Abstract
DNA damage by ultraviolet (UV) light poses a risk for mutagenesis and a potential hindrance for cell cycle progression. Cells cope with UV-induced DNA damage through two general strategies to repair the damaged nucleotides and to promote cell cycle progression in the presence of UV-damaged DNA. Defining the genetic pathways and understanding how they function together to enable effective tolerance to UV remains an important area of research. The structural maintenance of chromosomes (SMC) proteins form distinct complexes that maintain genome stability during chromosome segregation, homologous recombination, and DNA replication. Using a forward genetic screen, we identified two alleles of smc-5 that exacerbate UV sensitivity in Caenorhabditis elegans. Germ cells of smc-5-defective animals show reduced proliferation, sensitivity to perturbed replication, chromatin bridge formation, and accumulation of RAD-51 foci that indicate the activation of homologous recombination at DNA double-strand breaks. Mutations in the translesion synthesis polymerase polh-1 act synergistically with smc-5 mutations in provoking genome instability after UV-induced DNA damage. In contrast, the DNA damage accumulation and sensitivity of smc-5 mutant strains to replication impediments are suppressed by mutations in the C. elegans BRCA1/BARD1 homologs, brc-1 and brd-1. We propose that SMC-5/6 promotes replication fork stability and facilitates recombination-dependent repair when the BRC-1/BRD-1 complex initiates homologous recombination at stalled replication forks. Our data suggest that BRC-1/BRD-1 can both promote and antagonize genome stability depending on whether homologous recombination is initiated during DNA double-strand break repair or during replication stalling.
Keywords: SMC5/6, DNA replication, BRCA1, DNA repair, Caenorhabditis elegans
THE nuclear genome is constantly exposed to a variety of genotoxic insults. It has been estimated that tens of thousands damaging events attack the DNA of each cell on a daily basis (De Bont and Van Larebeke 2004). Genome stability is maintained by numerous specialized DNA repair systems that recognize and remove specific types of alterations in the DNA. During replication, obstructive DNA lesions, like those caused by ultraviolet (UV) irradiation, can lead to replication stalling and eventually to fork collapse (Lehmann 2011). Improper resolution of blocked replication forks can result in segregation errors during subsequent cell division, leading to chromosomal aberrations. Such genome instability comprises a hallmark of cancer development (Jackson and Bartek 2009). While the functions of DNA repair pathways have been investigated over several decades, it remains challenging to understand the complex interactions between functionally overlapping repair pathways at sites of replication fork collapse.
To overcome the replicative impasse that is posed by DNA lesions, cells can employ two general strategies: either halt cell cycle progression to allow time for repair or read-through and bypass the damaged template. The nucleotide excision repair (NER) pathway removes UV-induced cyclobutane pyrimidine dimers (CPDs) or 6-4 photoproducts upon detection through two distinct damage recognition pathways (Cleaver et al. 2009). The global genome (GG-) NER pathway is important in surveying the entire genome for UV lesions and in removing them before they block replication fork progression, whereas transcription-coupled (TC-) NER recognizes lesions when RNA polymerase II stalls during transcription elongation. In contrast to the removal and repair of the DNA lesions, cells may be capable of continuing DNA replication in spite of the presence of DNA lesions either by switching to error-prone translesion synthesis (TLS) polymerases or by employing homologous recombination (HR). DNA polymerase η (POLH) is particularly important for reading through UV-induced lesions when the replication fork stalls (Sale et al. 2012). Homologous recombination (HR) plays an important role in resolving collapsed replication forks that require recombination repair for restart (Petermann and Helleday 2010). HR is promoted by several proteins, including BRCA1, which forms a heterodimeric complex with BARD1 (Silver and Livingston 2012) and has recently been implicated in promoting recombination repair at collapsed replication forks (Pathania et al. 2011). Mutations in BRCA1 are associated with increased susceptibility to breast and ovarian cancers (Silver and Livingston 2012). In C. elegans the BRC-1/BRD-1 complex functions during repair of meiotic or ionizing radiation (IR)-induced double-strand breaks (DSBs) (Boulton et al. 2004; Adamo et al. 2008). How these distinct response pathways are coordinated to recover perturbed replication forks at DNA lesions remains incompletely understood.
The structural maintenance of chromosomes (SMC) complexes maintain genome stability through various mechanisms. The six subtypes of eukaryotic SMC proteins form three unique heterodimers that associate with specific sets of non-SMC subunits (reviewed in Nasmyth and Haering 2005 and Hirano 2006). The SMC-1/3 “cohesin” complex establishes cohesion between sister chromatids, while the SMC-2/4 “condensin” complex mediates chromosome condensation and resolution. Cohesin and condensin also function in DNA repair (Wu and Yu 2012). Cohesin is recruited to sites of DNA DSBs to facilitate HR through sister-chromatid cohesion and to elicit an efficient DNA damage checkpoint response (Kim et al. 2002; Ström et al. 2004; Unal et al. 2004). Condensin is implicated in repair of single- and double-strand breaks and in ribosomal DNA stability (Aono et al. 2002; Heale et al. 2006; Tsang et al. 2007; Wood et al. 2008). The function of the SMC-5/6 complex is less well characterized. The first smc6 mutations were identified in Schizosaccharomyces pombe where they confer hypersensitivity to UV and IR (Lehmann et al. 1995). smc6 is genetically epistatic with S. pombe rad51, rhp51, indicating a function in HR (Lehmann et al. 1995). Subsequent studies indeed implicated Smc5/6 in the resolution of HR structures (Ampatzidou et al. 2006; Branzei et al. 2006; De Piccoli et al. 2006; Sollier et al. 2009). Moreover, yeast Smc5/6 is important for restarting collapsed replication forks, likely by resolving recombination intermediates when HR complexes initiate template switches amid obstructing DNA damage (Ampatzidou et al. 2006; Santa Maria et al. 2007). Consistent with a conserved function during HR, C. elegans SMC-5/6 promotes recombination repair during meiosis (Bickel et al. 2010).
