation and difficulty in understanding the role of PPARβ/δ in breast cancer and illustrate the need for additional models to dissect the signaling components or co-factors of this nuclear receptor.
The present study describes the creation and characterizations of two stable human breast cancer cell lines over-expressing PPARβ/δ, the ER−, MDA-MB-231-hPPARβ/δ and ER+, MCF7-hPPARβ/δ cell lines. These cell lines were used to examine the effects of over-expression and/or ligand-mediated activation of PPARβ/δ on cell proliferation, cell cycle, apoptosis, clonogenicity and tumorigenicity using both in vitro, ex vivo and in vivo approaches.
Materials and Methods
Cell lines
The human breast cancer cell line MDA-MD-231 and MCF7 were obtained from American Type Culture Collection (ATCC, Manassas, VA). The cell lines were tested for authenticity in October 2010 using Short-Tandem Repeat Analysis (IDEXX-RADIL, Columbia, MO). The alleles for 9 different markers were determined and confirmed to be of human origin and non-mammalian inter-species contamination was detected. The MCF7 cells matched the genetic profile reported for that cell line. The MDA-MB-231 cells matched the genetic profile reported for this cell line for 7 of the 9 alleles, but were missing an allele at TPOX and had an extra allele at vWA when compared to the profile reported for cell line HTB-26 (MDA-MB-231). Cells were cultured in Dulbecco’s Modified Eagle’s Media (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% non-essential amino acid and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA) at 37°C with 5% carbon dioxide. The human breast cancer cell lines were cultured with or without different concentrations of the highly specific PPARβ/δ agonist GW0742 (kindly provided by Drs. Andrew Billin and Timothy Willson, GlaxoSmithKline, Research Triangle Park, NC) at concentrations ranging from 0.01 – 10.0 μM for 24 – 96 hours.
Stable human MDA-MD-231 and MCF7 breast cancer cell lines over-expressing PPARβ/δ
Stable human MDA-MD-231 and MCF7 breast cancer cell lines over-expressing PPARβ/δ were generated using the MigR1 retroviral vector that uses a mouse stem cell virus promoter to drive expression of PPARβ/δ, followed by an internal ribosome entry site (IRES) and a sequence encoding enhanced green fluorescent protein (eGFP) (19) as previously described (20, 21). This vector allows for expression of a protein of interest and/or eGFP (the latter served as a control), which facilitates identification and sorting of cells that have stably integrated the MigR1 retroviral vector as previously described (20, 21).
Western blotting
Soluble protein lysates were prepared as previously described (20). Thirty micrograms of protein per independent sample was resolved using SDS-polyacrylamide gels (10 or 12%) and transferred to PVDF membranes using an electroblotting method. The membranes were incubated with blocking buffer (5% dried milk in Tris buffered saline with Tween-20 (TBST)) at room temperature, washed with TBST and then incubated with primary antibodies against either, human PPARβ/δ, cellular retinoic acid binding protein II (CRABP-II; Abcam, Cambridge, MA), p65, proliferating cellular nuclear antigen (PCNA), β-ACTIN (Santa Cruz Biotechnologies, Santa Cruz, CA), fatty acid binding protein 5 (FABP5; Biovendor, Chandler, NC), poly (ADP-ribose) polymerase (PARP; Cell Signaling Technology, Danvers, MA), or lactate dehydrogenase (LDH, Rockland, Gilbertsville, PA) at 4°C overnight. After washing three times with TBST at room temperature for 10 minutes each, the membranes were incubated with biotin-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) at room temperature and then with 125I-streptavidin. The membranes were exposed to phosporimager plates and the level of radioactivity quantified with filmless autoradiographic analysis (Packard Phosphorimager, PerkinElmer, Waltham, MA). Hybridization signals for specific proteins were normalized to that of LDH or β-ACTIN.
Quantitative real-time polymerase chain reaction (qPCR)
qPCR was used to measure the PPARβ/δ target gene angiopoietin-like protein 4 (ANGPTL4) mRNA expression as previously described (22). Each assay included a standard curve and a non-template control performed in triplicate. The relative mRNA level of ANGPTL4 was normalized to the relative mRNA level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Cell proliferation assay
To determine the effect of over-expression and/or ligand activation of PPARβ/δ on cell proliferation, the xCELLigence system (Roche, Indianapolis, IN) was used as previously described (14). Briefly, MDA-MB-231, MDA-MB-231-MigR1 (control cells containing the MigR1 vector expressing eGFP), and MDA-MB-231-hPPARβ/δ cells (cells over-expressing human PPARβ/δ and eGFP) or MCF7, MCF7-MigR1, and MCF7-hPPARβ/δ cells were seeded on 16-well E-plates. Twenty-four hours after plating, the cells were cultured with medium with or without GW0742 for another 72 h. Cell proliferation was monitored in real-time and growth curves recorded using the Real-Time Cell Analyzer (RTCA) DP Analyzer and RTCA Software (Roche, Indianapolis, IN).
Clonogenicity assay
Cells were plated on 60 mm culture dishes (800 cells/plate) and cultured in medium with or without GW0742 for 12 days to assess clonogenicity as previously described (18). The plating efficiency and surviving fraction were calculated as previously described (18).
