Abstract
Development in Bacillus subtilis involves the formation of two cell types with activation of the transcription factors sigma F in the forespore and sigma E in the mother cell. Activation of sigma E is due to the processing of the inactive precursor pro-sigma E, which requires the putative protease SpoIIGA and the presence of active sigma F. We have introduced missense mutations altering the promoter recognition properties of sigma F. These mutations abolish pro-sigma E processing, suggesting that sigma F is involved through its transcriptional activity and that the processing machinery responds to a signal generated by the product(s) of some unidentified gene(s) transcribed in the forespore. The role of the septum in transducing this signal was investigated. Induction of sigma F during exponential growth in cells producing SpoIIGA and pro-sigma E led to a high level of processing and sigma E activity. Moreover, pro-sigma E was efficiently processed in a mutant strain blocked prior to septation and synthesizing sigma F in active form at the onset of sporulation. Therefore, the sporulation septum is not required for induction of pro-sigma E processing and pro-sigma E can be processed in the same cell in which sigma F is active. These results suggest that some unknown mechanism must exist to prevent sigma E from becoming active in the forespore.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alper S., Duncan L., Losick R. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell. 1994 Apr 22;77(2):195–205. doi: 10.1016/0092-8674(94)90312-3. [DOI] [PubMed] [Google Scholar]
- Beall B., Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991 Mar;5(3):447–455. doi: 10.1101/gad.5.3.447. [DOI] [PubMed] [Google Scholar]
- Beall B., Lutkenhaus J. Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J Bacteriol. 1992 Apr;174(7):2398–2403. doi: 10.1128/jb.174.7.2398-2403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson A. K., Haldenwang W. G. Bacillus subtilis sigma B is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund J. E., Zhang L., Haines M. A., Higgins M. L., Piggot P. J. Analysis by fluorescence microscopy of the development of compartment-specific gene expression during sporulation of Bacillus subtilis. J Bacteriol. 1994 May;176(10):2898–2905. doi: 10.1128/jb.176.10.2898-2905.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson H. C., Haldenwang W. G. The sigma E subunit of Bacillus subtilis RNA polymerase is present in both forespore and mother cell compartments. J Bacteriol. 1989 Apr;171(4):2216–2218. doi: 10.1128/jb.171.4.2216-2218.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppolecchia R., DeGrazia H., Moran C. P., Jr Deletion of spoIIAB blocks endospore formation in Bacillus subtilis at an early stage. J Bacteriol. 1991 Nov;173(21):6678–6685. doi: 10.1128/jb.173.21.6678-6685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederich B., Wilkinson J. F., Magnin T., Najafi M., Erringston J., Yudkin M. D. Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor sigma F of Bacillus subtilis. Genes Dev. 1994 Nov 1;8(21):2653–2663. doi: 10.1101/gad.8.21.2653. [DOI] [PubMed] [Google Scholar]