In C. elegans, mutations in NER genes and polh-1 confer UV hypersensitivity at distinct developmental stages. In adult worms, only the germline contains actively proliferating cells while somatic tissues are postmitotic. In response to UV irradiation, germ cells in mitosis transiently halt the cell cycle, while germ cells in pachytene (prophase I) of meiosis undergo apoptosis (Stergiou et al. 2007). In the germline and early embryos, GG-NER is particularly important for UV resistance. In contrast, TC-NER is most important for UV resistance during early larval stages. Complete inactivation of NER confers strongly elevated UV sensitivity in all cell types (Lans et al. 2010). The TLS polymerase POLH-1 is important for UV resistance during the rapid cell divisions that take place during early embryonic development (Holway et al. 2006; Roerink et al. 2012).
Here, we have employed the metazoan C. elegans as a model system to investigate how the various DNA repair systems interact to ensure genome stability in proliferating germ cells. Using forward genetics, we isolated two alleles of smc-5 that confer UV sensitivity in the germline. Similarly to S. pombe (Lehmann et al. 1995), C. elegans smc-5 functions in parallel to NER to maintain genome stability in the presence of UV lesions. Consistent with facilitating replication fork restart, smc-5 mutants exhibit synthetic lethality with a mutation in the DNA primase div-1 that functions in DNA replication. Inactivation of polh-1-mediated TLS in smc-5 mutants synergistically enhanced UV sensitivity. Mutations in smc-5 lead to accumulation of RAD-51 foci and enhanced chromosomal BRD-1 recruitment. Inactivation of the BRC-1/BRD-1 complex suppressed accumulation of RAD-51 foci and chromosome bridge formation as well as the DNA damage sensitivity in smc-5 mutants. Our results support a model in which the BRC-1/BRD-1 complex initiates the recruitment of HR factors to stalled replication forks, where their presence is toxic when the SMC-5/6 complex is dysfunctional.
Materials and Methods
Worm strains
Worms were maintained at 20° on nematode growth medium (NGM) agar plates with Escherichia coli strain OP50 as food source according to standard protocols (Brenner 1974). All experiments were performed at 20°. Strains used are listed in Supporting Information, Table S1. Given that loss of the SMC-5/6 complex leads to transgenerational sterility in C. elegans (Bickel et al. 2010), all strains used in this study were stabilized by maintaining them in a heterozygous state using a GFP-marked variant of the genetic balancer mIn1 (II) (Edgley and Riddle 2001).
EMS mutagenesis
Synchronized L1 wild-type worms were plated on NGM plates seeded with E. coli OP50, grown until L4, and treated with 30 μM EMS in M9 buffer for 4 hr at room temperature. Residual EMS was neutralized with 1 M NaOH and removed by two washes with 4 ml M9 buffer, and the worms were then plated on OP50-seeded NGM plates.
CPD repair assay using Slotblot
Day 1 adult worms were treated with 60 mJ/cm2 ultraviolet B and either processed directly or maintained at 20° for 24 hr to allow time for repair. After washing with ice-cold M9 buffer samples were quick-frozen in liquid nitrogen. Genomic DNA was prepped using Gentra Puregene Tissue Kit (Qiagen). Three milliliters of cell lysis solution and 15 μl of Proteinase K were added, and the mix was incubated for 3 hr at 55°. Samples were allowed to cool down to room temperature before adding 15 μl of RNAse A solution and 30 min of incubation at 37°. Samples were cooled for 3 min on ice and 1 ml of protein precipitation solution was added. After vortexing for 20 sec and centrifugation for 10 min at 2000 × g, supernatant was transferred to a new tube. For DNA precipitation, 3 ml of isopropanol was added and tubes were gently inverted 50 times. Then samples were centrifuged for 3 min at 2000 × g, and the pellet was allowed to air dry for 10 min at room temperature. Genomic DNA was dissolved in DNA rehydration buffer and concentration measured using a Nanodrop 8000. A 1:2 dilution series starting with 1 μg was prepared, and dilutions were denatured for 5 min at 95°, put directly on ice, and blotted onto an Hybond nylon membrane (Amersham) using a Whatman 96-well slot blotting device at 300 mbar vacuum. Cross-linking of the DNA was carried out for 2 hr at 80°. The membrane was blocked for 30 min in 3% milk/PBS. Anti-cyclobutane pyrimidine dimers (clone TDM-2, Cosmo Bio) were diluted 1:15,000 in PBS containing 0.1% Tween20 (PBST). The membrane was incubated in antibody solution overnight at 4°, washed 3× in PBST, and incubated 1 hr with 1:10,000 secondary antibody solution peroxidase-conjugated AffiniPure Goat Anti-Mouse IgG + IgM (H+L) (JacksonImmuno Research) in PBST at room temperature and washed 3× in PBST. DNA lesions were visualized by ECL plus Western blotting reagent (Amersham) and exposing CL-Xposure Film (Thermo Scientific).