Ectopic xenografts
Six-week-old female immunodeficient athymic nude (nu/nu) mice (Frederick National Laboratory for Cancer Research, Frederick, MD) were injected subcutaneously with 5 × 106 cells. The MDA-MB-231-MigR1 cells were injected in the left rear flank and the MDA-MB-231-hPPARβ/δ cells were injected in the right rear flank. For analysis of the MCF7 cells mice were implanted with a pellet containing 17β-estradiol (dose = 0.72 mg/kg) one day before injecting cells. The MCF7-MigR1 cells were injected in the left rear flank and the MCF7-hPPARβ/δ cells were injected in the right rear flank. Groups of mice were then treated with or without GW0742 (2.5 mg/kg/day) for up to 36 days. The GW0742 was provided by daily dosing with a pellet made with Bacon-flavored Transgenic Dough Diet (Bioserv, Inc., Frenchtown, NJ) mixed with either vehicle control (0.02% dimethylsulfoxide) or GW0742. Body weight and tumor volumes were measured three times a week. Mice were euthanized by over-exposure to carbon dioxide, and tumors harvested. Half of each tumor was fixed in 10% formalin, and the other half was snap frozen in liquid nitrogen for subsequent analysis of proteins by western blot or mRNA expression by qPCR as described above.
Statistical Analysis
All in vitro experimental groups were performed in triplicate and repeated using three independent samples of cells. The xenograft studies were performed twice using a total of 10 mice per treatment group. The data were analyzed for statistical significance using analysis of variance followed by the post-hoc Tukey test. Statistical significance was considered when P ≤ 0.05. Values are presented as the mean ± S.E.M..
Results
Confirmation of functional over-expression of PPARβ/δ in MDA-MD-231 and MCF7 breast cancer cell lines
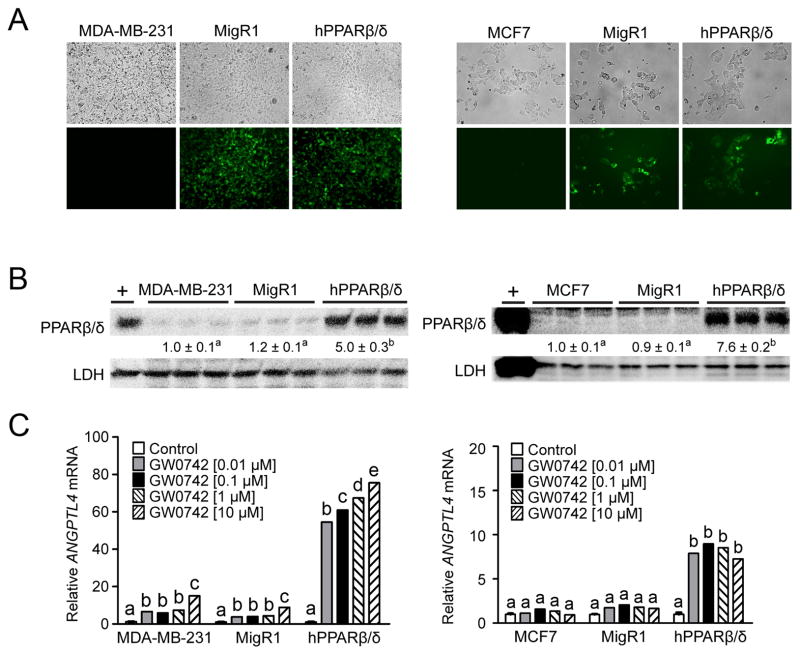
Fluorescent microscopic examination of control cells confirmed the lack of eGFP expression in both MDA-MB-231 and MCF7 cells whereas both cell lines containing the MigR1 vector expressed eGFP (Fig. 1A). Similarly, eGFP was expressed in both MDA-MB-231 and MCF7 cells over-expressing hPPARβ/δ (Fig. 1A). Increased expression of PPARβ/δ was confirmed by western blot analysis in both MDA-MB-231-hPPARβ/δ and MCF7-hPPARβ/δ cells by 5-fold and ~8-fold, respectively (Fig. 1A and B). Ligand activation of PPARβ/δ increased expression of the PPARβ/δ target gene ANGPTL4 in MDA-MB-231 cells and MDA-MB-231-MigR1 cells compared to controls, and the extent of induction was markedly higher in MDA-MB-231-hPPARβ/δ cells (Fig. 1C). In contrast, ligand activation of PPARβ/δ did not influence expression of ANGPTL4 mRNA in normal MCF7 and MCF7-MigR1 cells compared to controls, but did markedly increase expression of this PPARβ/δ target gene ANGPTL4 in MCF7-hPPARβ/δ cells (Fig. 1C). The lack of a statistically significant increase in ANGPTL4 mRNA in MCF7 and MCF7-MigR1 cells by ligand activation of PPARβ/δ could be due to the fact that expression of PPARβ/δ was not detectable in MCF7 cells compared to low but measureable expression of MDA-MB-231 cells (Fig. 1B).
Figure 1.
Characterization of human breast cancer cell lines (MDA-MB-231 or MCF7) over-expressing PPARβ/δ. (A) Representative photomicrographs of MDA-MB-231 cells, MDA-MB-231-MigR1 (MigR1) or MDA-MB-231-hPPARβ/δ (hPPARβ/δ; left panels) or MCF7 cells, MCF7-MigR1 (MigR1) or MCF7-hPPARβ/δ (hPPARβ/δ; right panels) examined by light microscopy (upper panels) or fluorescent microscopy (lower panels). (B) Western blot analysis of PPARβ/δ. Hybridization signals for each protein were normalized to that of LDH and represent the mean ± S.E.M.. + = positive control, cell lysate from COS cells transfected with hPPARβ/δ expression vector. (C) qPCR analysis of ANGPTL4 mRNA, normalized to GAPDH mRNA. Values represent the mean ± S.E.M.. Values with different letters are significantly different, P ≤ 0.05.