- Driks A., Losick R. Compartmentalized expression of a gene under the control of sporulation transcription factor sigma E in Bacillus subtilis. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):9934–9938. doi: 10.1073/pnas.88.22.9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L., Losick R. SpoIIAB is an anti-sigma factor that binds to and inhibits transcription by regulatory protein sigma F from Bacillus subtilis. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2325–2329. doi: 10.1073/pnas.90.6.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993 Mar;57(1):1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P., Piggot P. J. Nucleotide sequence of sporulation locus spoIIA in Bacillus subtilis. J Gen Microbiol. 1984 Aug;130(8):2147–2153. doi: 10.1099/00221287-130-8-2147. [DOI] [PubMed] [Google Scholar]
- Gonzy-Tréboul G., Karmazyn-Campelli C., Stragier P. Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon. J Mol Biol. 1992 Apr 20;224(4):967–979. doi: 10.1016/0022-2836(92)90463-t. [DOI] [PubMed] [Google Scholar]
- Guzmán P., Westpheling J., Youngman P. Characterization of the promoter region of the Bacillus subtilis spoIIE operon. J Bacteriol. 1988 Apr;170(4):1598–1609. doi: 10.1128/jb.170.4.1598-1609.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins M. L., Piggot P. J. Septal membrane fusion--a pivotal event in bacterial spore formation? Mol Microbiol. 1992 Sep;6(18):2565–2571. doi: 10.1111/j.1365-2958.1992.tb01433.x. [DOI] [PubMed] [Google Scholar]
- Illing N., Errington J. Genetic regulation of morphogenesis in Bacillus subtilis: roles of sigma E and sigma F in prespore engulfment. J Bacteriol. 1991 May;173(10):3159–3169. doi: 10.1128/jb.173.10.3159-3169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing N., Errington J. The spoIIIA operon of Bacillus subtilis defines a new temporal class of mother-cell-specific sporulation genes under the control of the sigma E form of RNA polymerase. Mol Microbiol. 1991 Aug;5(8):1927–1940. doi: 10.1111/j.1365-2958.1991.tb00816.x. [DOI] [PubMed] [Google Scholar]
- Jonas R. M., Haldenwang W. G. Influence of spo mutations on sigma E synthesis in Bacillus subtilis. J Bacteriol. 1989 Sep;171(9):5226–5228. doi: 10.1128/jb.171.9.5226-5228.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman P. A., DeGrazia H., Kellner E. M., Moran C. P., Jr Forespore-specific disappearance of the sigma-factor antagonist spoIIAB: implications for its role in determination of cell fate in Bacillus subtilis. Mol Microbiol. 1993 May;8(4):663–671. doi: 10.1111/j.1365-2958.1993.tb01610.x. [DOI] [PubMed] [Google Scholar]
- LaBell T. L., Trempy J. E., Haldenwang W. G. Sporulation-specific sigma factor sigma 29 of Bacillus subtilis is synthesized from a precursor protein, P31. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1784–1788. doi: 10.1073/pnas.84.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P. A., Losick R. Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J Bacteriol. 1994 Mar;176(5):1451–1459. doi: 10.1128/jb.176.5.1451-1459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. J., Partridge S. R., Errington J. Sigma factors, asymmetry, and the determination of cell fate in Bacillus subtilis. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3849–3853. doi: 10.1073/pnas.91.9.