Immunofluorescence
Extruded germlines and whole larvae were fixed on polylysine-coated slides using 1.5–3.7% paraformaldehyde. After 5 min of incubation at room temperature, the worms were freeze-cracked and then incubated in a 1:1 mixture of methanol and acetone or solely methanol at −20°. Some of the adult germline samples were permeabilized by washing in PBS 1% TritonX100, followed by washing with PBS 0.1% Tween20 (washing buffer). To saturate unspecific binding sites, the slides were incubated with washing buffer containing 10% donkey serum (blocking buffer) or 0.5% BSA for 30 min at room temperature. Primary antibodies were diluted in blocking buffer and incubated on slides in a humid chamber at 4° overnight. After washing, secondary antibodies were diluted in blocking buffer and allowed to bind at room temperature for 2 hr in the dark. Excess antibody was removed by washing, and slides were mounted using DAPI Fouromount-G (SouthernBiotech). Primary antibodies were diluted with the following: rabbit anti-RAD-51 antibody (SDIX) 1:300; rabbit anti-BRD-1 (kindly provided by Simon Boulton) (Boulton et al. 2004) 1:400; and rabbit anti-phospho-Chk1 (Ser345) antibody (Cell Signaling Technology) 1:50. Blocking was not applied for SMC-6 immunostaining. Secondary antibodies Alexa Fluor 594 Goat Anti-Rabbit and Alexa Fluor 488 Goat Anti-Rabbit IgG (Invitrogen) were used at 1:300 to 1:500 dilutions for detection of the respective primary antibodies.
Germline development assay
Day 1 adults were bleached and eggs were hatched overnight at 20° shaking. In case of balanced mutants, homozygous F1 from heterozygous mothers were used for synchronization by bleaching. L1 larvae were transferred to OP50-seeded NGM plates and treated with the indicated dose of radiation. Three days after treatment the number of worms with normal germline, malformed germline, or no germline was documented using the Leica M 165 C stereomicroscope. Micrographs of germline development were taken using the Axio Imager A1 (Carl Zeiss).
Quantification of germ cells in the proliferative zone
The proliferative zone starts at the distal tip of the gonad arm and continues until the appearance of nuclei with crescent-shape DNA morphology, characteristic of leptotene/zygotene (transition zone) germ cells as defined previously (Crittenden et al. 2006).
Hydroxyurea treatment
Worms were synchronized at early L1 development by hatching eggs in the absence of a bacterial food source. The starved L1 larvae were transferred to NGM plates seeded with OP50 bacteria to resume development. For hydroxyurea (HU) treatment, L1 larvae were transferred to NGM plates containing the indicated concentration of HU. After growth for 46–48 hr, the late-L4 stage larvae were harvested for analyses. Experiments using different lots of HU show variability in the severity of the HU-induced chromatin bridge defect in the smc-5(ok2421) mutant, ranging from 60 to >90% frequency for this defect.
5-Ethylnyl-2′-deoxyuridine labeling
L4 larvae (46–48 hr post-L1) were fed on 5-ethylnyl-2′-deoxyuridine (EdU)-labeled bacteria for the indicated time periods and then dissected to extrude the germline for fixation. EdU detection was performed using Click-IT EdU Alexa Fluor-555 labeling kit (Invitrogen) as described earlier (Dorsett et al. 2009). Fixed and DAPI-stained samples were incubated with two rounds of freshly prepared Click-IT cocktail for 30 min each. Germ cells at the distal-most 50 μm of the proliferative region were examined for the presence of EdU labeling, defined as nuclear Alexa 555 fluorescence. As negative controls, we performed the conjunction reaction on male and hermaphrodite worms that did not receive EdU, and we also dissected males lacking EdU with adult hermaphrodites that were fed EdU on the same slide. The latter internal control ensured specificity of EdU detection to germ cells exposed to EdU. Images shown in Figure 3C were deconvolved with Huygens Essential (SVI).
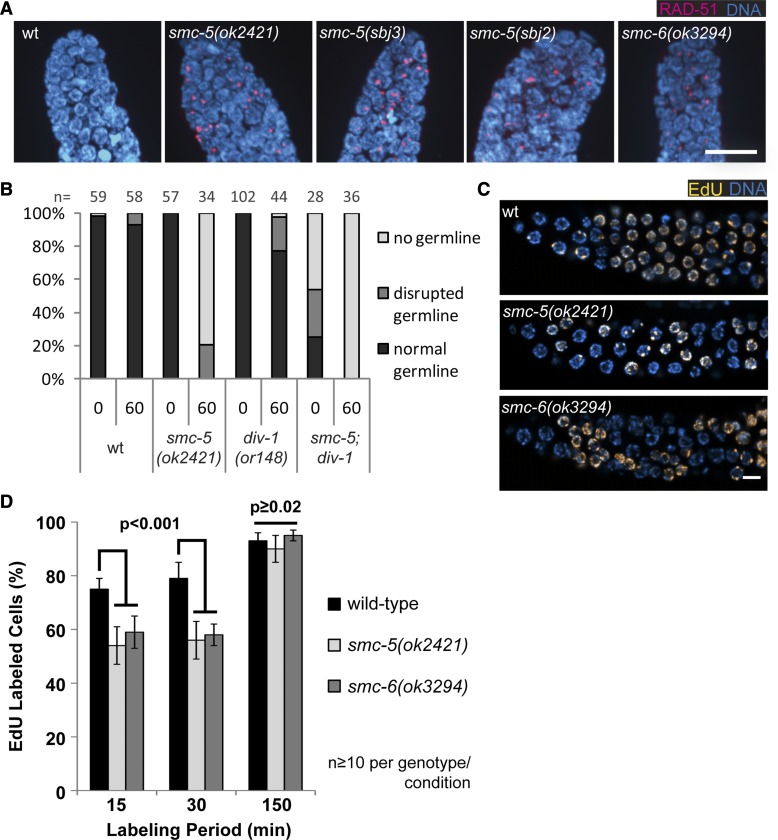
Figure 3.
smc-5 and smc-6 are hypersensitive to replication stress. (A) Representative images showing αRAD-51 and DAPI staining of mitotic zone germline of indicated genotypes. Bar, 10 µm. (B) Germline development quantification of worms 3 days after irradiation with UVB at L1 stage or untreated control worms. Worms were categorized into groups of “normal,” “disrupted,” and “no germline” by inspection on a dissection microscope. “n” indicates number of animals assessed. A representative experiment is shown. (C) EdU incorporation in L4 germ cell DNA detected by Click-It Alexa555 conjugation and DAPI staining of DNA. Germlines were isolated after feeding worms for 15 min on EdU-containing bacteria. Bar, 5 µm. (D) Mean percentage of germ cells with EdU incorporation at defined time periods of labeling. A minimum of 10 germlines per genotype and labeling period were analyzed. Statistical analyses used the two-tailed Fisher’s exact test comparing the total number of EdU-positive and negative germ cells, and P-values are indicated. Error bars represent 95% confidence interval.
