Abstract
Hormonal induction of the mouse mammary tumour virus (MMTV) promoter is mediated by interactions between hormone receptors and other transcription factors bound to a complex array of sites. Previous results suggested that access to these sites is modulated by their precise organization into a positioned regulatory nucleosome. Using genomic footprinting, we show that MMTV promoter DNA is rotationally phased in intact cells containing either episomal or chromosomally integrated proviral fragments. Prior to induction there is no evidence for factors bound to the promoter. Following progesterone induction of cells with high levels of receptor, genomic footprinting detects simultaneous protection over the binding sites for hormone receptors, NF-I and the octamer binding proteins. Glucocorticoid or progestin induction leads to a characteristic chromatin remodelling that is independent of ongoing transcription. The centre of the regulatory nucleosome becomes more accessible to DNase I and restriction enzymes, but the limits of the nucleosome are unchanged and the 145 bp core region remains protected against micrococcal nuclease digestion. Thus, the nucleosome covering the MMTV promoter is neither removed nor shifted upon hormone induction, and all relevant transcription factors bind to the surface of the rearranged nucleosome. Since these factors cannot bind simultaneously to free DNA, maintainance of the nucleosome may be required for binding of factors to contiguous sites.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almer A., Rudolph H., Hinnen A., Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986 Oct;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer T. K., Cordingley M. G., Wolford R. G., Hager G. L. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991 Feb;11(2):688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer T. K., Lefebvre P., Wolford R. G., Hager G. L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992 Mar 20;255(5051):1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- Ausio J., van Holde K. E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986 Mar 25;25(6):1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- Becker P. B., Gloss B., Schmid W., Strähle U., Schütz G. In vivo protein-DNA interactions in a glucocorticoid response element require the presence of the hormone. Nature. 1986 Dec 18;324(6098):686–688. doi: 10.1038/324686a0. [DOI] [PubMed] [Google Scholar]
- Becker P., Renkawitz R., Schütz G. Tissue-specific DNaseI hypersensitive sites in the 5'-flanking sequences of the tryptophan oxygenase and the tyrosine aminotransferase genes. EMBO J. 1984 Sep;3(9):2015–2020. doi: 10.1002/j.1460-2075.1984.tb02084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Ewel A., McKeon J., Chovav M., Juan E. Characterization and purification of Adh distal promoter factor 2, Adf-2, a cell-specific and promoter-specific repressor in Drosophila. Nucleic Acids Res. 1992 Sep 11;20(17):4481–4489. doi: 10.1093/nar/20.17.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bode J., Gómez-Lira M. M., Schröter H. Nucleosomal particles open as the histone core becomes hyperacetylated. Eur J Biochem. 1983 Feb 15;130(3):437–445. doi: 10.1111/j.1432-1033.1983.tb07170.x. [DOI] [PubMed] [Google Scholar]
