Abstract
Rhabdomyosarcomas (RMS) are the most frequent soft-tissue sarcoma in children and characteristically show features of developing skeletal muscle. The alveolar subtype is frequently associated with a PAX3-FOXO1 fusion protein that is known to contribute to the undifferentiated myogenic phenotype of RMS cells. Histone methylation of lysine residues controls developmental processes in both normal and malignant cell contexts. Here we show that JARID2, that encodes a protein known to recruit various complexes with histone methylating activity to their target genes, is significantly overexpressed in RMS with PAX3-FOXO1 compared to fusion gene negative RMS (t test p<0.0001). Multivariate analyses showed higher JARID2 levels are also associated with metastases at diagnosis, independent of fusion gene status and RMS subtype (n= 120; p=0.039). JARID2 levels were altered by silencing or over-expressing PAX3-FOXO1 in RMS cell lines with and without the fusion gene, respectively. Consistent with this, we demonstrated that JARID2 is a direct transcriptional target of the PAX3-FOXO1 fusion protein. Silencing JARID2 resulted in reduced cell proliferation coupled with myogenic differentiation including increased expression of MYOGENIN (MYOG) and MYOSIN LIGHT CHAIN (MYL1) in RMS cell lines representative of both the alveolar and embryonal subtypes. Induced myogenic differentiation was associated with a decrease in JARID2 levels and this phenotype could be rescued by overexpressing JARID2. Furthermore, we that showed JARID2 binds to and alters the methylation status of histone H3 lysine 27 in the promoter regions of MYOG and MYL1 and that the interaction of JARID2 at these promoters is dependent upon EED, a core component of the Polycomb Repressive Complex 2 (PRC2). Therefore JARID2 is a downstream effector of PAX3-FOXO1 that maintains an undifferentiated myogenic phenotype that is characteristic of RMS. JARID2 and other components of PRC2 may represent novel therapeutic targets for treating RMS patients.
Keywords: Rhabdomyosarcoma, PAX3-FOXO1, histone methylation, Polycomb Repressive Complex 2, JARID2, myogenic differentiation
Introduction
Rhabdomyosarcomas (RMS) are the most frequent pediatric soft tissue sarcomas and resemble developing skeletal muscle. There are two main histological subtypes, alveolar (ARMS) and embryonal (ERMS). The majority of ARMS are associated with characteristic fusion genes, either PAX3-FOXO1 or, less frequently, PAX7-FOXO1 and rarer variants. Patients with PAX3-FOXO1 fusion gene positive tumors are generally considered to have a poor prognosis1-4. The fusion genes encode potent transcriptional activators that contribute to the pathogenesis of these tumors through aberrantly driving the expression of multiple genes5-8. Silencing the PAX3-FOXO1 fusion gene results in myogenic differentiation in RMS cell lines9. PAX3-FOXO1 has been shown to suppress the transcriptional activity of MyoD-target genes and direct downstream targets of the fusion gene may also be involved in suppressing differentiation9-11.
Histone methylation is a key element of chromatin-based modifications that regulates a number of cellular processes including DNA replication, DNA repair, and gene transcription. Histone demethylase gene family members (HDMs) regulate gene expression by removing the methyl marks on histone tails to either activate or repress transcription12. There are two main families of HDMs; the KDM1 family, which demethylate mono- and dimethylated lysines, and the Jumonji (JmjC) domain-containing demethylases, which demethylate mono-, di- and tri-methyl marks. Several HDMs have been shown to be involved in cancer, including KDM5B, which is overexpressed in breast and prostate cancer13, 14, and LSD1, which is overexpressed in neuroblastomas and sarcomas15, 16. HDM gene family members are involved in normal developmental and differentiation processes and recently an isoform of the KDM4A subfamily has been implicated in myogenic differentiation17.
Transcriptional control via histone methylation, particularly in the process of differentiation, has been shown to be under tight regulation by the methyltransferase-containing Polycomb Repressive Complex 2 (PRC2)18-20. PRC2 contains 4 core subunits; EED and RbAp46/48, two WD40 domain proteins and EZH1/EZH2 and SUZ12 that confer methylating activity to the complex18-21. PRC2 may also contain the jumonji domain-containing interacting protein JARID2, although JARID2 lacks critical active site residues required for demethylase catalytic activity12, 22, 23. EZH2 in particular has been linked to various cancer types and has been the focus of studies to target PRC2 as a potential therapeutic strategy24.
Here we identified several HDM gene family members as highly expressed in primary RMS relative to normal skeletal muscle including JARID2, which correlated with metastatic behavior and showed highest levels in the fusion positive alveolar subtype. We demonstrate that JARID2 expression is modulated by, and is a direct downstream transcriptional target of, PAX3-FOXO1. JARID2-containing complexes include Rb and Nkx2.5/GATA4 confer methyltransferase activity at H3K9, and PRC2 that has been reported to both activate and repress H3K27 methylation19, 20, 25-27. As we also identified high expression levels of multiple components of PRC2-EZH2 in RMS, we further investigated the biological roles for JARID2 and its association with PRC2 in RMS cells. We demonstrate that JARID2 binds to the promoter region of specific myogenic genes in RMS cells and, in conjunction with a PRC2 protein, regulates H3K27 tri-methylation at these promoters. Critically, this is associated with maintaining the undifferentiated myogenic phenotype of RMS cells.
Results
Histone demethylases are highly expressed in rhabdomyosarcomas
To identify HDM gene family members that may be involved in maintaining the undifferentiated myogenic phenotype of RMS, we assessed mRNA levels of 32 HDMs in a panel of 101 RMS patient samples relative to 30 skeletal muscle samples using our previously published patient ITCC/CIT dataset (Innovative Therapies for Children with Cancer/Carte d’Identité des Tumeurs)28 and publicly available Affymetrix expression profiling data for skeletal muscle (SkM), embryonic (ESC) and mesenchymal (MSC) stem cells. These comparators were included because MSCs are a putative cell of origin for RMS29 and RMS show features of skeletal muscle differentiation, with potential for RMS cell lines to be induced to form muscle myotubes30. Table 1 shows the HDM gene family members most highly expressed in RMS relative to skeletal muscle and the expression levels of all HDMs are represented in a heatmap (Supplementary Figure S1). Eleven HDMs were overexpressed in the RMS patient sample set, whereas none were significantly under-expressed (Expression < 0.5, p<0.0016; Supplementary Table S1). As fusion gene status appears highly relevant to the biology and clinical behaviour of RMS1, 2, 28, levels of HDM gene family members were also analyzed in samples according to their fusion gene status. FBXL19, KDM2A and KDM3A were under-expressed in fusion gene positive (PAX3-FOXO1 and PAX7-FOXO1) versus negative samples, whereas JARID2 and JMJD1C were overexpressed in all RMS samples, but most highly in the fusion gene positive subset (p<0.0001, Supplementary Tables S2 and S3). The HR and KDM6B genes were overexpressed in the PAX3-FOXO1 (p=0.0005) and PAX7-FOXO1 (p<0.0001) subsets, respectively.