Influence of over-expressed PPARβ/δ in MDA-MD-231 and MCF7 breast cancer cell line proliferation
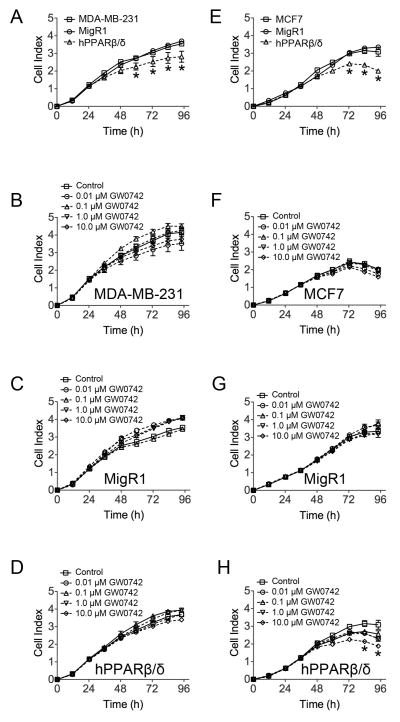
Over-expression of PPARβ/δ in MDA-MD-231 and MCF7 breast cancer cell lines inhibited cell proliferation after 48–72 of culture as compared to controls (Fig. 2A and E). Ligand activation of PPARβ/δ in MDA-MD-231, MDA-MD-231-MigR1 or MDA-MD-231-hPPARβ/δ cells did not further influence this effect (Fig. 2B, C and D) whereas ligand activation of PPARβ/δ in MCF7-hPPARβ/δ did inhibit cell proliferation as compared to controls, but this effect was only observed with the highest dose of 10 μM GW0742 (Fig. 2F, G and H). None of these changes in cell proliferation resulting from over-expression and/or ligand activation of PPARβ/δ in MDA-MD-231 and MCF7 breast cancer cell lines were associated with alterations in cell cycle progression (Supplementary Fig. S1).
Figure 2.
The effect of over-expressing PPARβ/δ and/or ligand activation of PPARβ/δ on cell proliferation in MDA-MB-231 and MCF7 cells. Cell proliferation was examined in real time in (A) MDA-MB-231 cells, MDA-MB-231-MigR1 (MigR1) or MDA-MB-231-hPPARβ/δ (hPPARβ/δ) or (E) MCF7 cells, MCF7-MigR1 (MigR1) or MCF7-hPPARβ/δ (hPPARβ/δ). Cell proliferation was examined in real time for (B) MDA-MB-231, (C) MDA-MB-231-MigR1 (MigR1) or (D) MDA-MB-231-hPPARβ/δ (hPPARβ/δ) cells with or without the indicated concentration of GW0742. Cell proliferation was examined in real time for (F) MCF7, (G) MCF7-MigR1 (MigR1) or (H) MCF7-hPPARβ/δ (hPPARβ/δ) cells with or without the indicated concentration of GW0742. *Significantly different than control, P ≤ 0.05.
Over-expression and/or ligand activation of PPARβ/δ in MDA-MD-231 and MCF7 breast cancer cell lines has no effect on inducible apoptosis
As previous studies proposed a link between ligand activation of PPARβ/δ and inhibition of apoptosis (reviewed in (4)), the effect of over-expression and/or ligand activation of PPARβ/δ was examined using two different approaches to induce apoptosis: staurosporine and UV treatment. Staurosporine induced apoptosis in MDA-MD-231, MDA-MD-231-MigR1 and MDA-MD-231-hPPARβ/δ cells but no differences in the concentration of staurosporine required for this effect, or the timing of PARP cleavage following staurosporine was observed between the MDA-MD-231 cell lines (Supplementary Fig. 2A and B). Further, the ligand activation did not influence staurosporine-induced PARP cleavage between any of the MDA-MD-231 cell lines (Supplementary Fig. 2C). A similar lack of effect was observed in MCF7, MCF7-MigR1 or MCF7-hPPARβ/δ cell lines (Supplementary Fig. 2A, B and C). Over-expression of PPARβ/δ did not influence UVB-induced PARP cleavage in either MDA-MD-231, MDA-MD-231-MigR1 and MDA-MD-231-hPPARβ/δ cells or MCF7, MCF7-MigR1 or MCF7-hPPARβ/δ cells (Supplementary Fig. 3A). Further, ligand activation of PPARβ/δ did not alter PARP cleavage in any of the MDA-MB-231 or MCF7 cells lines as compared to controls (Supplementary Fig. 3B).
Ligand activation of PPARβ/δ in MDA-MD-231 and MCF7 breast cancer cell lines over-expressing PPARβ/δ inhibits clonogenicity
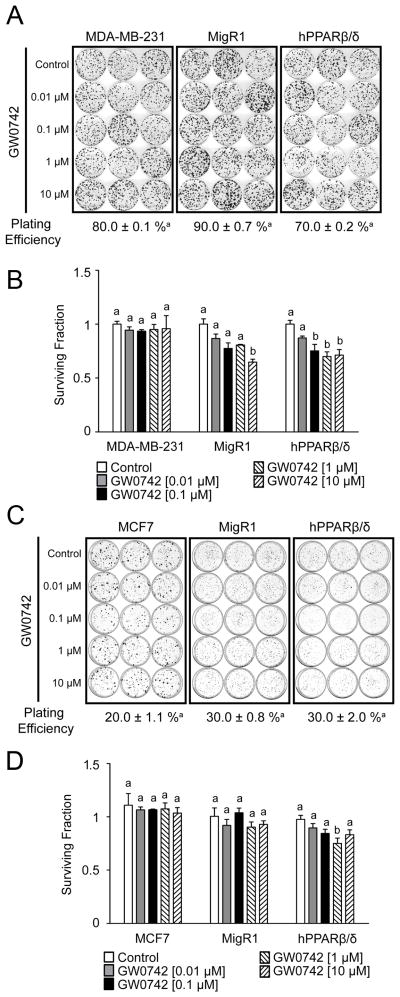
To begin to determine if over-expression and/or ligand activation of PPARβ/δ influenced relative tumorigenicity of these cells, anchorage-dependent clonogenicity was examined. No difference in plating efficiency was observed between MDA-MD-231, MDA-MD-231-MigR1 and MDA-MD-231-hPPARβ/δ cells or MCF7, MCF7-MigR1 and MCF7-hPPARβ/δ cells (Fig. 3A and B). While ligand activation of PPARβ/δ had no effect on the relative clonogenicity of either MDA-MB-231 or MCF7 cells, inhibition of clonogenicity was observed in MDA-MD-231-MigR1 cells treated with 10 μM GW0742 as compared to controls (Fig. 3C). Moreover, ligand activation of PPARβ/δ more markedly inhibited clonogenicity in MDA-MD-231-hPPARβ/δ between 0.1 and 10 μM GW0742 as compared to controls (Fig. 3C). Ligand activation of PPARβ/δ had no effect on the relative clonogenicity of MCF7-MigR1 cells but was inhibited with 1 μM GW0742 in MCF7-hPPARβ/δ cells as compared to controls (Fig. 3D).