- Bresnick E. H., Bustin M., Marsaud V., Richard-Foy H., Hager G. L. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992 Jan 25;20(2):273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick E. H., John S., Hager G. L. Histone hyperacetylation does not alter the positioning or stability of phased nucleosomes on the mouse mammary tumor virus long terminal repeat. Biochemistry. 1991 Apr 9;30(14):3490–3497. doi: 10.1021/bi00228a020. [DOI] [PubMed] [Google Scholar]
- Bresnick E. H., Rories C., Hager G. L. Evidence that nucleosomes on the mouse mammary tumor virus promoter adopt specific translational positions. Nucleic Acids Res. 1992 Feb 25;20(4):865–870. doi: 10.1093/nar/20.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüggemeier U., Kalff M., Franke S., Scheidereit C., Beato M. Ubiquitous transcription factor OTF-1 mediates induction of the MMTV promoter through synergistic interaction with hormone receptors. Cell. 1991 Feb 8;64(3):565–572. doi: 10.1016/0092-8674(91)90240-y. [DOI] [PubMed] [Google Scholar]
- Brüggemeier U., Rogge L., Winnacker E. L., Beato M. Nuclear factor I acts as a transcription factor on the MMTV promoter but competes with steroid hormone receptors for DNA binding. EMBO J. 1990 Jul;9(7):2233–2239. doi: 10.1002/j.1460-2075.1990.tb07393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buetti E., Kühnel B., Diggelmann H. Dual function of a nuclear factor I binding site in MMTV transcription regulation. Nucleic Acids Res. 1989 Apr 25;17(8):3065–3078. doi: 10.1093/nar/17.8.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buetti E. Stably integrated mouse mammary tumor virus long terminal repeat DNA requires the octamer motifs for basal promoter activity. Mol Cell Biol. 1994 Feb;14(2):1191–1203. doi: 10.1128/mcb.14.2.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttinelli M., Di Mauro E., Negri R. Multiple nucleosome positioning with unique rotational setting for the Saccharomyces cerevisiae 5S rRNA gene in vitro and in vivo. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9315–9319. doi: 10.1073/pnas.90.20.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B. R., Kim Y. J., Sayre M. H., Laurent B. C., Kornberg R. D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Campbell V. W., Jackson D. A. The effect of divalent cations on the mode of action of DNase I. The initial reaction products produced from covalently closed circular DNA. J Biol Chem. 1980 Apr 25;255(8):3726–3735. [PubMed] [Google Scholar]
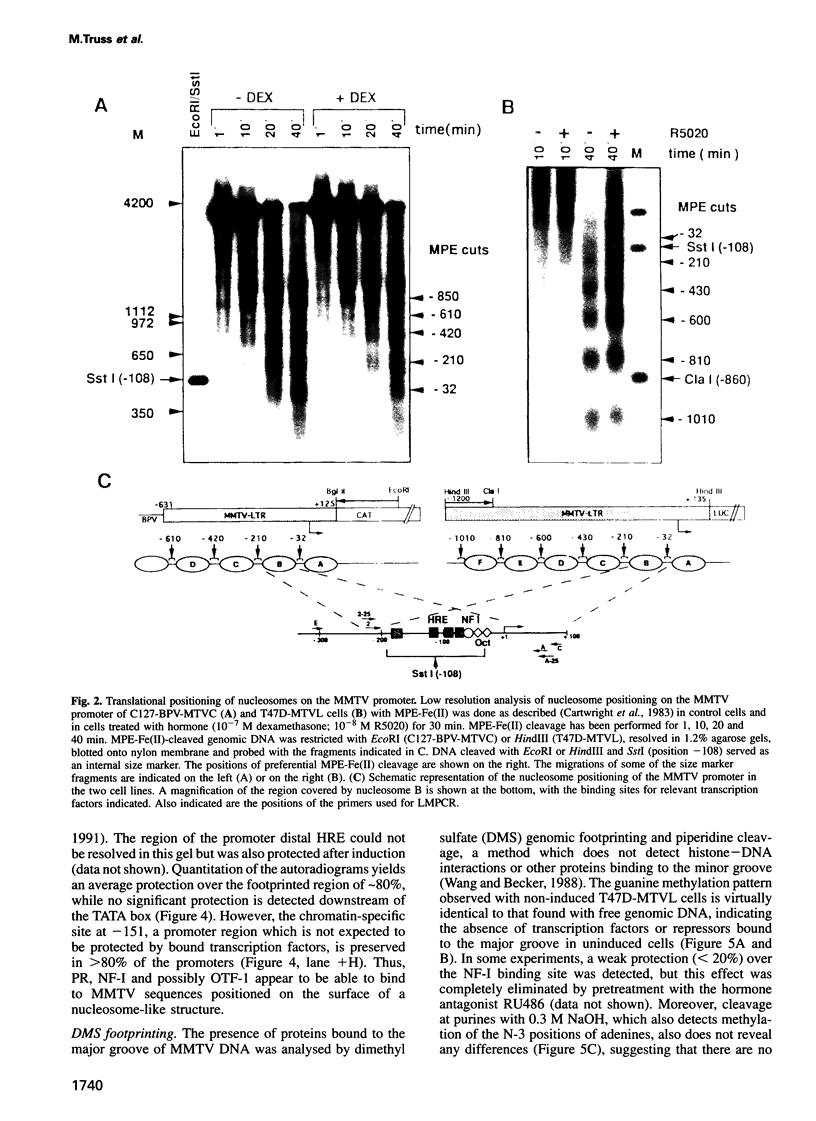
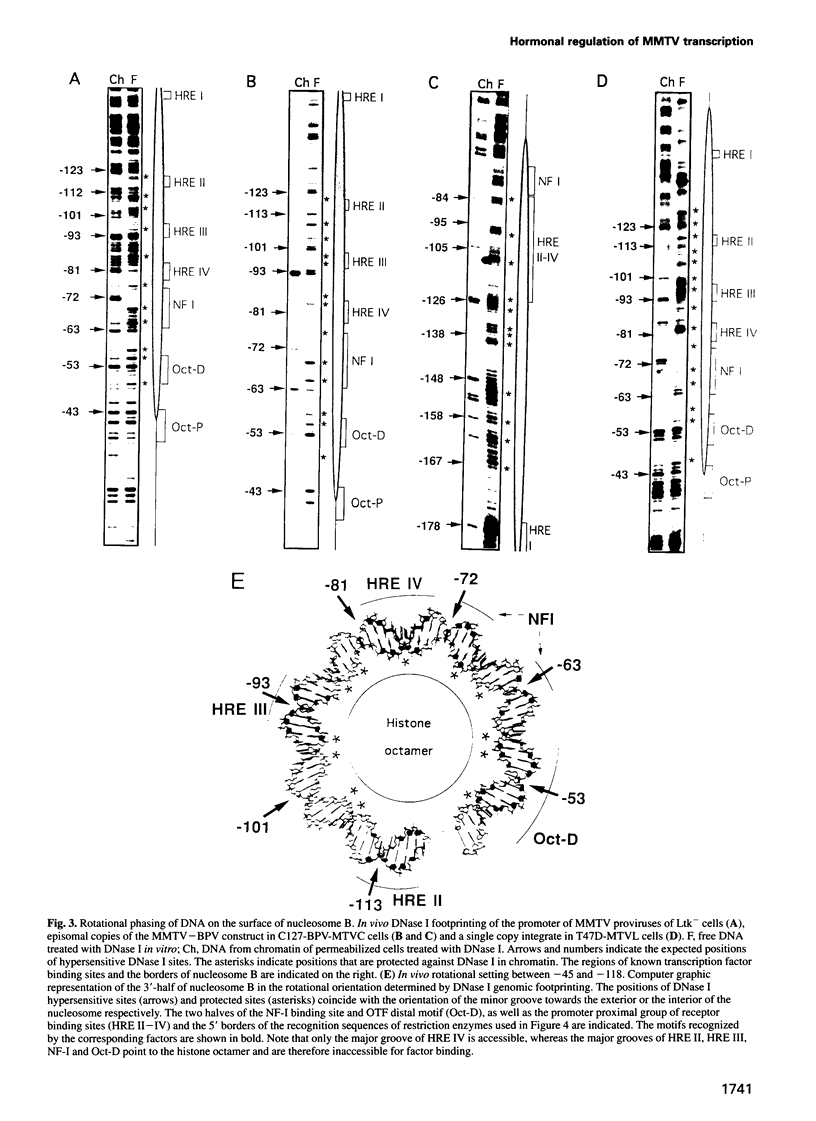
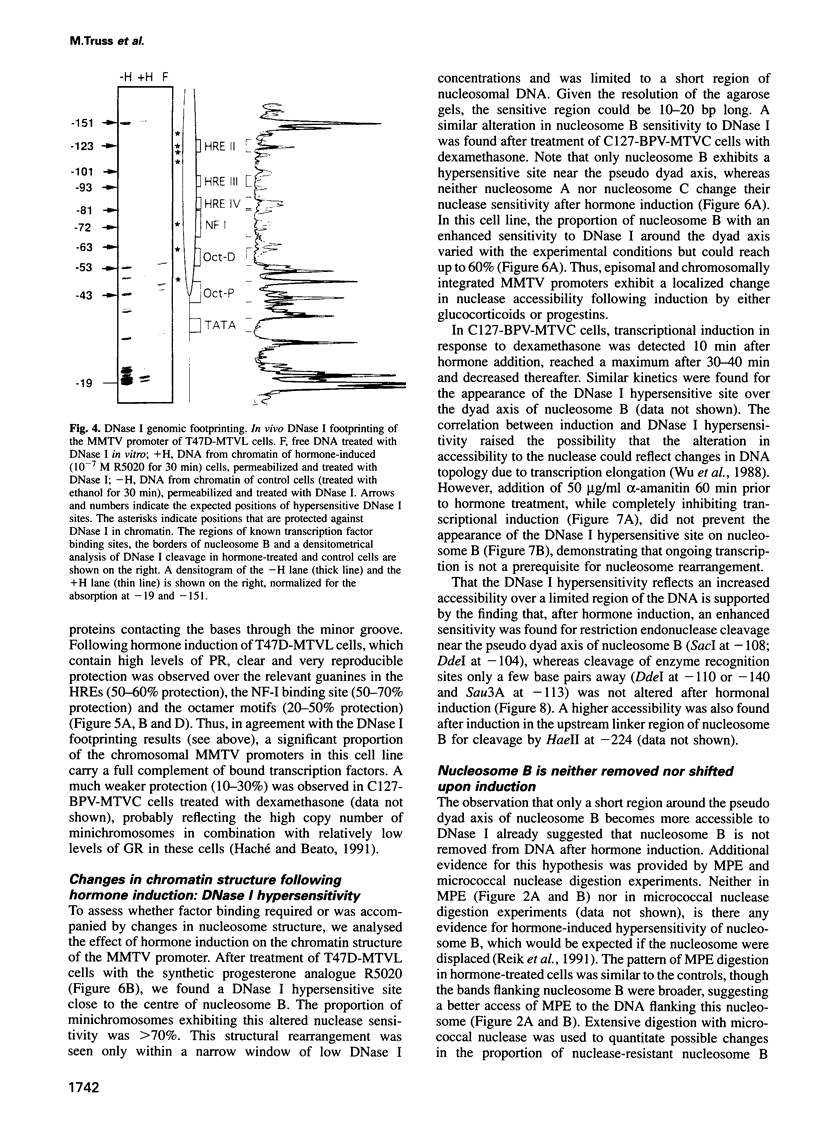