Table 1. Histone demethylase (HDM) gene family members identified in gene expression profiling data as overexpressed in rhabdomyosarcomas (RMS) relative to skeletal muscle (see also supplementary tables).
| ENTREZ | Gene | Probe | Fold change over SkM | p-value |
|---|---|---|---|---|
| 10765 | KDM5B | 201549_x_at | 6.5927 | 9.98E-14 |
| 221037 | JMJD1Ca | 221763_at | 5.3574 | 1.94E-39 |
| 55818 | KDM3A | 212689_s_at | 3.2628 | 1.47E-27 |
| 84678 | KDM2B | 226215_s_at | 2.9793 | 5.89E-38 |
| 3720 | JARID2a | 203297_s_at | 2.8843 | 1.31E-21 |
| 5927 | KDM5A | 226371_at | 2.6749 | 6.20E-26 |
| 23030 | KDM4B | 212496_s_at | 2.3355 | 9.68E-08 |
| 23210 | JMJD6 | 212723_at | 2.1731 | 1.83E-10 |
| 84864 | MINA | 213188_s_at | 2.0883 | 4.45E-10 |
| 7403 | KDM6A | 203992_s_at | 2.0847 | 6.51E-12 |
| 23133 | PHF8 | 212916_at | 2.0529 | 6.57E-20 |
Also overexpressed in fusion positive versus fusion negative RMS
JARID2 is highly expressed in fusion positive ARMS and correlates with metastasis at diagnosis
As PAX3-FOXO1 plays a key role in the biology and clinical behavior of ARMS3, 28, 31 we identified HDM gene family members that might be directly regulated by the fusion protein. As JARID2 and JMJD1C were most highly overexpressed in fusion positive patient samples from analysis of the ITCC/CIT gene expression data (Supplementary Tables S2 and S3) we focused on these. JARID2 was more highly differentially expressed between fusion gene positive and negative samples (JARID2: fold difference=2.6249, p<0.0001, R2=0.4293; JMJD1C: fold difference=2.0359, p<0.0001, R2=0.1666; Figure 1A). Quantitative (q)RT-PCR analysis of 120 samples, including 45 overlapping samples from the ITCC/CIT dataset (39 PAX3/7-FOXO1 fusion gene positive and 81 fusion gene negative, detailed in Supplementary Table S4), confirmed a significant difference in levels of JARID2 expression associated with fusion gene status (n=120, p<0.0001) (Figure 1B). JARID2 RNA and protein levels in RMS cell lines broadly correlated indicating that mRNA quantification is likely representative of protein levels. It was not possible to directly assess protein expression in primary tumors due to insufficient material for Western analysis and the lack of JARID2 antibodies suitable for reliable immunohistochemical analysis.
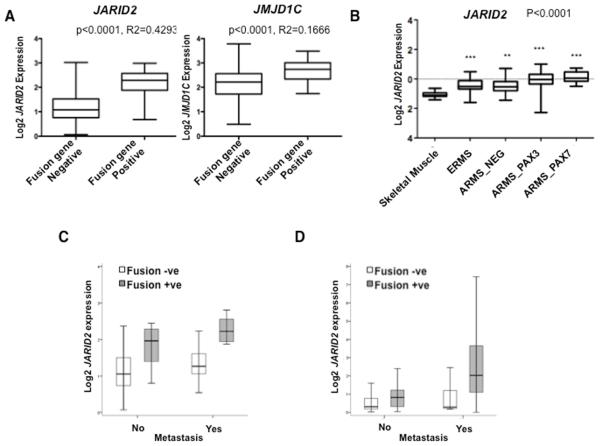
Figure 1. JARID2 is more highly expressed in fusion positive RMS patients and correlates with metastasis and poor clinical outcome.
(A) ITCC/CIT affymetrix microarray expression profiling data for RMS fusion gene positive and fusion gene negative patient samples (n=101). These were analyzed for fold differences (FD) by T-test (p<0.0001) for both JARID2 (FD=2.6249, p<0.0001 and R2=0.4293) and JMJD1C (FD=2.0359, p<0.0001, R2=0.1666). (B) Confirmation of JARID2 expression levels in RMS patient samples (alveolar RMS with a PAX3-FOXO1 fusion gene; ARMS_PAX3, ARMS with PAX7-FOXO1; ARMS_PAX7, embryonal RMS; ERMS and ARMS without a fusion gene; ARMS_NEG) (n=120) by TaqMan quantitative RT-PCR analyses. All samples were compared to normal skeletal muscle (p<0.0001, one-way ANOVA). Three stars represent p-values <0.001; two stars represent p-values <0.01. (C) Box-plots illustrate higher levels of JARID2 expression in fusion gene positive and metastatic RMS by analyses of ITCC/CIT gene expression profiling data (p=0.0002, n=101) and (D) by quantitative RT-PCR analyses (p=0.039, n=120).
Analysis of the ITCC/CIT gene expression profiling data found a strong correlation with metastasis at diagnosis (p=0.0002, Figure 1C), which was confirmed in the qRT-PCR dataset (p=0.008, Figure 1D). To test whether this was independent of fusion gene status, multivariate analysis was performed using a stepwise logistic regression model of the qRT-PCR data, considering factors including histological subtype and fusion gene status (Supplementary Table S5). This showed an independent association between high JARID2 expression and metastasis at diagnosis (p=0.039). High JARID2 expression also correlated with metastasis at diagnosis within fusion gene positive cases (p=0.0073, n=39) and within the PAX3-FOXO1 positive subgroup (p=0.0189, n=37). However, Kaplan-Meier analysis showed that higher JARID2 expression was not significantly associated with overall survival (OS) in either fusion gene positive RMS (p=0.386, n=39) or fusion gene negative RMS (p=0.089, n=81), although the latter may be considered a trend towards significance.
PRC2-EZH2 complex members are overexpressed in RMS
JARID2 is associated with PRC2 in ESC and has been linked to the differentiation of myoblasts19, 20, 25-27, 32. Therefore, we also analyzed our ITCC/CIT gene expression profiling data for expression of other known members of PRC2, including AEBP2 and PCL2 along with the core subunits. EZH2 was found most highly overexpressed relative to skeletal muscle in all RMS and also in RMS grouped according to histology and fusion gene status (Fold change in all RMS samples=24.03, p<0.0001, Supplementary Tables S6 and S7). EED, SUZ12, AEBP2 and PCL2 were also overexpressed in all RMS (Supplementary Tables S6 and S7). EZH1 has also been associated with PRC2 in differentiated skeletal myoblasts32. However, EZH1 was under-expressed in RMS (Supplementary Table S7), consistent with predominance of an EZH2-containing PRC2 complex in RMS.