Western blotting
For each genotype, 150 adult worms were collected and boiled for 5 min in 1× SDS buffer supplemented with 3.8 M urea, frozen and thawed once, and boiled again with 2% β-mercaptoethanol immediately prior to SDS-PAGE. SMC-5 and AMA-1 (RNA polymerase II) were detected by rabbit anti-SMC-5 antibodies (Bickel et al. 2010) at 1:500 dilution and rabbit anti-phosphorylated CTD (RNA polymerase II) antibodies (Abcam ab5131) at 1:1000 dilution, respectively.
Statistical analysis
To valuate statistical differences, tests were applied as mentioned in the figure legends, and, for germline development assays, χ2 and two-tailed Fisher’s exact tests were utilized to determine P-values (Table S2). For execution of Fisher’s exact test, categories of normal and disrupted germline were summed up and compared to the group of worms without germline.
Results
smc-5 is required for resistance to UV-induced DNA lesions in the C. elegans germline
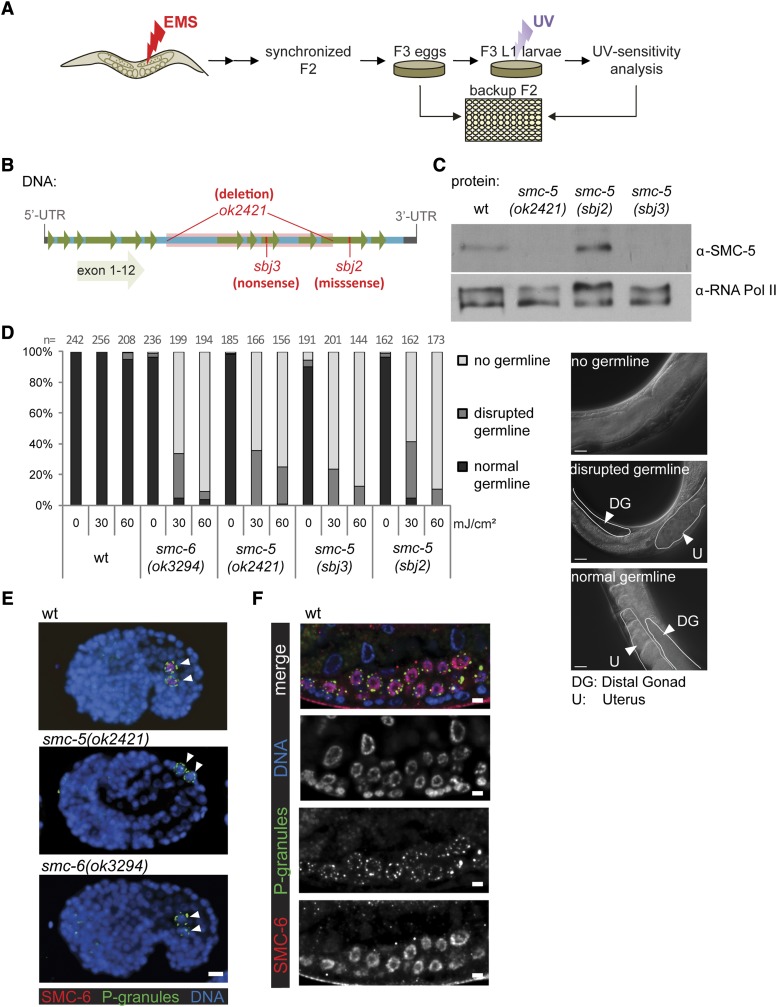
We performed a forward mutagenesis screen to identify genes involved in the response to UV-induced DNA damage. We mutagenized C. elegans with EMS and recovered mutant worms that are hypersensitive to UVB irradiation (Figure 1A). We UV-treated populations of L1 larvae and followed their developmental growth. Worms can be readily synchronized at the L1 stage, which is the earliest of four larval stages preceding adulthood. We identified one mutant that exhibited a complete L1 arrest after treatment with low doses of UV containing a novel allele of the NER endonuclease xpg-1, thus providing a proof of principle that our screening strategy was effective at discovering mutations in DNA repair genes (data not shown). We also isolated two mutant strains that displayed hypersensitivity to UV treatment specifically in the germline, reminiscent of the UV-sensitivity phenotypes of GG-NER xpc-1 and rad-23 mutants (Figure 2, B and C) (Lans et al. 2010). Noncomplementation analysis for UV hypersensitivity indicated that the mutations are allelic. Subsequent SNP mapping and whole-genome sequencing revealed two different mutations in smc-5 (Figure 1B). The sbj2 allele has a missense mutation in the ABC transporter signature motif of smc-5 (Figure S1A). Previous studies in yeast indicate that the ATPase activity of the Smc5/6 complex is essential for its function (Verkade et al. 1999; Fousteri and Lehmann 2000). The sbj2 allele does not reduce SMC-5 protein or messenger RNA (mRNA) level (Figure 1C and Figure S1B). The sbj3 allele introduces a premature stop codon and disrupts the expression of the full-length protein (Figure 1, B and C). Similar to sbj2 and sbj3, we found that the smc-5(ok2421) and smc-6(ok3294) mutations also confer UV hypersensitivity (Figure 1D). Noncomplementation test for UV hypersensitivity found sbj2 and sbj3 to be allelic to the smc-5(ok2421) deletion mutant (data not shown). These results further corroborate the causal role of the smc-5 mutation in the UV hypersensitivity.