- Cartwright I. L., Hertzberg R. P., Dervan P. B., Elgin S. C. Cleavage of chromatin with methidiumpropyl-EDTA . iron(II). Proc Natl Acad Sci U S A. 1983 Jun;80(11):3213–3217. doi: 10.1073/pnas.80.11.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato A. C., Henderson D., Ponta H. The hormone response element of the mouse mammary tumour virus DNA mediates the progestin and androgen induction of transcription in the proviral long terminal repeat region. EMBO J. 1987 Feb;6(2):363–368. doi: 10.1002/j.1460-2075.1987.tb04763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato A. C., Miksicek R., Schütz G., Arnemann J., Beato M. The hormone regulatory element of mouse mammary tumour virus mediates progesterone induction. EMBO J. 1986 Sep;5(9):2237–2240. doi: 10.1002/j.1460-2075.1986.tb04490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalepakis G., Arnemann J., Slater E., Brüller H. J., Gross B., Beato M. Differential gene activation by glucocorticoids and progestins through the hormone regulatory element of mouse mammary tumor virus. Cell. 1988 May 6;53(3):371–382. doi: 10.1016/0092-8674(88)90157-2. [DOI] [PubMed] [Google Scholar]
- Chandler V. L., Maler B. A., Yamamoto K. R. DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell. 1983 Jun;33(2):489–499. doi: 10.1016/0092-8674(83)90430-0. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., Riegel A. T., Hager G. L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987 Jan 30;48(2):261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- Dong F., van Holde K. E. Nucleosome positioning is determined by the (H3-H4)2 tetramer. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10596–10600. doi: 10.1073/pnas.88.23.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasel N., Pearson K., Buetti E., Diggelmann H. The region of mouse mammary tumor virus DNA containing the long terminal repeat includes a long coding sequence and signals for hormonally regulated transcription. EMBO J. 1982;1(1):3–7. doi: 10.1002/j.1460-2075.1982.tb01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Subramani S. Firefly luciferase as a tool in molecular and cell biology. Anal Biochem. 1988 Nov 15;175(1):5–13. doi: 10.1016/0003-2697(88)90353-3. [DOI] [PubMed] [Google Scholar]
- Haché R. J., Beato M. Generation of C127-derived cell lines expressing estrogen or progesterone receptors for studying gene regulation on bovine papilloma virus minichromosomes. Receptor. 1991;1(4):217–228. [PubMed] [Google Scholar]
- Hayes J. J., Clark D. J., Wolffe A. P. Histone contributions to the structure of DNA in the nucleosome. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. J., Wolffe A. P. Histones H2A/H2B inhibit the interaction of transcription factor IIIA with the Xenopus borealis somatic 5S RNA gene in a nucleosome. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1229–1233. doi: 10.1073/pnas.89.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. J., Wolffe A. P. The interaction of transcription factors with nucleosomal DNA. Bioessays. 1992 Sep;14(9):597–603. doi: 10.1002/bies.950140905. [DOI] [PubMed] [Google Scholar]
- Hebbes T. R., Thorne A. W., Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988 May;7(5):1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz K. B., Zava D. T., Thilagar A. K., Jensen E. M., McGuire W. L. Steroid receptor analyses of nine human breast cancer cell lines. Cancer Res. 1978 Aug;38(8):2434–2437. [PubMed] [Google Scholar]