JARID2 is a direct transcriptional target of PAX3-FOXO1
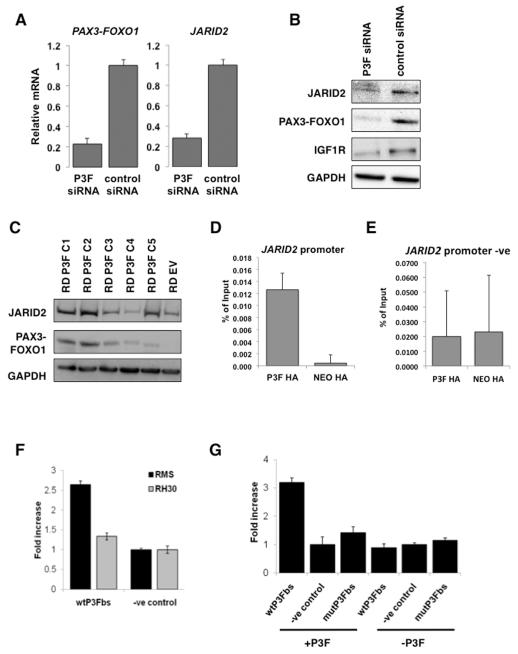
To determine whether the association of higher JARID2 levels with fusion positive ARMS is because JARID2 is a downstream target of the PAX3-FOXO1 fusion protein we used a previously validated PAX3-FOXO1 specific siRNA that targets the fusion junction to specifically silence PAX3-FOXO1 expression in the RH30 cell line9. This showed a reduction in JARID2 RNA and protein levels by TaqMan and Western blot analysis, respectively (Figure 2A,B). IGF1R, a known target of the fusion gene10, showed similar reduction to JARID2 at the protein level (Figure 2B). Conversely, constitutive overexpression of PAX3-FOXO1 using a hemaglutinin (HA)-tagged PAX3-FOXO1 construct (HA-PAX3-FOXO1) in multiple clones of the fusion negative RD cell line resulted in variable increases in JARID2 expression at the RNA (data not shown) and protein levels (Figure 2C). We then tested whether JARID2 is a direct downstream target of the fusion protein. A PAX3 consensus binding sequence CGTGAC[CT] has been described as most frequently bound by the PAX3-FOXO1 fusion protein10. We identified a potential PAX3-FOXO1 binding motif of CGTGACT −187/−180 base pairs upstream of the translation start site in the JARID2 promoter. To determine whether the PAX3-FOXO1 binds to this sequence in ARMS cell lines we first introduced the HA-PAX3-FOXO1 construct into the RH30 cell line and performed chromatin immunoprecipitation (ChIP) pull down using a HA antibody (Supplementary Figure S2A). Enrichment of PAX3-FOXO1 bound DNA was confirmed using primers surrounding the previously described IGF1R PAX3-FOXO1 binding site (10, Supplementary Figure S2B). ChIP-PCR of the putative PAX3-FOXO1 binding motif in the JARID2 promoter confirmed binding of PAX3-FOXO1 (Figure 2D). A region upstream of the putative PAX3-FOXO1 binding site was also analyzed as a negative control and IGF1R as a positive control (Figure 2E, Supplementary Figure S2B). Using a luciferase reporter assay surrounding the −187/−180 consensus motif (wtP3Fbs) we observed a greater than two fold increase in luciferase activity in vitro in the RMS-01 cell line (Figure 2F). Only a small increase in reporter activity was seen in RH30, likely due to the lower levels of PAX3-FOXO1 fusion gene protein in RH30 compared to RMS-01 as we have previously shown33. Co-transfection of this construct with a HA-PAX3-FOXO1 into the fusion gene negative ERMS line RD resulted in a 3-fold increase in luciferase activity over the negative control (Figure 2G). To further confirm these findings, we co-transfected a reporter construct containing a mutated binding sequence CATTTCT (mutP3Fbs) along with HA-PAX3-FOXO1 into RD and showed negligible luciferase activity with this mutant construct compared to the wild type (Figure 2G). Taken together these data are consistent with direct PAX3-FOXO1-mediated JARID2 expression.
Figure 2. JARID2 is a direct downstream target of the PAX3-FOXO1 fusion protein.
(A) Silencing of PAX3-FOXO1 (P3F) by siRNA results in a decrease in expression of JARID2 at the RNA level by quantitative RT-PCR analyses of the RH30 cell line at 24hrs post transfection. Error bars represent SD of 3 replicates. (B) Western blot demonstrating reduction in JARID2 protein levels after PAX3-FOXO1 siRNA reduction in the RH30 cell line at 48hrs post transfection. IGF1R is included as a positive downstream control target of PAX3-FOXO1. (C) Overexpression of PAX3-FOXO1 in the fusion gene negative embryonal RMS cell line RD results in an increase in expression of JARID2 at the protein level. Western blot showing 5 PAX3-FOXO1 clones (RD P3F C1-5) and an empty vector control clone (EV). (D) ChIP-PCR on RH30 HA-tagged PAX3-FOXO1 over-expressing clones demonstrating that PAX3-FOXO1 binds to the JARID2 promoter sequence by quantified enrichment at the −180/−187 motif binding fragment over the vector only (NEO) controls. Error bars represent SD of 3 replicates. (E) Negative region in the JARID2 promoter for PAX3-FOXO1 bound DNA upstream of the JARID2 PAX3-FOXO1 binding site by Sybr Green PCR as a negative control. Error bars represent the SD of 3 replicates. (F) Luciferase enhancer activity for the −180/−187 motif binding fragment in PAX3-FOXO1 positive RMS cell lines, RMS-01 and RH30. Fold increase corresponds to luciferase enhancer activity. Error bars represent SD of 3 replicates. (G) Luciferase enhancer activity for the −180/−187 motif binding fragment in the fusion negative RD cell line, with and without PAX3-FOXO1. Fold increase corresponds to luciferase enhancer activity. Error bars represent SD of 3 replicates.