Figure 1.
Screening for UV-sensitive mutants identified two novel alleles of smc-5. (A) Worms were mutagenized with EMS and F2 generation synchronized by bleaching. After egg laying for 2–3 hr, F2 adults were backed up in 96-well plate liquid culture. F3 larvae were irradiated with 60 mJ/cm2 UVB and screened for impaired development and reproduction 48 hr post-irradiation. Phenotype was confirmed by using the worms from the 96-well backup plate. (B) Scheme of smc-5 genomic region (chromosome II: 5,062,121–5,067,121 bp) with exon location. sbj2, sbj3, and ok2421 alleles are indicated. sbj2 is a guanine-to-adenine missense mutation changing glycine to arginine, and sbj3 is a cytosine-to-thymine mutation transforming glutamine into a stop codon. (C) Western blot indicating protein levels in wild-type and smc-5 mutants. RNA Pol II was used as loading control. (D) Quantification from a representative experiment examining germline development of worms 3 days after mock or UVB irradiation at the L1 stage. Worms were grouped into categories of “normal,” “disrupted,” and “no germline.” “n” indicates number of animals assessed. Representative DIC micrographs from each of the three categories show the middle one-third of smc-5(ok2421) adult worms with the germline and uterus outlined. Bar, 20 µm. (E and F) Immunofluorescence of SMC-6 and DAPI staining of DNA. Bar, 5 µm. (E) Staining of embryos post 100-cell stage. P-granule immunofluorescence marks primordial germ cells Z2 and Z3 (arrows). (F) Proliferating germ cells from a L2/L3 larva with costaining of SMC-6 and P-granules as a marker for germ cells. All micrographs were processed by deconvolution as described in Materials and Methods.
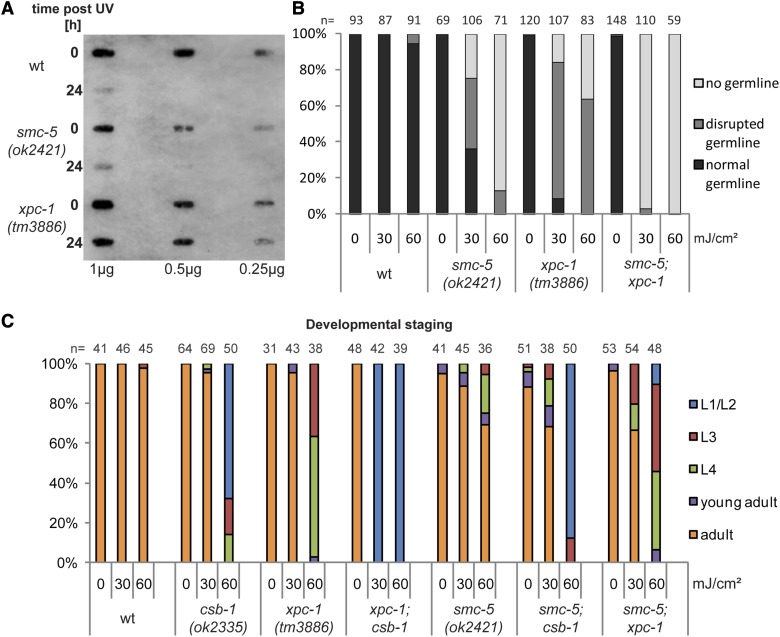
Figure 2.
smc-5 acts in parallel to NER in genome maintenance upon UV irradiation. (A) Slot blot stained with αCPD antibody of whole genomic DNA of young adult worms immediately (0 hr) or 1 day after (24 hr) irradiation with 60 mJ/cm2 UVB. From left to right, decreasing amounts of DNA were blotted on the membrane. (B) Germline development quantification of worms 3 days after irradiation with UVB at L1 stage or untreated control worms. Worms were categorized into groups of “normal,” “disrupted,” and “no germline” by inspection on a dissection microscope. (C) Percentage of larval stages 72 hr after UVB irradiation at L1 larval stage of the indicated genotypes. “n” indicates number of animals assessed. A representative experiment is shown.
The requirement of the SMC-5/6 complex during development has not been closely examined. Immunostaining for SMC-6 revealed tissue-specific enrichment in the germ-cell lineage with high levels of staining detected in the primordial germ cells in embryos (Figure 1E) and proliferating germ cells in larvae (Figure 1F), which are disrupted in the smc-5(ok2421) and the smc-6(ok3294) deletion mutant embryos (Figure 1E). Low levels of immunostaining are also detected in somatic blastomeres of young embryos with 100 cells or less (data not shown), which suggests that SMC-6 expression is ubiquitous but enriched in the germline, similar to the mRNA expression of human SMC5 and SMC6 that is ubiquitous but highly enriched in the testis (Taylor et al. 2001).