Figure 3.
The effect of over-expressing PPARβ/δ and/or ligand activation of PPARβ/δ on clonogenicity in MDA-MB-231 and MCF7 cells. Anchorage-dependent clonogenicity was examined in (A) MDA-MB-231, MDA-MB-231-MigR1 (MigR1) and MDA-MB-231-hPPARβ/δ (hPPARβ/δ) cells or (B) MCF7, MCF7-MigR1 (MigR1) and MCF7-hPPARβ/δ (hPPARβ/δ) cells. The surviving fraction for each treatment of (C) MDA-MB-231, MDA-MB-231-MigR1 (MigR1) or MDA-MB-231-hPPARβ/δ (hPPARβ/δ) cells or (D) MCF7, MCF7-MigR1 (MigR1) or MCF7-hPPARβ/δ (hPPARβ/δ) cells is presented. Values represent the mean ± S.E.M.. Values with different letters are significantly different, P ≤ 0.05.
Over-expression and/or ligand activation of PPARβ/δ in MDA-MD-231 and MCF7 breast cancer cell lines inhibits ectopic xenografts in part by enhancing necrosis
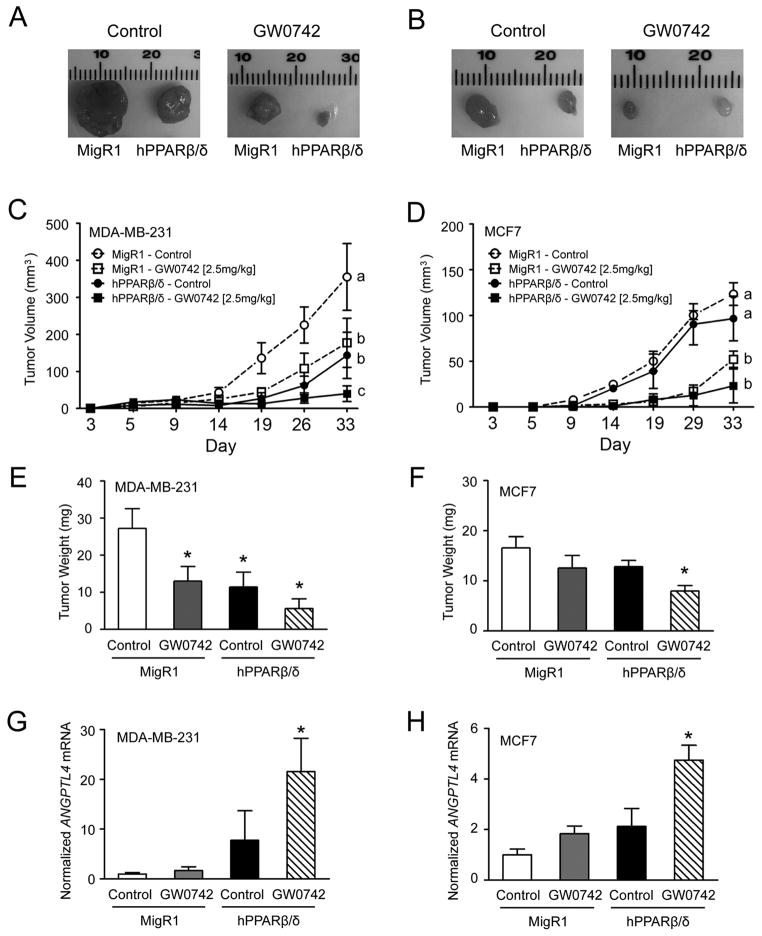
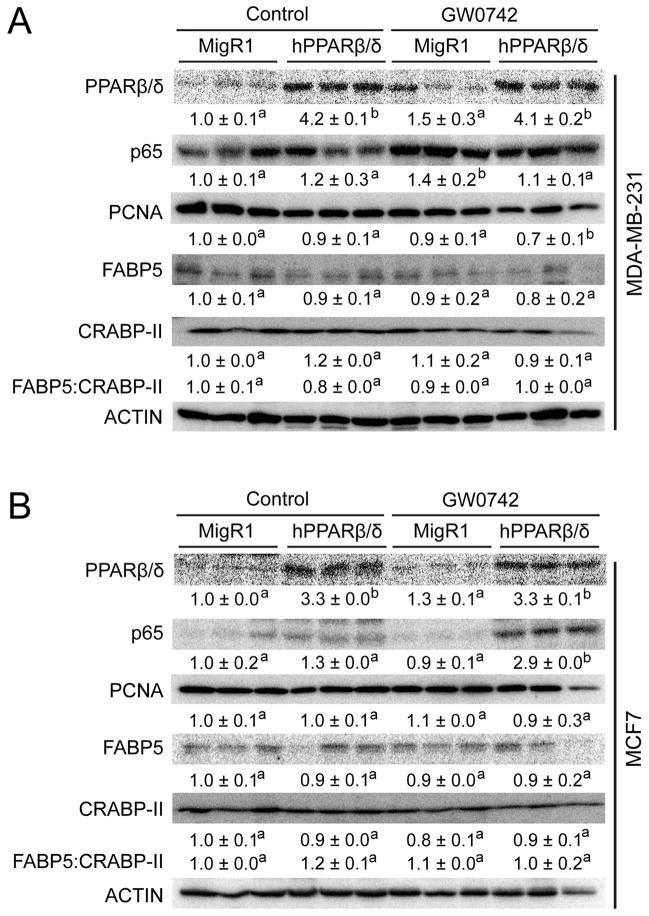
Over-expression of PPARβ/δ markedly reduced average tumor volume in both MDA-MD-231 (60%) and MCF7 (20%) xenografts as compared to control MDA-MD-231-MigR1 or MCF7-MigR1 xenografts (Fig. 4A, B, C and D, Supplementary Fig. 4A and B). Ligand activation of PPARβ/δ caused even further reduction in average tumor volume of MDA-MD-231-MigR1 (50%), MDA-MD-231-hPPARβ/δ (90%), MCF7-MigR1 (60%) and MCF7-hPPARβ/δ (80%) xenografts as compared to controls, and this effect was more pronounced in the MDA-MD-231-hPPARβ/δ and MCF7-hPPARβ/δ xenografts compared to their respective controls (Fig. 4A, B, C and D, Supplementary Fig. 4A and B). These changes were reflected by similar changes in relative tumor weight (Fig. 4E and F). Expression of PCNA was also decreased by ligand activation of PPARβ/δ in MDA-MD-231-hPPARβ/δ xenografts as compared to controls; an effect not found in similarly treated MCF7-hPPARβ/δ xenografts (Fig. 5A and B). By contrast, expression of p65 was higher in ligand treated MCF7-hPPARβ/δ xenografts compared to controls and this effect was not found in similarly treated MDA-MB-231-hPPARβ/δ xenografts (Fig. 5A and B). Increased expression of ANGPTL4 mRNA confirmed effective activation of PPARβ/δ in the xenografts following treatment with GW0742 (Fig. 4G and H). Histopathological analysis revealed that the necrotic index was greatest in xenografts from ligand-treated MDA-MD-231-hPPARβ/δ xenografts (Supplementary Fig. 4C), and interestingly, ligand activation of