JARID2 affects cell proliferation in RMS cell lines
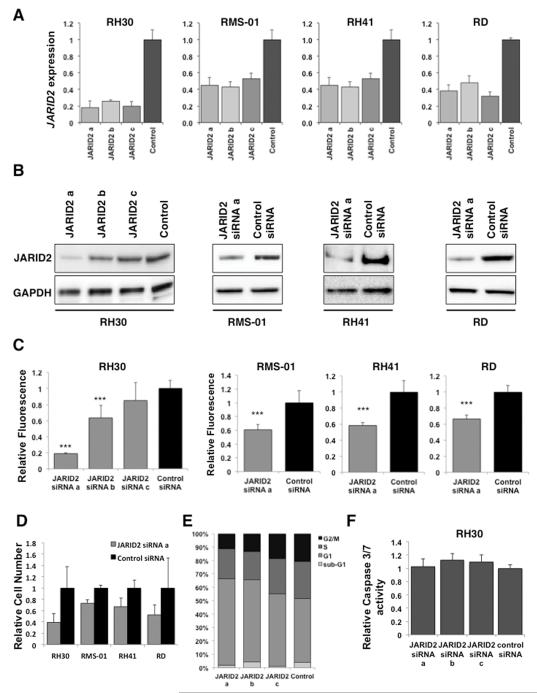
To investigate how JARID2 contributes to the phenotype of RMS, JARID2 mRNA levels were reduced in RH30, RMS-01, RH41 and RD cell lines using 3 siRNAs (siRNA a, b and c) (Figure 3A). Reduction in JARID2 at the protein level was confirmed by Western blot for 3 siRNAs in RH30, and with siRNA a in RMS-01, RH41 and RD cell lines (Figure 3B). This reduction of JARID2 expression resulted in a significant decrease in proliferation in cell lines at 96 hours post transfection (p<0.005, Figure 3C). These results were confirmed using a pool of siRNAs for JARID2 in RH30 (p<0.005, Supplementary Figure S3B). Decreased proliferation was confirmed for all cell lines using a cell count assay (CyQuant) following JARID2 knockdown with siRNA a (Figure 3D). We performed cell cycle analysis by flow cytometry on JARID2-silenced RH30 cells and observed an increase in the number of cells in the G1 fraction of the cell cycle, consistent with a reduction in proliferation (Figure 3E). No significant increase in Caspase 3/7 activity was seen after silencing JARID2 in RH30 suggesting that the reduction in cell proliferation was not due to apoptosis (Figure 3F, Supplementary Figure S3C).
Figure 3. Silencing JARID2 in RMS results in decreased cell viability without apoptosis.
Knockdown of JARID2 mRNA by 3 siRNAs (a, b and c) in the cell lines indicated (A) by quantitative RT-PCR analyses (at 24 hours post-transfection, error bars represent SD of 3 replicates) and (B) by Western blotting (72hours post-transfection). (C) Silencing of JARID2 by siRNA results in decreased viability by the MTS metabolic assay. The values of the absorbance are normalised to the control siRNA and are shown as mean of 6 replicates ± SD (***p< 0.005, Student’s t-test). (D) Cell count (CyQuant) analysis after JARID2 silencing in all 4 cell lines at 72 hours post transfection. Fluorescence values were normalised to the control siRNA and are shown as mean of 6 replicates ± SD. (E) Cell cycle analysis of JARID2-silenced RH30 cells by Propidium Iodide staining followed by flow cytometry, n=3. (F) Lack of caspase activity in JARID2 silenced RH30 cells at 72hrs post transfection using the 3 JARID2 siRNAs. The values of the absorbance were normalised to the control siRNA and are shown as mean of 6 replicates ± SD.
JARID2 maintains the undifferentiated myogenic phenotype of RMS
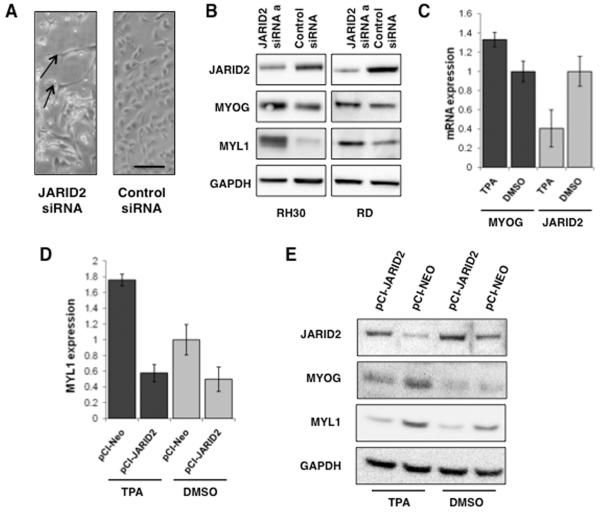
We observed that JARID2 knockdown resulted in an elongated cell morphology and appearance of cross-striations in some RH30 cells, consistent with a more differentiated myogenic phenotype (Figure 4A). Myogenic differentiation was confirmed by the detection of increased levels of MYOGENIN (MYOG), an early marker of differentiation, and MYOSIN LIGHT CHAIN (MYL1) protein expression, a marker of terminal differentiation, at 72hrs post siRNA transfection in both RH30 and RD cell lines (Figure 4B).
Figure 4. Silencing JARID2 results in myogenic differentiation and TPA-induced differentiation can be reversed by increasing JARID2 levels in RMS cells.
(A) Phase contrast image of RH30 cells with and without JARID2 siRNA silencing 72 hours post transfection (scale bar, 100 microns). Arrows show elongated cells consistent with myogenic differentiation. (B) Myogenin (MYOG) and Myosin Light Chain (MYL1) expression in RH30 and RD by Western blotting 72hrs after transfection with siRNAa against JARID2. (C) JARID2 expression is reduced in TPA-induced differentiation of RMS cell lines. MYOG and JARID2 mRNA expression in RD cells at 72hrs post TPA addition relative to DMSO control. Dark bars represent expression of MYOG in TPA-induced and DMSO control samples. Light grey bars represent JARID2 expression in these samples. Error bars represent SD of 3 replicates. (D) JARID2 overexpression in the RD cell line reduced expression of MYL1 expression in TPA-induced cells relative to DMSO vector-only control (pCI-Neo). Error bars represent SD of 3 replicates. (E) Confirmation of rescue of myogenic marker expression with overexpression of JARID2 in the RD cell line by Western blot. JARID2 overexpression in the RD cell line reduces expression of MYOG and MYL1 in TPA-induced cells relative to DMSO vector-only control (pCI-Neo).
To further investigate the role of JARID2 in myogenic differentiation, we analyzed the expression levels of JARID2 in RD cells induced to differentiate with 12-O-tetradecanoylphorbol-1-acetate (TPA). Addition of TPA to RD cells was associated with MYOG induction, consistent with differentiation, and a decrease in JARID2 RNA expression levels (Figure 4C). Addition of Differentiation Media (DM) to human myoblasts was associated with the induction of MYOG, MYOD and MYL1 and a decrease in JARID2 RNA expression (Supplementary Figure S4).
To determine whether JARID2 overexpression could reverse the effects of TPA-induced differentiation we transiently overexpressed a constitutively active wild type JARID2 clone in control and TPA treated RD cells and analyzed the effects on MYOG and MYL1. Overexpression of JARID2 (Supplementary Figure S5) in TPA-induced cells resulted in decreased MYL1 RNA and protein levels (Figure 4D,E), consistent with the high JARID2 levels in RMS contributing to its undifferentiated phenotype.