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto M., Gribskov M., Gross C. A. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992 Jun;174(12):3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R., Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992 Feb 13;355(6361):601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- Margolis P., Driks A., Losick R. Establishment of cell type by compartmentalized activation of a transcription factor. Science. 1991 Oct 25;254(5031):562–565. doi: 10.1126/science.1948031. [DOI] [PubMed] [Google Scholar]
- Min K. T., Hilditch C. M., Diederich B., Errington J., Yudkin M. D. Sigma F, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell. 1993 Aug 27;74(4):735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- Partridge S. R., Foulger D., Errington J. The role of sigma F in prespore-specific transcription in Bacillus subtilis. Mol Microbiol. 1991 Mar;5(3):757–767. doi: 10.1111/j.1365-2958.1991.tb00746.x. [DOI] [PubMed] [Google Scholar]
- Peters H. K., 3rd, Haldenwang W. G. Synthesis and fractionation properties of SpoIIGA, a protein essential for pro-sigma E processing in Bacillus subtilis. J Bacteriol. 1991 Dec;173(24):7821–7827. doi: 10.1128/jb.173.24.7821-7827.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Margolis P., Duncan L., Coppolecchia R., Moran C. P., Jr, Losick R. Control of developmental transcription factor sigma F by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9221–9225. doi: 10.1073/pnas.87.23.9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43(1-2):85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
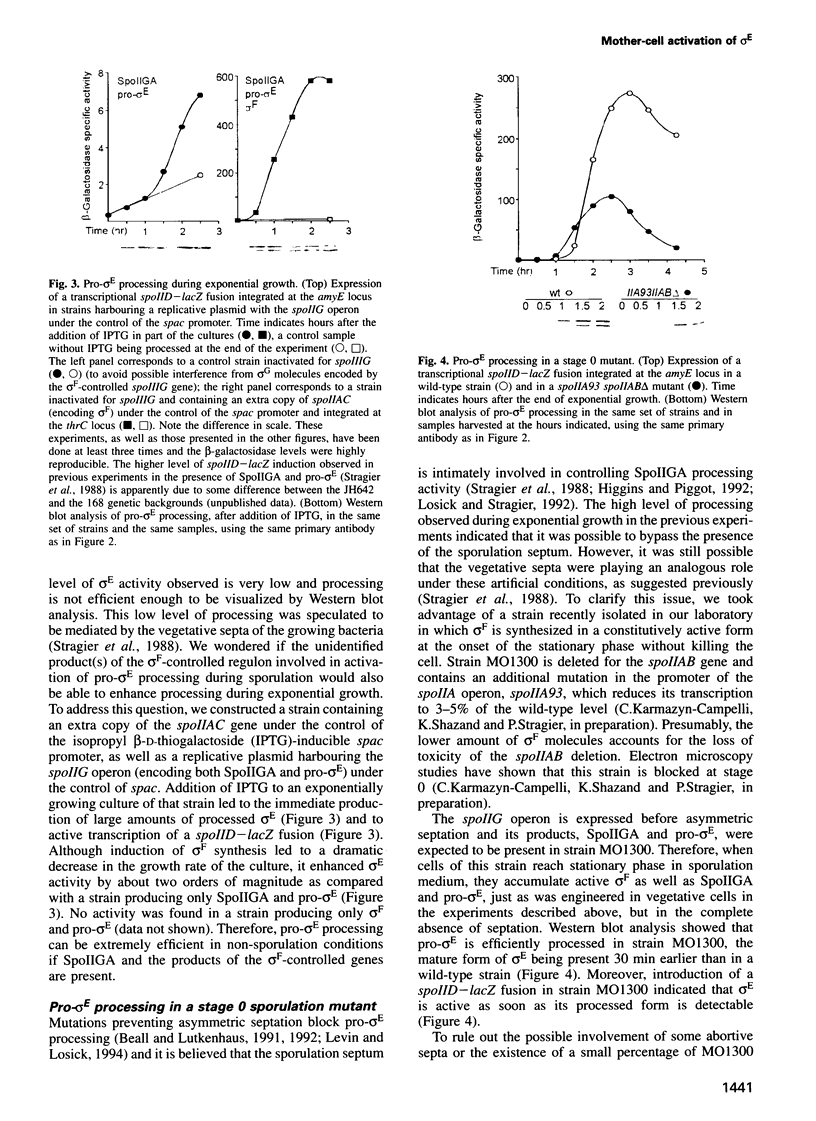
- Stragier P., Bonamy C., Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988 Mar 11;52(5):697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- Sun D., Fajardo-Cavazos P., Sussman M. D., Tovar-Rojo F., Cabrera-Martinez R. M., Setlow P. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by E sigma F: identification of features of good E sigma F-dependent promoters. J Bacteriol. 1991 Dec;173(24):7867–7874. doi: 10.1128/jb.173.24.7867-7874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempy J. E., Bonamy C., Szulmajster J., Haldenwang W. G. Bacillus subtilis sigma factor sigma 29 is the product of the sporulation-essential gene spoIIG. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4189–4192. doi: 10.1073/pnas.82.12.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. J., Piggot P. J., Tatti K. M., Moran C. P., Jr Transcription of the Bacillus subtilis spoIIA locus. Gene. 1991 May 15;101(1):113–116. doi: 10.1016/0378-1119(91)90231-y. [DOI] [PubMed] [Google Scholar]
- York K., Kenney T. J., Satola S., Moran C. P., Jr, Poth H., Youngman P. Spo0A controls the sigma A-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J Bacteriol. 1992 Apr;174(8):2648–2658. doi: 10.1128/jb.174.8.2648-2658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkin M. D. Structure and function in a Bacillus subtilis sporulation-specific sigma factor: molecular nature of mutations in spoIIAC. J Gen Microbiol. 1987 Mar;133(3):475–481. doi: 10.1099/00221287-133-3-475. [DOI] [PubMed] [Google Scholar]