SMC-5 confers tolerance to UV damage in parallel to NER-mediated repair of CPD lesions
Given the similarity of UV-induced defects in smc-5 mutants compared to GG-NER mutants, we tested whether SMC-5 is needed for the repair of UV-induced DNA lesions. The smc-5(ok2421) mutants showed equivalent capacity to remove CPD lesions compared to wild-type, while xpc-1 mutants that are defective in GG-NER failed to remove CPDs after UV irradiation (Figure 2A). This suggests that SMC-5 does not mediate DNA damage tolerance through direct repair of DNA lesions. To further test this prediction, we examined the genetic interactions between the smc-5(ok2421) deletion mutant and loss-of-function mutations in the GG and the TC branches of the NER pathway. If SMC-5 functions in parallel to NER to promote tolerance to UV damage, then the combination of the smc-5(ok2421) null mutant with null mutations in the NER pathway should enhance the UV-damage sensitivity. In agreement with this prediction, the smc-5(ok2421);xpc-1(tm3886) double mutant exhibited greater disruption in germline development from UV damage compared to the single mutants of smc-5(ok2421) and xpc-1(tm3886); this is especially apparent for the 30 mJ/cm2 dose at which the fraction of worms lacking a germline increases from <25% in the single mutants to nearly 100% in the double mutant (Figure 2B). Combining the smc-5(ok2421) mutation with the GG-NER xpc-1 mutant or the TC-NER csb-1 mutant also enhances UV-induced delay in somatic development compared to the single mutants (Figure 2C), indicating that SMC-5 functions in parallel to both branches of the NER pathway in the soma. It should be noted that the smc-5(ok2421) mutant showed higher UV sensitivity in the germline than in the soma, with 100% of animals exhibiting germline defects (Figure 2B) compared to only ∼30% of animals with somatic developmental delay (Figure 2C) after a 60 mJ/cm2 UV treatment. Together, these findings indicate that SMC-5 is required for an additional DNA damage tolerance mechanism other than NER-mediated DNA repair.
smc-5/6 mutants are sensitive to replicative stress
The smc-5 and smc-6 mutant strains exhibit several defects suggestive of impaired DNA replication. The three smc-5 mutant strains and the smc-6(ok3294) strain all exhibited ectopic RAD-51 foci in the mitotic germline in adults (Figure 3A) (Bickel et al. 2010) and in L4 larvae (Figure S2). RAD-51 foci in germ cells can form following replication stress (Ward et al. 2007) and following the creation of meiotic DSBs (Alpi et al. 2003). Unlike meiotic RAD-51 foci, the RAD-51 foci in the mitotic germline of smc-5(ok2421) mutants do not require SPO-11, a nuclease involved in DSB formation as evidenced by RAD-51 staining in the smc-5(ok2421);spo-11(ok79) double mutant (Figure S2B). The mitotic germline of smc-5(ok2421) and smc-6(ok3294) mutants also had chromatin bridges between germ cells (Figure S3A), which were specifically enhanced in the smc-5(ok2421) and smc-6(ok3294) mutants compared to wild-type by prolonged exposure to 5 mM HU, which causes replication stress during larval development (Figure S3B). Likewise, germline development of smc-5 mutant animals is impaired when worms are grown on plates containing HU (Figure 5B). Moreover, pronounced chromatin bridges were also observed in the intestine (Figure S3C).
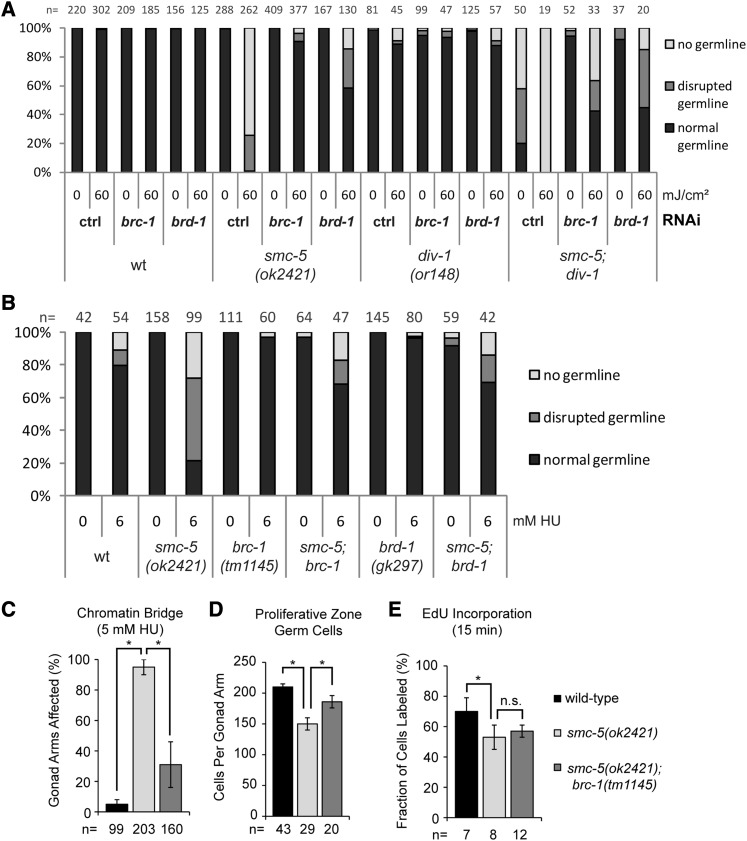
Figure 5.
Inactivation of the BRC-1/BRD-1 complex alleviates sensitivity of smc-5 mutants to DNA damage and replicative impairments. (A) Graph shows germline development quantification of worms 3 days after irradiation with UVB at L1 stage or untreated control worms. As read-out, worms were categorized into groups of “normal,” “disrupted,” and “no germline” by inspection on a dissection microscope. “n” indicates number of animals assessed. A representative experiment is shown. (B) Germline development quantification of worms raised on NGM plates containing indicated concentration of HU starting from L1 larval stage. Worms were categorized into groups of “normal,” “disrupted,” and “no germline” by inspection on a dissection microscope. “n” indicates number of worms inspected. A representative experiment is shown. (C) Fraction of gonad arms with chromatin bridges is averaged from three independent experiments with error bars indicating 95% confidence interval (C.I.) of the mean. The asterisk represents a P-value < 0.001 calculated by two-tailed Fisher’s exact test comparing the total chromatin-bridge positive and negative gonad arms combined from the three experiments. “n” represents total number of gonad arms examined. (D) Number of germ cells in the proliferative zone. The number of gonad arms analyzed (“n”) is indicated at the base of each column. P-values calculated by two-tailed Student’s t-test are indicated. Error bars indicate the 95% C.I. (E) Mean fraction of germ cells with EdU incorporation after 15 min of labeling is presented with error bars indicating 95% C.I. Number of germ lines examined (“n”) is indicated. The asterisk represents a P-value < 0.001 calculated by two-tailed Fisher’s exact test comparing the total number EdU-positive and -negative germ cells per genotype.