JARID2 binds to the MYOG and MYL1 promoters in RMS cells as part of PRC2
Since JARID2 has been associated with transcriptional repression via H3K27me3 at the myogenin promoter in mouse myoblasts18, we tested whether it is directly involved in repression of the MYOG gene in RMS cells through a mechanism involving this methyl mark. ChIP-qPCR using a JARID2 or H3K27me3 antibody confirmed binding at a region in the MYOG promoter previously described as being associated with H3K27me3 in the mouse myogenin promoter18 (Figure 5A). This demonstrates that JARID2 binds to the MYOG promoter in RMS cells at a region associated with the H3K27 tri-methyl mark.
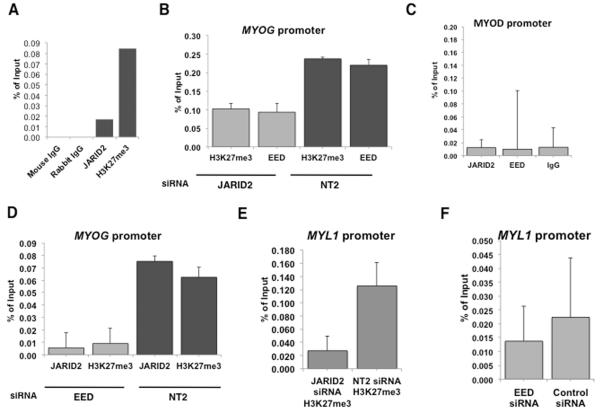
Figure 5. JARID2 binds to the MYOG and MYL1 promoters and alters H3K27me in association with a component of PRC2.
(A) ChIP qPCR showing fold enrichment at −1.5kb in the MYOG promoter over the IgG ChIP sample using JARID2 and H3K27me3 antibodies in RMS-01 cell line. (B) siRNA silencing of JARID2 reduces H3K27me3 and binding of EED at the MYOG promoter. Light grey bars represent JARID2 knockdown followed by ChIP with either an H3K27me3 or EED antibody, dark grey bars a non-targeting control siRNA (NT2). Error bars represent SD of 3 replicates. (C) JARID2 and EED do not bind to the MYOD promoter. ChIP pull down using a JARID2 antibody, EED antibody or IgG control antibody, followed by Sybr Green PCR of a region encompassing a CpG island in the MYOD promoter (−94 to +64). (D) Silencing of EED also reduces JARID2 binding and H3K27me3 at the at the MYOG promoter. Light grey bars represent EED knockdown, dark grey bars a non-targeting control siRNA (NT2). Error bars represent SD of 3 replicates. (E) Silencing of JARID2 versus control siRNA (NT2) reduces H3K27me3 at the MYL1 promoter. Error bars represent SD of 3 replicates. (F) Silencing of EED versus control siRNA (NT2) reduces JARID2 binding at the MYL1 promoter. Error bars represent SD of 3 replicates.
To determine whether JARID2 contributes to repression of MYOG transcription via H3K27me3 we performed ChIP pull down on JARID2-silenced and control siRNA RH30 cells using an H3K27me3 antibody. H3K27me3 was reduced at the MYOG promoter with JARID2 knockdown, thus indicating a role for JARID2 in repressing MYOG transcription (Figure 5B). ChIP-qPCR using an EED antibody on JARID2-silenced cells showed a reduction in binding at the H3K27me3 region in the MYOG promoter (Figure 5B). We used an EED antibody as representative of PRC2 rather than EZH2 to prevent possible EZH1 compensation confounding the experiment34. A region in the MYOD promoter was used as a negative control in these ChIP experiments 18 (Figure 5C). Results were supported by performing ChIP on EED silenced RH30 cells using JARID2 and H3K27me3 antibodies (Figure 5D).
Consistent with silencing of JARID2 in RMS cell lines inducing MYL1, ChIP analysis also demonstrated that JARID2 binds to the MYL1 promoter region, and that silencing of JARID2 results in a reduction of H3K27me3 at this promoter (Figure 5E). ChIP pull down on EED silenced RH30 cells using JARID2 antibody confirmed that JARID2 binds at the MYL1 promoter as part of PRC2 (Figure 5F). Taken together, these data show that JARID2, in conjunction with PRC2, affects H3K27me3 status at the promoter regions of MYOG and MYL1 that is associated with repressing MYOG and MYL1 expression in RMS cells.
Discussion
In this study we have shown that a number of HDM gene family members are highly expressed in RMS patient samples and that, of these, JARID2 plays a key role in maintaining the undifferentiated phenotype of RMS cells. Although JARID2 is highly expressed in all RMS, levels were significantly higher in fusion-gene positive alveolar samples, in-keeping with its inclusion in a PAX3-FOXO1-dependent gene signature1, 28. We demonstrated that JARID2 levels were altered through modulating PAX3-FOXO1 expression and that it is a direct downstream target of the PAX3-FOXO1 fusion protein, which binds to the JARID2 promoter in ARMS. Silencing JARID2 reduced the proliferative rate of RMS cells through inducing myogenic differentiation. As reduction of PAX3-FOXO1 levels also results in a more differentiated skeletal muscle cell morphology9, 35, JARID2 can be considered an effector of PAX3-FOXO1 and is involved in maintaining the undifferentiated phenotype that is characteristic of ARMS.
In multivariate analyzes of RMS patient samples, we found that high JARID2 expression levels were an independent factor that correlated with metastasis at diagnosis in all RMS and in the fusion gene subgroup. The degree of myogenic differentiation in RMS has been previously described to be inversely related to the rate of cellular proliferation, migratory behaviour and invasiveness2, 36-38. Therefore our findings are consistent with JARID2 levels adversely affecting the clinical behavior of tumors through its impact on maintaining an undifferentiated phenotype.
JARID2, the founding member of the Jumonji family, has been shown to be required for developmental processes such as cardiac myocyte differentiation and neural tube formation25, 39-41. ESCs lacking JARID2 fail to respond to differentiation signals due to a delay in the repression of genes usually down-regulated during differentiation of ESCs26. However, contrary to our findings in RMS cells, silencing JARID2 in ESCs was not sufficient to reduce cell growth or induce differentiation26. This may be due to JARID2 acting within a more differentiated cell context in RMS. JARID2 expression levels were lower in MSCs than in both ESCs and RMS, which showed similarly high levels. This may reflect recapitulation of embryonic features in RMS.
Although JARID2 lacks critical active site residues required for demethylase catalytic activity22, it is known to be involved in the repression of transcriptional activation at specific loci via its association with Rb and Nkx2.5/GATA4 that confer methyltransferase activity at H3K9 and PRC2 which methylates H3K2719, 20, 25-27. Here we have shown that multiple components of the PRC2-EZH2 complex are expressed at high levels in primary RMS, including EED and, as previously noted in RMS cell lines42, EZH2. This and the low levels of EZH1 gene expression we observed in RMS suggests that the PRC2-EZH2 complex contributes to the undifferentiated phenotype of RMS through repression of myogenic regulatory factors (MRFs). This is consistent with the switch described from a PRC2-Ezh2 to a PRC2-Ezh1 complex controlling myogenin activation in mouse myoblasts32. There are interesting parallels with the fusion protein EWS-FLI1 in Ewing sarcoma, which binds to the EZH2 promoter region and enhances expression that contributes to the undifferentiated and malignant phenotype of these tumors43.