PPARβ/δ caused an increase in the total necrotic region of xenografts from control MDA-MD-231-MigR1 cells compared to control (Supplementary Fig. 4D). Additionally, the percentage of necrotic area was increased by ligand activation of PPARβ/δ in control MDA-MD-231-MigR1 xenografts as compared to control MDA-MD-231-MigR1 xenografts. This increase in the percentage of necrotic area was higher in both control and ligand treated xenografts from MDA-MD-231-hPPARβ/δ cells (Supplementary Fig. 4E). These changes in necrosis were not observed in xenografts from MCF7 cells. Thus, despite the increased expression of PPARβ/δ in two human breast cancer cell line-derived tumors, the ratio of FABP5:CRABP-II (Fig. 5A and B) had no influence on relative tumorigenicity in vivo as suggested by others (23).
Figure 4.
The effect of over-expressing PPARβ/δ and/or ligand activation of PPARβ/δ on human breast cancer cell line xenografts. Morphology of representative xenografts from (A) MDA-MB-231-MigR1 (MigR1), and MDA-MB-231-hPPARβ/δ (hPPARβ/δ) in host mice treated with or without GW0742 or (B) MCF7-MigR1 (MigR1), and MCF7-hPPARβ/δ (hPPARβ/δ) in host mice treated with or without GW0742. Average tumor volume over time from the (C) MDA-MB-231 cell-derived xenograft groups or (D) MCF7 cell-derived xenograft groups. Average tumor weight at the end of the study from the (E) MDA-MB-231 cell-derived xenograft groups or (F) MCF7 cell-derived xenograft groups. qPCR analysis of ANGPTL4 mRNA from the (G) MDA-MB-231 cell-derived xenografts or (H) MCF7 cell-derived xenografts. Values represent the mean ± S.E.M.. Values with different letters are significantly different, P ≤ 0.05. *Significantly different than control, P ≤ 0.05.
Figure 5.
The effect of over-expressing PPARβ/δ and/or ligand activation of PPARβ/δ on tumor-marker proteins. Representative western blot analysis of xenografts from (A) MDA-MB-231-MigR1 (MigR1), MDA-MB-231-hPPARβ/δ (hPPARβ/δ) cells in host mice treated with or without GW0742 or (B) MCF7-MigR1 (MigR1), MCF7-hPPARβ/δ (hPPARβ/δ) cells in host mice treated with or without GW0742. Expression of PPARβ/δ, p65, PCNA, FABP5 and CRABP-II proteins were examined. Signals for each protein were normalized to that of ACTIN and represent the mean ± S.E.M.. Values represent the mean ± S.E.M.. Values with different letters are significantly different, P ≤ 0.05.