- Härtig E., Nierlich B., Mink S., Nebl G., Cato A. C. Regulation of expression of mouse mammary tumor virus through sequences located in the hormone response element: involvement of cell-cell contact and a negative regulatory factor. J Virol. 1993 Feb;67(2):813–821. doi: 10.1128/jvi.67.2.813-821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. R., Benyajati C. DNA-histone interactions are sufficient to position a single nucleosome juxtaposing Drosophila Adh adult enhancer and distal promoter. Nucleic Acids Res. 1993 Feb 25;21(4):957–967. doi: 10.1093/nar/21.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990 Jan 23;29(3):719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- Jantzen K., Fritton H. P., Igo-Kemenes T., Espel E., Janich S., Cato A. C., Mugele K., Beato M. Partial overlapping of binding sequences for steroid hormone receptors and DNaseI hypersensitive sites in the rabbit uteroglobin gene region. Nucleic Acids Res. 1987 Jun 11;15(11):4535–4552. doi: 10.1093/nar/15.11.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kalff M., Gross B., Beato M. Progesterone receptor stimulates transcription of mouse mammary tumour virus in a cell-free system. Nature. 1990 Mar 22;344(6264):360–362. doi: 10.1038/344360a0. [DOI] [PubMed] [Google Scholar]
- Klehr D., Maass K., Bode J. Scaffold-attached regions from the human interferon beta domain can be used to enhance the stable expression of genes under the control of various promoters. Biochemistry. 1991 Feb 5;30(5):1264–1270. doi: 10.1021/bi00219a015. [DOI] [PubMed] [Google Scholar]
- Langer S. J., Ostrowski M. C. Negative regulation of transcription in vitro by a glucocorticoid response element is mediated by a trans-acting factor. Mol Cell Biol. 1988 Sep;8(9):3872–3881. doi: 10.1128/mcb.8.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. Y., Hayes J. J., Pruss D., Wolffe A. P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993 Jan 15;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Lee H. L., Archer T. K. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol Cell Biol. 1994 Jan;14(1):32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Garrard W. T. Transcription-induced nucleosome 'splitting': an underlying structure for DNase I sensitive chromatin. EMBO J. 1991 Mar;10(3):607–615. doi: 10.1002/j.1460-2075.1991.tb07988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Garrard W. T. Uncoupling gene activity from chromatin structure: promoter mutations can inactivate transcription of the yeast HSP82 gene without eliminating nucleosome-free regions. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9166–9170. doi: 10.1073/pnas.89.19.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wrange O. Translational positioning of a nucleosomal glucocorticoid response element modulates glucocorticoid receptor affinity. Genes Dev. 1993 Dec;7(12A):2471–2482. doi: 10.1101/gad.7.12a.2471. [DOI] [PubMed] [Google Scholar]