Control of differentiation in both ESCs and mouse embryos has been linked with the presence of histone methyl marks at the promoter regions of genes that control differentiation18-20, 26, 27, 44. DNA promoter methylation prevents premature expression of myogenin and demethylation is required for skeletal muscle differentiation in mouse embryos44. Furthermore, recent work into the control of differentiation in mouse myoblasts and myotubes has linked transcriptional repression at the promoter of genes that are required for myogenic differentiation with specific methyl marks, notably H3K27me318. We have shown that JARID2 binds directly to the MYOG promoter in human RMS cells at the same genomic region as the H3K27me3 mark in mouse and that silencing of JARID2 results in abrogation of this mark, demonstrating that JARID2 is controlling transcriptional repression of the MYOG promoter by acting as a scaffold to direct and/or maintain methylation of the repressive H3K27 mark. We also demonstrate for the first time that JARID2 binds to the MYL1 promoter and that silencing of JARID2 removes a H3K27me3 mark within the binding region. These interactions were shown to be dependent on the association of JARID2 with EED, which is a representative component of PRC2.
The methyl marks H3K9me3 on the myoD promoter and H3K27me3 on the myogenin promoter have been shown to be under the control of the histone methyl transferase KMT1A and the HDM KDM4A, respectively, during normal myogenesis17, 45. In addition, KMT1A has recently been shown to play a role in ARMS by inhibiting myogenic differentiation17. Although not shown directly, the authors speculated that KMT1A levels may be regulated by PAX3-FOXO1, as KMT1A expression was only increased on induction of differentiation in PAX3-FOXO1 positive cell lines17. We also identified JMJD1C as strongly associated with PAX3-FOXO1 and PAX7-FOXO1 expression in ARMS. Therefore this and other HDMs highly expressed in RMS may also contribute to the tumor phenotype. Taken together, maintenance of repressive histone methyl marks on MRFs appears critically important for the undifferentiated phenotype of RMS.
As expression of HDM gene family members is limited in non-embryonic tissues, these may represent tumor-specific targets that could be manipulated to induce terminal differentiation. Differentiation therapy has been successfully exploited in the clinic for the treatment of several pediatric and adult tumors including neuroblastoma and acute promyelocytic leukemia46-48. Despite the lack of evidence for any demethylase catalytic activity of JARID212, 22, 23, this protein contains the methyl-lysine binding pocket that could mediate a scaffolding role of JARID2. The substrate binding pocket of members of the Jumonji demethylase family with known structures is predicted to be druggable using structural methods (https://www.ebi.ac.uk/chembl/drugebility/structure) and evidence of modulation with a small molecule exists for some members of the family49, 50. This, together with our work, makes JARID2 and other members of PRC2 potential therapeutic targets for the treatment of RMS.
Materials and Methods
Cell culture
Human RMS cell lines are described previously33 Cell lines were cultured in Dulbecco Modified Eagle Medium (DMEM) (RD, RH30, RMS-01) or Roswell Park Memorial Institute 1640 (RPMI) Medium (RH41) supplemented with 10% Fetal Calf Serum (FCS), 2mM L-glutamine and 1% penicillin/streptomycin. Cells were maintained at 37°C at 5% CO2.
Gene Expression analysis
Affymetrix U133A plus 2.0 profiling data from 101 previously described patient samples28, 24 mesenchymal stem cells samples (GEO accessions: GSE6460, GSE7637, GSE9451, GSE9520, GSE9593, GSE10315, GSE13604), 19 embryonal stem cell samples (GEO accessions: GSE7896, GSE8884, GSE9440, GSE9510, GSE9940, GSE13828) and 30 skeletal muscle samples were normalised and polished using the Robust Multiarray Average in RMAExpress. A representative probeset was chosen for each HDM gene family member and the median value calculated across skeletal muscle samples. Tumor and normal samples were assessed relative to this. Gene expression levels were deemed overexpressed if the median across the RMS sample set was 2-fold above the median of the skeletal muscle samples using a two-tailed T-test and Bonferroni Correction (Supplementary Table S1). Control and sample sets were tested for equal variance using an F-Test. If equal variance was found then an unpaired t-test was conducted. Where the variances were significantly different an unpaired t-test with Welch’s correction was used. Bonferroni correction was used to account for multiple testing.
Total RNA was extracted from patients and skeletal muscle samples as detailed in our previous publication28. Clinicopathological features of 120 patients samples used in quantitative RT-PCR analyses are summarized in Supplementary Table S4. Total RNA from cell lines was extracted using TRIzol Reagent according to the manufacturers instructions (Invitrogen, Carlsbad, CA, USA). cDNA was synthesised using SuperScript II (Invitrogen) and quantitative RT-PCR was performed using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Primer and probe sets for β-ACTIN, JARID2, PAX3, MYOG and MYL1 were from Applied Biosystems (4310881E, Hs01004460_m1, Hs00992437_m1, Hs01072232_m1, Hs00984901_m1). Primers and probes designed for analysis of PAX3-FOXO1 were as follows: Forward 5′-GAACCCACCATTGGCAAT-3′, Probe 5′-CCTCTCACCTCAGAATTCAATTCGTCATAATCTG-3′, Reverse 5′-TCTGCACACGAATGAACTTGCT-3′.
Analyses of clinicopathological correlates
Association of gene expression with clinicopathological characteristics was analyzed using the t-student or one-way ANOVA test when appropriate. Genes whose expression was significantly associated (p<0.05) with aggressive RMS characteristics were considered in a multivariate analysis (MVA) using a stepwise log-regression together with other significant clinicopathological characteristics. The Kaplan–Meier method was used to estimate median survival times, and the log-rank test was used to compare survival curves using SPSS program (Version 19.0, SPSS Inc, Chicago, IL, USA).
RNA interference
siRNAs were synthesised by Sigma (sequences available on request). For RNA analyzes, Western blotting and cell viability assays, cell lines were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions with 15nM siRNA and cultured for 24h (RNA) and 48h (protein) unless otherwise stated.
Cell viability assays
Cell proliferation was measured using a metabolic assay, MTS (Promega, Madison, WI, USA), and cell count assay, CyQuant (Invitrogen). Cells were plated in 96-well plates and transfected 24 hours post plating. Cells were assayed 24 to 96 hours post transfection unless otherwise stated. MTS was carried out according to manufacturer’s instructions and measured using ELx800 plate reader (BioTek Instruments, Winooski, VT, USA). CyQuant NF assay was carried out according to manufacturer’s instructions and results were analyzed on the VICTOR2 D fluorometer plate reader (PerkinElmer, Waltham, MA, USA). The Caspase-Glo 3/7 assay (Promega) was used according to the manufacturer’s instructions to assess apoptosis at 72hrs post-transfection.