Discussion
Over-expression and/or ligand activation of PPARβ/δ in both an ER+ and ER− human breast cancer cell line has marked effects on cell growth, anchorage-dependent clonogenicity, and relative tumorigenicity in an ectopic xenograft model. The fact that ligand activation of PPARβ/δ inhibited ER+, MCF7 cell proliferation is consistent with a previous study (13) as this effect was only observed with 10 μM GW0742 in MCF7 cells cultured in medium with FCS. The inhibitory effect of ligand activation on cell proliferation was not observed in the ER−, MDA-MB-231 cells. Interestingly, when PPARβ/δ was over-expressed in both MDA-MB-231 and MCF7 cells, inhibition of cell proliferation was observed in both breast cancer cell lines, but the only cells that exhibited inhibition of cell proliferation following ligand activation of PPARβ/δ were the ER+, MCF7 cells. These results are in direct contrast to a previous study showing that ligand activation of PPARβ/δ increased proliferation of ER+ human breast cancer cell lines (MCF7 and T47-D) but not ER−, MDA-MB-231 cells, and that inducible expression of PPARβ/δ enhanced the increase in cell proliferation following ligand activation of PPARβ/δ (10). The reason for these opposing results cannot be clearly established from the present studies. However, it is important to note that the present study confirmed that the increase in expression of PPARβ/δ ranged from 5 to ~8-fold using quantitative western blotting, included a positive control for PPARβ/δ protein, and also validated a functional increase in expression of the PPARβ/δ target gene ANGPTL4, whereas the former study relied on expression of mRNA and qualitative immunofluorescence to demonstrate relative expression of PPARβ/δ. Further, of the two studies showing that ligand activation of PPARβ/δ inhibited human breast cancer cell lines, one study used Coulter counting and examined cell growth over time (13), while the present study used real-time assessment of cell proliferation. By contrast, Stephen and colleagues used an enzyme-linked assay that could be influenced by the induction of mitochondrial dehydrogenase enzymes, which are known to be induced by PPARβ/δ ligands (24). Thus, it remains possible that differences in the relative level of PPARβ/δ expression and/or the approach used to assess cell proliferation could explain the differences observed between these studies.
Results from the present studies are the first to demonstrate that ligand activation of PPARβ/δ inhibited anchorage-dependent clonogenicity in human breast cancer cells lines. It is also important to note that this effect was greater in ER−, MDA-MB-231 cells that over-expressed PPARβ/δ as compared to controls, and that this effect was not as marked in the ER+, MCF7 cells that over-expressed PPARβ/δ. These data are consistent with a previous study showing that anchorage-dependent clonogenicity was dose-dependently decreased by ligand activation of PPARβ/δ in a mouse mammary gland cancer cell line (18). Results from the present studies are also consistent with the observed inhibition of ectopic xenograft growth in vivo. For example, ligand activation of PPARβ/δ inhibited tumor growth in xenografts developing from injected ER−, MDA-MB-231 cells and ER+, MCF7 cells. While others have suggested that GW0742 can weakly interact with other nuclear receptors other than PPARβ/δ (25), the dose of GW0742 used for the in vivo studies in the present experiments likely resulted in average serum concentrations less than 1 μM as shown by others (26). Since this concentration is markedly lower than that required to weakly interact with other nuclear receptors in vitro, and the in vitro models over-estimate the concentration required to interact with a nuclear receptor in vivo, it is unlikely that the effects mediated by GW0742 are mediated by interactions with other receptors other than PPARβ/δ given the high affinity for GW0742 for this receptor. Additionally, over-expression of PPARβ/δ had a greater inhibitory effect on xenograft tumor growth in ER–, MDA-MB-231 cells as compared to ER+, MCF7 cells. The findings that over-expression of PPARβ/δ inhibited both anchorage-dependent clonogenicity and ectopic xenograft growth is of particular interest because expression of PPARβ/δ mRNA has been reported to be significantly lower in ductal breast carcinomas as compared to normal ductal epithelium in humans (27–29).
The mechanisms that underlie the observed inhibitory effect on xenograft tumor growth appear to be qualitatively different between ER− and ER+ human breast cancer cells. For example, decreased expression of PCNA was observed in ER−, MDA-MB-231 xenografts following ligand activation of PPARβ/δ and this effect was not found in similarly treated xenografts developing from ER+, MCF7 cells. There was also a marked increase in necrosis found in ER−, MDA-MB-231 xenografts that was enhanced by ligand activation of PPARβ/δ. This effect was not found in ER+, MCF7 xenografts. While the mechanisms that induce apoptosis and necrosis are not divergent, there is evidence suggesting that drugs that promote necrosis in tumors may be more efficacious for chemoprevention/chemotherapy as compared to drugs that induce apoptosis (30). This hypothesis deserves further evaluation with GW0742 and other PPARβ/δ agonists as this phenotype has never been observed to date. Combined, these data suggest that ER− breast cancer cells could be effectively targeted by developing an approach to increase expression of PPARβ/δ and by treatment with PPARβ/δ agonists. This hypothesis deserves further experimentation. In contrast, increased expression of p65 was noted in PPARβ/δ ligand treated ER+, MCF7 xenografts. This effect was not observed in other treatment groups of ER+, MCF7 xenografts or in the ER−, MDA-MB-231 xenografts. The reason why p65 expression was increased in PPARβ/δ ligand treated ER+, MCF7 xenografts cannot be determined by the current studies. However, it is worth noting that there are many reports showing that PPARβ/δ can bind p65 and interfere with NF-kB-dependent pro-inflammatory signaling (reviewed in (4, 6)). This suggests that the higher expression of p65 could be the result of its stabilization through its increased binding with PPARβ/δ and the inhibition of xenograft growth may be due to reduced p65-dependent pro-inflammatory signaling. Further studies are needed to examine this hypothesis.