- Lowy D. R., Rands E., Scolnick E. M. Helper-independent transformation by unintegrated Harvey sarcoma virus DNA. J Virol. 1978 May;26(2):291–298. doi: 10.1128/jvi.26.2.291-298.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckow B., Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987 Jul 10;15(13):5490–5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight G. S., Hager L., Palmiter R. D. Butyrate and related inhibitors of histone deacetylation block the induction of egg white genes by steroid hormones. Cell. 1980 Nov;22(2 Pt 2):469–477. doi: 10.1016/0092-8674(80)90357-8. [DOI] [PubMed] [Google Scholar]
- McPherson C. E., Shim E. Y., Friedman D. S., Zaret K. S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993 Oct 22;75(2):387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- Miksicek R., Borgmeyer U., Nowock J. Interaction of the TGGCA-binding protein with upstream sequences is required for efficient transcription of mouse mammary tumor virus. EMBO J. 1987 May;6(5):1355–1360. doi: 10.1002/j.1460-2075.1987.tb02375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C., Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993 Nov;12(11):4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möws C. C., Preiss T., Slater E. P., Cao X., Verrijzer C. P., van Der Vliet P. C., Beato M. Two independent pathways for transcription from the MMTV promoter. J Steroid Biochem Mol Biol. 1994 Oct;51(1-2):21–32. doi: 10.1016/0960-0760(94)90111-2. [DOI] [PubMed] [Google Scholar]
- Nowock J., Borgmeyer U., Püschel A. W., Rupp R. A., Sippel A. E. The TGGCA protein binds to the MMTV-LTR, the adenovirus origin of replication, and the BK virus enhancer. Nucleic Acids Res. 1985 Mar 25;13(6):2045–2061. doi: 10.1093/nar/13.6.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks W. P., Hubbell E. S., Goldberg R. J., O'Neill F. J., Scolnick E. M. High frequency variation in mammary tumor virus expression in cell culture. Cell. 1976 May;8(1):87–93. doi: 10.1016/0092-8674(76)90189-6. [DOI] [PubMed] [Google Scholar]
- Pederson D. S., Fidrych T. Heat shock factor can activate transcription while bound to nucleosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1994 Jan;14(1):189–199. doi: 10.1128/mcb.14.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmann T. Glucocorticoid receptor DNA-binding specificity is increased by the organization of DNA in nucleosomes. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3884–3888. doi: 10.1073/pnas.89.9.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmann T., Wrange O. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 1988 Oct;7(10):3073–3079. doi: 10.1002/j.1460-2075.1988.tb03172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry C. A., Annunziato A. T. Histone acetylation reduces H1-mediated nucleosome interactions during chromatin assembly. Exp Cell Res. 1991 Oct;196(2):337–345. doi: 10.1016/0014-4827(91)90269-z. [DOI] [PubMed] [Google Scholar]
- Peterson C. L., Dingwall A., Scott M. P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992 Feb 7;68(3):573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Riggs A. D. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev. 1991 Jun;5(6):1102–1113. doi: 10.1101/gad.5.6.1102. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Piña B., Barettino D., Truss M., Beato M. Structural features of a regulatory nucleosome. J Mol Biol. 1990 Dec 20;216(4):975–990. doi: 10.1016/S0022-2836(99)80015-1. [DOI] [PubMed] [Google Scholar]