Flow Cytometry
For cell cycle analysis, cells were harvested at 72hrs post transfection, fixed in 70% ethanol and stained for 30mins with propidium iodide (Invitrogen). Cells were then subject to cell cycle distribution analysis on an LSR II (BD, Oxford, UK).
Overexpression studies
PAX3-FOXO1 cDNA was amplified from RMS-01 cell line RNA with an additional haemagglutinin (HA) tag and cloned into pCI-neo vector (Promega). JARID2 cDNA was cloned into pCI-neo from the I.M.A.G.E. clone 4520786 (Source Bioscience, Nottingham, UK). Stable transfections were carried out using Lipofectamine 2000 according to the manufacturer’s instructions. Selection was carried out using G-418 sulphate (PAA, Pasching Austria) and single colonies were picked, grown and checked for expression by Western blot and qRT-PCR.
Chromatin Immunoprecipitation (ChIP) studies
ChIP was carried out using the MAGnify ChIP kit (Invitrogen) according to manufacturer’s instructions. ChIP was carried out using anti-HA antibody (Roche Diagnostics, Burgess Hill, UK). Anti-JARID2, -Histone3K27me3 and -EED were from Abcam (Cambridge, MA, USA). DNA was quantified by PicoGreen (Thermo Scientific, Waltham, MA, USA).
qPCR AND Sybr Green PCR of ChIP DNA
ChIP DNA was measured for enrichment by either qPCR or Sybr green PCR using primers spanning the IGF1R, JARID2, MYOG, and MYOD promoter regions as specified in the text (primer sequences available on request).
Western blotting
Total cell lysates were extracted from siRNA experiments in RMS cell lines using Cell Lysis Buffer at 72hrs post-transfection (Cell Signalling Technology, Danvers, MA, USA). 20μg protein were resolved in a 3-8% Tris-Acetate acrylamide gel and transferred onto PVDF membranes using the X-Cell II and iBlot systems according to manufacturer’s instructions (Invitrogen). Primary antibodies used: anti-JARID2 (Abcam), anti-GAPDH (Millipore, Billerica, MA, USA), anti-HA (Roche), anti-FOXO1 (Cell signaling Technology), anti-MYOGENIN (Developmental Studies Hybridoma Bank), anti-MYL1 (Abcam). Antibodies were detected using ECL Western blot analysis system (GE Healthcare UK Ltd) and signals viewed using the Molecular Imager ChemiDoc XRS System (Bio-Rad, Hemel Hempstead, UK).
Luciferase Assays
pGL3 Promoter, pGL3 Control and pRL-CMV (Promega) were used for luciferase assays. DNA containing the putative PAX3-FOXO1 binding site was amplified from RMS-01 genomic DNA and cloned into pGL3 Promoter. Mutant enhancers were generated using the QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies UK Ltd., Berkshire, UK) using DNA polymerase PfuUltra II fusion HS (Agilent) (primer sequences available on request). Cells were cultured for 24h in 96-well plates, and transfections were carried using Lipofectamine 2000 according to manufacturer’s protocol. Dual luciferase assays were performed at 72h using Dual-Glo luciferase reporter assay system (Promega). For each assay, 3 separate experiments were performed.
TPA assay
Differentiation was induced using 12-O-tetradecanoylphorbol-13-acetate (TPA) in RMS cells over a period of 8 days similar to the procedure previously described30. Cells were collected and RNA and protein extracted at 144 hours post TPA addition.
Supplementary Material
Acknowledgement
We are very grateful for help and support with tumor collection and annotation from the Children’s Cancer and Leukemia Group. Thanks also to David Landeira for help with the JARID2 antibody. This work was supported by grants from Cancer Research UK (C5066/A10399); Sarcoma UK; the Chris Lucas Trust; Rob’s ARTTT Charity; Medical Research Council for a studentship; the Spanish Society of Medical Oncology (SEOM) Translational Research Fellowship (D.O.). We acknowledge NHS funding to the NIHR Biomedical Research Centre.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66(14):6936–46. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 2.Davicioni E, Anderson MJ, Finckenstein FG, Lynch JC, Qualman SJ, Shimada H, et al. Molecular classification of rhabdomyosarcoma--genotypic and phenotypic determinants of diagnosis: a report from the Children’s Oncology Group. Am J Pathol. 2009;174(2):550–64. doi: 10.2353/ajpath.2009.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Missiaglia E, Williamson D, Chisholm J, Wirapati P, Pierron G, Petel F, et al. PAX3/FOXO1 Fusion Gene Status Is the Key Prognostic Molecular Marker in Rhabdomyosarcoma and Significantly Improves Current Risk Stratification. J Clin Oncol. 2012 doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- 4.Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children’s oncology group. J Clin Oncol. 2002;20(11):2672–9. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 5.Bennicelli JL, Edwards RH, Barr FG. Mechanism for transcriptional gain of function resulting from chromosomal translocation in alveolar rhabdomyosarcoma. Proc Natl Acad Sci U S A. 1996;93(11):5455–9. doi: 10.1073/pnas.93.11.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein JA, Song B, Lakkis M, Wang C. Tumor-specific PAX3-FKHR transcription factor, but not PAX3, activates the platelet-derived growth factor alpha receptor. Mol Cell Biol. 1998;18(7):4118–30. doi: 10.1128/mcb.18.7.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fredericks WJ, Galili N, Mukhopadhyay S, Rovera G, Bennicelli J, Barr FG, et al. The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Mol Cell Biol. 1995;15(3):1522–35. doi: 10.1128/mcb.15.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Wang C. Identification of a new class of PAX3-FKHR target promoters: a role of the Pax3 paired box DNA binding domain. Oncogene. 2007;26(11):1595–605. doi: 10.1038/sj.onc.1209958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikuchi K, Tsuchiya K, Otabe O, Gotoh T, Tamura S, Katsumi Y, et al. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem Biophys Res Commun. 2008;365(3):568–74. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Cao L, Yu Y, Bilke S, Walker RL, Mayeenuddin LH, Azorsa DO, et al. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010;70(16):6497–508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calhabeu F, Hayashi S, Morgan JE, Relaix F, Zammit PS. Alveolar rhabdomyosarcoma-associated proteins PAX3/FOXO1A and PAX7/FOXO1A suppress the transcriptional activity of MyoD-target genes in muscle stem cells. Oncogene. 2012 doi: 10.1038/onc.2012.73. [DOI] [PubMed] [Google Scholar]
- 12.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22(9):1115–40. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S, et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem. 1999;274(22):15633–45. doi: 10.1074/jbc.274.22.15633. [DOI] [PubMed] [Google Scholar]