Over-expression and/or ligand activation of PPARβ/δ had no influence on either staurosporine or UVB-induced apoptosis. Previous studies by others have suggested that PPARβ/δ promotes anti-apoptotic signaling through a variety of mechanisms (reviewed in (4, 6)). One hypotheses is that FABP5 can deliver ligands directly to PPARβ/δ and promote anti-apoptotic activity; this has been suggested in a breast cancer model (23). However, results from the present studies and other work have shown that despite increasing the intracellular concentration of PPARβ/δ and in the presence of FABP5, FABP5 does not function to deliver ligands to PPARβ/δ, and activating PPARβ/δ does not result in anti-apoptotic activities (20–22, 31). These findings re-emphasize the need for further studies examining the role of PPARβ/δ in apoptosis to clarify these disparities.
The results from the present studies can also not explain why other studies using mouse models suggest that PPARβ/δ promotes breast cancer. This has been suggested by studies showing that mammary tumorigenesis is mitigated in Cox2-null mice crossed with Pparβ/δ-null mice (15), studies showing that DMBA-induced mammary tumorigenesis is enhanced by ligand activation of PPARβ/δ (16), and studies using a transgenic mouse model showing enhanced mammary tumorigenesis by over-expression of mouse PPARβ/δ (17). One possible explanation is that there are species differences between the mouse and human PPARβ/δ, whereby the mouse PPARβ/δ promotes mammary tumorigenesis while the human PPARβ/δ inhibits mammary tumorigenesis. However, there are also many examples from other models suggesting that PPARβ/δ promotes cancer in mice and other examples suggesting that PPARβ/δ inhibits cancer in mice (reviewed in (1, 2, 4, 6)). For example, ligand activation of PPARβ/δ inhibits proliferation of a mouse mammary gland cancer cell line by increasing apoptosis and inhibits anchorage-dependent clonogenicity (18). Further, there is recent evidence from human studies suggesting that PPARβ/δ is a tumor suppressor. For example, others have shown that expression of PPARβ/δ mRNA is lower in ductal breast carcinomas as compared to non-transformed tissue (27–29, 32). Further, analysis of protein expression of human breast cancer as compared to control non-transformed tissue (The Human Protein Atlas, version 12, www.proteinatlas.org) indicates that expression of PPARβ/δ is low or not detectable in 11 of 12 samples (33). This is similar to analysis of PPARβ/δ protein expression of 195 tumors compared to control non-transformed tissue where expression of PPARβ/δ is markedly lower in tumors, including colon, prostate, ovarian, cervical, endometrial, head and neck, thymus, glioma, lymphoma, lung, melanoma, skin, testis, urothelial, renal, stomach, pancreatic and liver (33). This suggests that the idea of a species difference is unlikely and the confusion in the literature is more likely due to differences in experimental approaches and conditions employed to study the role of PPARβ/δ in cancer. Whether the immunocompromised nature of the mice used for the present studies influenced the outcome is also uncertain. Thus, the new stable ER−, MDA-MB-231 and ER+, MCF7 human breast caner cell lines characterized in the present study represent invaluable tools for future studies to more definitively determine the role of PPARβ/δ in breast cancer; an area of research that clearly requires more investigation.
Supplementary Material
Acknowledgments
Financial information: National Institutes of Health (CA124533, CA141029, CA140369, AA018863) J.M. Peters, and the National Cancer Institute Intramural Research Program (ZIABC005561, ZIABC005562, ZIABC005708) F.J. Gonzalez.
We gratefully acknowledge the Center for Quantitative Cell Analysis Facility at the Huck Institutes of Life Sciences of The Pennsylvania State University for their technical support with flow cytometry and data analysis.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Peters JM, Gonzalez FJ. Sorting out the functional role(s) of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in cell proliferation and cancer. Biochim Biophys Acta. 2009;1796:230–41. doi: 10.1016/j.bbcan.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters JM, Hollingshead HE, Gonzalez FJ. Role of peroxisome-proliferator-activated receptor β/δ (PPARβ/δ) in gastrointestinal tract function and disease. Clin Sci (Lond) 2008;115:107–27. doi: 10.1042/CS20080022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters JM, Morales JL, Gonzales FJ. Modulation of gastrointestinal inflammation and colorectal tumorigenesis by peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) Drug Discovery Today: Disease Mechanisms. 2011;8:e85–e93. doi: 10.1016/j.ddmec.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters JM, Shah YM, Gonzales FJ. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat Rev Cancer. 2012;12:181–95. doi: 10.1038/nrc3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khozoie C, Borland MG, Zhu B, Baek S, John S, Hager GL, et al. Analysis of the peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) cistrome reveals novel co-regulatory role of ATF4. BMC genomics. 2012;13:665. doi: 10.1186/1471-2164-13-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters JM, Foreman JE, Gonzalez FJ. Dissecting the role of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in colon, breast and lung carcinogenesis. Cancer Metastasis Rev. 2011;30:619–40. doi: 10.1007/s10555-011-9320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girroir EE, Hollingshead HE, He P, Zhu B, Perdew GH, Peters JM. Quantitative expression patterns of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) protein in mice. Biochem Biophys Res Commun. 2008;371:456–61. doi: 10.1016/j.bbrc.2008.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kilgore KS, Billin AN. PPARβ/δ ligands as modulators of the inflammatory response. Curr Opin Investig Drugs. 2008;9:463–9. [PubMed] [Google Scholar]