- Piña B., Brüggemeier U., Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990 Mar 9;60(5):719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- Power R. F., Mani S. K., Codina J., Conneely O. M., O'Malley B. W. Dopaminergic and ligand-independent activation of steroid hormone receptors. Science. 1991 Dec 13;254(5038):1636–1639. doi: 10.1126/science.1749936. [DOI] [PubMed] [Google Scholar]
- Prior C. P., Cantor C. R., Johnson E. M., Littau V. C., Allfrey V. G. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 1983 Oct;34(3):1033–1042. doi: 10.1016/0092-8674(83)90561-5. [DOI] [PubMed] [Google Scholar]
- Reik A., Schütz G., Stewart A. F. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: induction leads to a reversible disruption of nucleosomes over an enhancer. EMBO J. 1991 Sep;10(9):2569–2576. doi: 10.1002/j.1460-2075.1991.tb07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985 Dec 16;4(13A):3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Foy H., Hager G. L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987 Aug;6(8):2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridsdale J. A., Hendzel M. J., Delcuve G. P., Davie J. R. Histone acetylation alters the capacity of the H1 histones to condense transcriptionally active/competent chromatin. J Biol Chem. 1990 Mar 25;265(9):5150–5156. [PubMed] [Google Scholar]
- Rigaud G., Roux J., Pictet R., Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991 Nov 29;67(5):977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Shimizu M., Johnson L., Grunstein M., Simpson R. T. Stable nucleosome positioning and complete repression by the yeast alpha 2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992 Mar;6(3):411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- Sarver N., Byrne J. C., Howley P. M. Transformation and replication in mouse cells of a bovine papillomavirus--pML2 plasmid vector that can be rescued in bacteria. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7147–7151. doi: 10.1073/pnas.79.23.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidereit C., Beato M. Contacts between hormone receptor and DNA double helix within a glucocorticoid regulatory element of mouse mammary tumor virus. Proc Natl Acad Sci U S A. 1984 May;81(10):3029–3033. doi: 10.1073/pnas.81.10.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidereit C., Geisse S., Westphal H. M., Beato M. The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus. Nature. 1983 Aug 25;304(5928):749–752. doi: 10.1038/304749a0. [DOI] [PubMed] [Google Scholar]