- 14.Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, et al. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci U S A. 2007;104(49):19226–31. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schildhaus HU, Riegel R, Hartmann W, Steiner S, Wardelmann E, Merkelbach-Bruse S, et al. Lysine-specific demethylase 1 is highly expressed in solitary fibrous tumors, synovial sarcomas, rhabdomyosarcomas, desmoplastic small round cell tumors, and malignant peripheral nerve sheath tumors. Hum Pathol. 2011;42(11):1667–75. doi: 10.1016/j.humpath.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 16.Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69(5):2065–71. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 17.Lee MH, Jothi M, Gudkov AV, Mal AK. Histone methyltransferase KMT1A restrains entry of alveolar rhabdomyosarcoma cells into a myogenic differentiated state. Cancer Res. 2011;71(11):3921–31. doi: 10.1158/0008-5472.CAN-10-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci U S A. 2011;108(22):E149–58. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12(6):618–24. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–14. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juan AH, Derfoul A, Feng X, Ryall JG, Dell’Orso S, Pasut A, et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25(8):789–94. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7(9):715–27. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 23.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–10. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 24.Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106(2):243–7. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim TG, Chen J, Sadoshima J, Lee Y. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol Cell Biol. 2004;24(23):10151–60. doi: 10.1128/MCB.24.23.10151-10160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24(4):368–80. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williamson D, Missiaglia E, de Reynies A, Pierron G, Thuille B, Palenzuela G, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28(13):2151–8. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- 29.Charytonowicz E, Cordon-Cardo C, Matushansky I, Ziman M. Alveolar rhabdomyosarcoma: is the cell of origin a mesenchymal stem cell? Cancer Lett. 2009;279(2):126–36. doi: 10.1016/j.canlet.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 30.Aguanno S, Bouche M, Adamo S, Molinaro M. 12-O-tetradecanoylphorbol-13-acetate-induced differentiation of a human rhabdomyosarcoma cell line. Cancer Res. 1990;50(11):3377–82. [PubMed] [Google Scholar]
- 31.Linardic CM. PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer Lett. 2008;270(1):10–8. doi: 10.1016/j.canlet.2008.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stojic L, Jasencakova Z, Prezioso C, Stutzer A, Bodega B, Pasini D, et al. Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenetics Chromatin. 2011;4:16. doi: 10.1186/1756-8935-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tonelli R, McIntyre A, Camerin C, Walters ZS, Di Leo K, Selfe J, et al. Antitumor activity of sustained N-myc reduction in rhabdomyosarcomas and transcriptional block by antigene therapy. Clin Cancer Res. 2012;18(3):796–807. doi: 10.1158/1078-0432.CCR-11-1981. [DOI] [PubMed] [Google Scholar]
- 34.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia SJ, Holder DD, Pawel BR, Zhang C, Barr FG. High expression of the PAX3-FKHR oncoprotein is required to promote tumorigenesis of human myoblasts. Am J Pathol. 2009;175(6):2600–8. doi: 10.2353/ajpath.2009.090192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lollini PL, De Giovanni C, Landuzzi L, Nicoletti G, Scotlandi K, Nanni P. Reduced metastatic ability of in vitro differentiated human rhabdomyosarcoma cells. Invasion Metastasis. 1991;11(2):116–24. [PubMed] [Google Scholar]
- 37.Merlino G, Helman LJ. Rhabdomyosarcoma--working out the pathways. Oncogene. 1999;18(38):5340–8. doi: 10.1038/sj.onc.1203038. [DOI] [PubMed] [Google Scholar]
- 38.Barlow JW, Wiley JC, Mous M, Narendran A, Gee MF, Goldberg M, et al. Differentiation of rhabdomyosarcoma cell lines using retinoic acid. Pediatr Blood Cancer. 2006;47(6):773–84. doi: 10.1002/pbc.20650. [DOI] [PubMed] [Google Scholar]
- 39.Jung J, Kim TG, Lyons GE, Kim HR, Lee Y. Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J Biol Chem. 2005;280(35):30916–23. doi: 10.1074/jbc.M414482200. [DOI] [PubMed] [Google Scholar]
- 40.Kim TG, Kraus JC, Chen J, Lee Y. JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J Biol Chem. 2003;278(43):42247–55. doi: 10.1074/jbc.M307386200. [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi T, Kojima M, Nakajima K, Kondo S. jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mech Dev. 1999;86(1-2):29–38. doi: 10.1016/s0925-4773(99)00100-8. [DOI] [PubMed] [Google Scholar]
- 42.Ciarapica R, Russo G, Verginelli F, Raimondi L, Donfrancesco A, Rota R, et al. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8(1):172–5. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
- 43.Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci U S A. 2009;106(13):5324–9. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palacios D, Summerbell D, Rigby PW, Boyes J. Interplay between DNA methylation and transcription factor availability: implications for developmental activation of the mouse Myogenin gene. Mol Cell Biol. 2010;30(15):3805–15. doi: 10.1128/MCB.00050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verrier L, Escaffit F, Chailleux C, Trouche D, Vandromme M. A new isoform of the histone demethylase JMJD2A/KDM4A is required for skeletal muscle differentiation. PLoS Genet. 2011;7(6):e1001390. doi: 10.1371/journal.pgen.1001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimwade D, Mistry AR, Solomon E, Guidez F. Acute promyelocytic leukemia: a paradigm for differentiation therapy. Cancer Treat Res. 2010;145:219–35. doi: 10.1007/978-0-387-69259-3_13. [DOI] [PubMed] [Google Scholar]
- 47.Matthay KK, Reynolds CP, Seeger RC, Shimada H, Adkins ES, Haas-Kogan D, et al. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a children’s oncology group study. J Clin Oncol. 2009;27(7):1007–13. doi: 10.1200/JCO.2007.13.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veal GJ, Cole M, Errington J, Pearson AD, Foot AB, Whyman G, et al. Pharmacokinetics and metabolism of 13-cis-retinoic acid (isotretinoin) in children with high-risk neuroblastoma - a study of the United Kingdom Children’s Cancer Study Group. Br J Cancer. 2007;96(3):424–31. doi: 10.1038/sj.bjc.6603554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo X, Liu Y, Kubicek S, Myllyharju J, Tumber A, Ng S, et al. A selective inhibitor and probe of the cellular functions of Jumonji C domain-containing histone demethylases. J Am Chem Soc. 2011;133(24):9451–6. doi: 10.1021/ja201597b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Upadhyay AK, Rotili D, Han JW, Hu R, Chang Y, Labella D, et al. An analog of BIX-01294 selectively inhibits a family of histone H3 lysine 9 Jumonji demethylases. J Mol Biol. 2012;416(3):319–27. doi: 10.1016/j.jmb.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.