- 9.Grimaldi PA. Metabolic and nonmetabolic regulatory functions of peroxisome proliferator-activated receptor β. Curr Opin Lipidol. 2010;21:186–91. doi: 10.1097/mol.0b013e32833884a4. [DOI] [PubMed] [Google Scholar]
- 10.Stephen RL, Gustafsson MC, Jarvis M, Tatoud R, Marshall BR, Knight D, et al. Activation of peroxisome proliferator-activated receptor δ stimulates the proliferation of human breast and prostate cancer cell lines. Cancer Res. 2004;64:3162–70. doi: 10.1158/0008-5472.can-03-2760. [DOI] [PubMed] [Google Scholar]
- 11.Aung CS, Faddy HM, Lister EJ, Monteith GR, Roberts-Thomson SJ. Isoform specific changes in PPARα and β in colon and breast cancer with differentiation. Biochem Biophys Res Commun. 2006;340:656–60. doi: 10.1016/j.bbrc.2005.12.061. [DOI] [PubMed] [Google Scholar]
- 12.Buttitta LA, Edgar BA. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr Opin Cell Biol. 2007;19:697–704. doi: 10.1016/j.ceb.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girroir EE, Hollingshead HE, Billin AN, Willson TM, Robertson GP, Sharma AK, et al. Peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) ligands inhibit growth of UACC903 and MCF7 human cancer cell lines. Toxicology. 2008;243:236–43. doi: 10.1016/j.tox.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palkar PS, Borland MG, Naruhn S, Ferry CH, Lee C, Sk UH, et al. Cellular and Pharmacological Selectivity of the PPARβ/δ Antagonist GSK3787. Mol Pharmacol. 2010;78:419–30. doi: 10.1124/mol.110.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh M, Ai Y, Narko K, Wang Z, Peters JM, Hla T. PPARδ is pro-tumorigenic in a mouse model of COX-2-induced mammary cancer. Prostaglandins Other Lipid Mediat. 2009;88:97–100. doi: 10.1016/j.prostaglandins.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin Y, Russell RG, Dettin LE, Bai R, Wei ZL, Kozikowski AP, et al. Peroxisome proliferator-activated receptor δ and γ agonists differentially alter tumor differentiation and progression during mammary carcinogenesis. Cancer Res. 2005;65:3950–7. doi: 10.1158/0008-5472.CAN-04-3990. [DOI] [PubMed] [Google Scholar]
- 17.Yuan H, Lu J, Xiao J, Upadhyay G, Umans R, Kallakury B, et al. PPARδ Induces Estrogen Receptor-Positive Mammary Neoplasia through an Inflammatory and Metabolic Phenotype Linked to mTOR Activation. Cancer Res. 2013;73:4349–61. doi: 10.1158/0008-5472.CAN-13-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foreman JE, Sharma AK, Amin S, Gonzalez FJ, Peters JM. Ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) inhibits cell growth in a mouse mammary gland cancer cell line. Cancer Lett. 2010;288:219–25. doi: 10.1016/j.canlet.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–92. [PubMed] [Google Scholar]
- 20.Borland MG, Khozoie C, Albrecht PP, Zhu B, Lee C, Lahoti TS, et al. Stable over-expression of PPARβ/δ and PPARγ to examine receptor signaling in human HaCaT keratinocytes. Cell Signal. 2011;23:2039–50. doi: 10.1016/j.cellsig.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foreman JE, Chang WC, Palkar PS, Zhu B, Borland MG, Williams JL, et al. Functional characterization of peroxisome proliferator-activated receptor-β/δ expression in colon cancer. Mol Carcinog. 2011;50:884–900. doi: 10.1002/mc.20757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borland MG, Foreman JE, Girroir EE, Zolfaghari R, Sharma AK, Amin SM, et al. Ligand Activation of Peroxisome Proliferator-Activated Receptor-β/δ (PPARβ/δ) Inhibits Cell Proliferation in Human HaCaT Keratinocytes. Mol Pharmacol. 2008;74:1429–42. doi: 10.1124/mol.108.050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schug TT, Berry DC, Toshkov IA, Cheng L, Nikitin AY, Noy N. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARβ/δ to RAR. Proc Natl Acad Sci U S A. 2008;105:7546–51. doi: 10.1073/pnas.0709981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka T, Yamamoto J, Iwasaki S, Asaba H, Hamura H, Ikeda Y, et al. Activation of peroxisome proliferator-activated receptor δ induces fatty acid β-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci U S A. 2003;100:15924–9. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nandhikonda P, Yasgar A, Baranowski AM, Sidhu PS, McCallum MM, Pawlak AJ, et al. Peroxisome proliferation-activated receptor δ agonist GW0742 interacts weakly with multiple nuclear receptors, including the vitamin D receptor. Biochemistry. 2013;52:4193–203. doi: 10.1021/bi400321p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham TL, Mookherjee C, Suckling KE, Palmer CN, Patel L. The PPARδ agonist GW0742X reduces atherosclerosis in LDLR−/− mice. Atherosclerosis. 2005;181:29–37. doi: 10.1016/j.atherosclerosis.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 27.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 28.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiaris H, Schally AV. Apoptosis versus necrosis: which should be the aim of cancer therapy? Proc Soc Exp Biol Med. 1999;221:87–8. doi: 10.1046/j.1525-1373.1999.d01-59.x. [DOI] [PubMed] [Google Scholar]
- 31.Rieck M, Meissner W, Ries S, Muller-Brusselbach S, Muller R. Ligand-mediated regulation of peroxisome proliferator-activated receptor (PPAR) β/δ: a comparative analysis of PPAR-selective agonists and all-trans retinoic acid. Mol Pharmacol. 2008;74:1269–77. doi: 10.1124/mol.108.050625. [DOI] [PubMed] [Google Scholar]
- 32.Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–50. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.