Given the technical challenge of directly examining DNA replication in an intact germline, we applied genetic assays to indirectly assess replication-associated phenotypes. Studies in budding and fission yeast have implicated the yeast Smc5/6 complexes in promoting the progression of replication forks via various mechanisms such as maintenance of stably stalled replication forks, altering DNA topology conducive to processivity of the replication fork, and restart of collapsed forks (Branzei et al. 2006; Irmisch et al. 2009; Kegel et al. 2011). We examined genetic interactions between smc-5 and div-1, which encodes the B-subunit of DNA polymerase α-primase (Encalada et al. 2000). Disruption in DIV-1 primase activity is expected to impede the progression of DNA replication forks. The temperature-sensitive div-1(or148) mutants developed germlines normally at the semipermissive temperature of 20° (Figure 3B). Upon irradiation with 60 mJ/cm2, only a slightly increased number of div-1(or148) mutants developed disrupted germline compared to wild-type worms (Figure 3B). In contrast, 80% of smc-5(ok2421);div-1(or148) double mutants developed no or formed only disrupted germlines even in the absence of exogenous DNA damage. Moreover, upon UV treatment, when ∼20% of smc-5(ok2421) mutant worms were still capable of forming disrupted germlines, all double-mutant worms completely lacked germlines (Figure 3B). The enhancement of the germ-cell proliferation defect in smc-5(ok2421);div-1(or148) mutants is consistent with a role for the SMC-5/6 complex in DNA replication.
To test whether DNA replication is impaired in the smc-5(ok2421) and smc-6(ok3294) mitotic germ cells, we compared the efficiency of incorporation of the thymidine analog EdU between mutant and wild-type strains. Previous studies that utilized the incorporation of EdU and bromodeoxyuridine (BrdU) to measure cell cycle progression in germ cells showed that EdU and BrdU incorporation occurs rapidly in the wild-type germline. Only 15 min of feeding on EdU/BrdU-containing bacteria was sufficient for measureable incorporation (Crittenden et al. 2006; Michaelson et al. 2010). We reasoned that if the smc-5 and smc-6 mutant germ cells had impaired progression in DNA replication, then they should be less efficient in EdU incorporation, especially during short labeling periods. As predicted, wild-type germ cells rapidly incorporated EdU after 15 and 30 min of exposure to EdU-containing bacteria, as ∼75% of cells showed EdU-associated fluorescence after 15 min of labeling (Figure 3, C and D; see Materials and Methods for EdU specificity controls). The germ cells in the smc-5(ok2421) and smc-6(ok3294) mutants had less EdU incorporation compared to wild-type (Figure 3, C and D), consistent with the requirement for SMC-5 and SMC-6 in promoting efficient DNA replication. To control for differences in the proportion of S-phase germ cells between wild-type and the smc-5(ok2421) and smc-6(ok3294) mutants, we extended the EdU labeling to 150 min to allow EdU detection even in S-phase cells that may have a reduced level of EdU incorporation. At the longer labeling period, we found minor-to-no significant differences in the fraction of EdU-labeled cells in the mutants compared to wild-type (Figure 3D), thus indicating that the proportion of replicating cells is similar between wild-type and the smc-5(ok2421) and smc-6(ok3294) mutants. Taken together, the EdU labeling results indicate that the smc-5(ok2421) and smc-6(ok3294) mutants have less nucleotide incorporation, consistent with impaired DNA replication. This conclusion is also consistent with smaller germlines observed in older smc-5 and smc-6 mutant adults (Bickel et al. 2010), which would be expected if progression through mitotic S phase were slowed.
As smc-5 and smc-6 mutant germ cells exhibited enhanced sensitivity to replication stress and ectopic RAD-51 foci, we tested whether DNA damage checkpoint signaling was activated. In response to replication fork stalling, the C. elegans homolog of ATR, ATL-1, is activated (Garcia-Muse and Boulton 2005). ATL-1, in turn, phosphorylates CHK-1 to induce cell cycle arrest. To test for CHK-1 activation, we stained with antibodies specific for phosphorylated CHK-1 (Lee et al. 2010). Despite RAD-51 foci formation (Figure 3A), there was no detectable CHK-1 phosphorylation in smc-5(ok2421) mutant germ cells in the absence of exogenous genotoxic insult (Figure S4A). However, upon UV irradiation, CHK-1 phosphorylation was readily detectable in smc-5(ok2421) mutant animals. The smc-5(ok2421) mutant worms showed CHK-1 activation similar to wild-type in response to UV treatment, suggesting that DNA damage checkpoint activation is normal in smc-5(ok2421) mutant worms (Figure S4A). Checkpoint activation in mitotic germ cells arrests cell cycle progression but not cellular growth, resulting in enlargement of the mitotic germ cells (Ahmed and Hodgkin 2000). In agreement with the phosphorylated CHK-1 staining data, we found that the average diameter of the smc-5(ok2421) and smc-6(ok3294) mutant mitotic germ cells was equivalent to wild-type (Figure S4B). In response to HU treatment, average diameter increased in the smc-5(ok2421) and smc-6(ok3294) mutants and in wild-type (Figure S4B). Thus, DNA damage checkpoints remain intact in SMC-5/6-deficient germ cells; however, they do not appear to be activated in the smc-5(ok2421) and smc-6(ok3294) mutants in the absence of exogenous genotoxic insults.