- Schild C., Claret F. X., Wahli W., Wolffe A. P. A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J. 1993 Feb;12(2):423–433. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Roth S. Y., Szent-Gyorgyi C., Simpson R. T. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 1991 Oct;10(10):3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Conneely O. M., O'Malley B. W. Modulation of the ligand-independent activation of the human estrogen receptor by hormone and antihormone. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6120–6124. doi: 10.1073/pnas.90.13.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straka C., Hörz W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 1991 Feb;10(2):361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J., Hörz W. Histones, nucleosomes and transcription. Curr Opin Genet Dev. 1993 Apr;3(2):219–225. doi: 10.1016/0959-437x(93)90026-l. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Dong Y., Li Q., Okret S., Gustafsson J. A. Identification and characterization of a cis-acting element that interferes with glucocorticoid-inducible activation of the mouse mammary tumor virus promoter. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5393–5397. doi: 10.1073/pnas.88.12.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I. C., Workman J. L., Schuetz T. J., Kingston R. E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991 Jul;5(7):1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- Thomas G. H., Elgin S. C. Protein/DNA architecture of the DNase I hypersensitive region of the Drosophila hsp26 promoter. EMBO J. 1988 Jul;7(7):2191–2201. doi: 10.1002/j.1460-2075.1988.tb03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toohey M. G., Lee J. W., Huang M., Peterson D. O. Functional elements of the steroid hormone-responsive promoter of mouse mammary tumor virus. J Virol. 1990 Sep;64(9):4477–4488. doi: 10.1128/jvi.64.9.4477-4488.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderbilt J. N., Miesfeld R., Maler B. A., Yamamoto K. R. Intracellular receptor concentration limits glucocorticoid-dependent enhancer activity. Mol Endocrinol. 1987 Jan;1(1):68–74. doi: 10.1210/mend-1-1-68. [DOI] [PubMed] [Google Scholar]
- Wallrath L. L., Lu Q., Granok H., Elgin S. C. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. Bioessays. 1994 Mar;16(3):165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- Wang Z., Becker M. M. Selective visualization of gene structure with ultraviolet light. Proc Natl Acad Sci U S A. 1988 Feb;85(3):654–658. doi: 10.1073/pnas.85.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992 Nov;8(11):387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. New insights into chromatin function in transcriptional control. FASEB J. 1992 Dec;6(15):3354–3361. doi: 10.1096/fasebj.6.15.1464369. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Buchman A. R. Multiple functions of nucleosomes and regulatory factors in transcription. Trends Biochem Sci. 1993 Mar;18(3):90–95. doi: 10.1016/0968-0004(93)90160-o. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Kingston R. E. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science. 1992 Dec 11;258(5089):1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Yoshinaga S. K., Peterson C. L., Herskowitz I., Yamamoto K. R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992 Dec 4;258(5088):1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Yamamoto K. R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984 Aug;38(1):29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]