SMC-5 functions in parallel to POLH-1-mediated TLS
During DNA replication, TLS DNA polymerases can incorporate nucleotides at sites of UV-induced lesions, preventing replication fork blockage. POLH-1, the C. elegans homolog of TLS polymerase Pol η, bypasses lesions induced by UV radiation, particularly during the fast embryonic cell divisions (Holway et al. 2006; Roerink et al. 2012). Because POLH-1 is important in mitotic cells to counteract fork stalling, we explored its genetic interactions with smc-5. We tested two alleles of polh-1: the previously described ok3317 deletion allele (Roerink et al. 2012) and the premature stop allele mn156. The mn156 allele was initially identified as rad-2 in a screen for radiation-sensitive mutants in C. elegans (Hartman and Herman 1982). We identified a nonsense mutation in polh-1 in the mn156 allele (Figure S5A). A complementation test analyzing egg laying and hatching rate between polh-1(ok3317) and rad-2(mn156) showed that the two mutants failed to complement, suggesting that they are defective in the same gene (Figure S5, B and C).
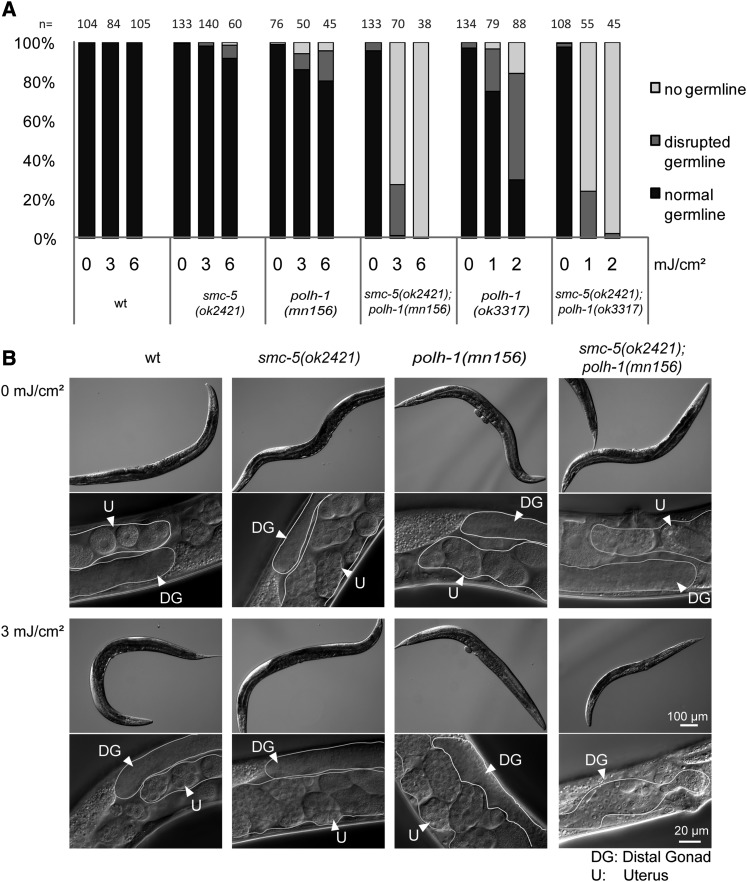
Given the high UV sensitivity of polh-1 mutants, less than one-tenth the UV dose used in NER or the smc-5 mutant analysis was applied. Interestingly, irradiating L1 larvae in the smc-5(ok2421);polh-1(mn156) and smc-5(ok2421);polh-1(ok3317) double-mutant strains with a UV dose of 2–3 mJ/cm2 impeded germline development. This dose showed no or hardly any effect on germline development in smc-5 and polh-1 single mutants, respectively (Figure 4A). Notably, some smc-5;polh-1 double mutants displayed synthetic somatic defects upon UV irradiation characterized by a reduction in size (Figure 4B and Figure S6) and additional somatic defects (data not shown).
Figure 4.
smc-5 and polh-1 act in parallel pathways to overcome UV lesions in mitotic germ cells. (A) Germline development quantification of worms 3 days after irradiation with UVB at L1 stage or untreated control worms. Worms were categorized into groups of “normal,” “disrupted,” and “no germline” using a dissection microscope. “n” indicates number of animals assessed. A representative experiment is shown. (B) DIC images of whole worms and magnified view on mid-body 72 hr after L1 stage. Synchronized worms were irradiated at L1 larval stage and kept at 20 °C until reaching adulthood. Bar, 100 µm (top) and 20 µm (bottom), of the untreated and treated panels, respectively.
In response to UV-induced DNA damage, cells in the mitotic germline of adult worms respond with rapid RAD-51 foci formation and transient cell cycle arrest that becomes evident as a drop in cell number and enlargement of the nucleoplasm (Gartner et al. 2004). To further characterize the role of polh-1 and smc-5 in the DNA damage response, we followed the persistence of RAD-51 foci and cell cycle arrest in the adult germline. We treated animals with UV at the L4 larval stage and 24 hr later assessed the level of UV sensitivity in the young adult germline. RAD-51 was rapidly loaded on DNA following UV treatment (Figure 6A). After 24 hr, RAD-51 staining resembles the level seen in untreated worms of the same genotype for both wild-type and smc-5 mutants (Figure S7). In contrast, cells in the mitotic region of polh-1 mutants retained RAD-51 foci 24 hr post-treatment, with some cells displaying extensive RAD-51 staining, indicative of high loads of unprocessed DNA breaks. The size of the germ cells also increased upon UV treatment in polh-1 mutants, indicative of cell cycle arrest in the mitotic germline (Figure S7B).
Figure 6.
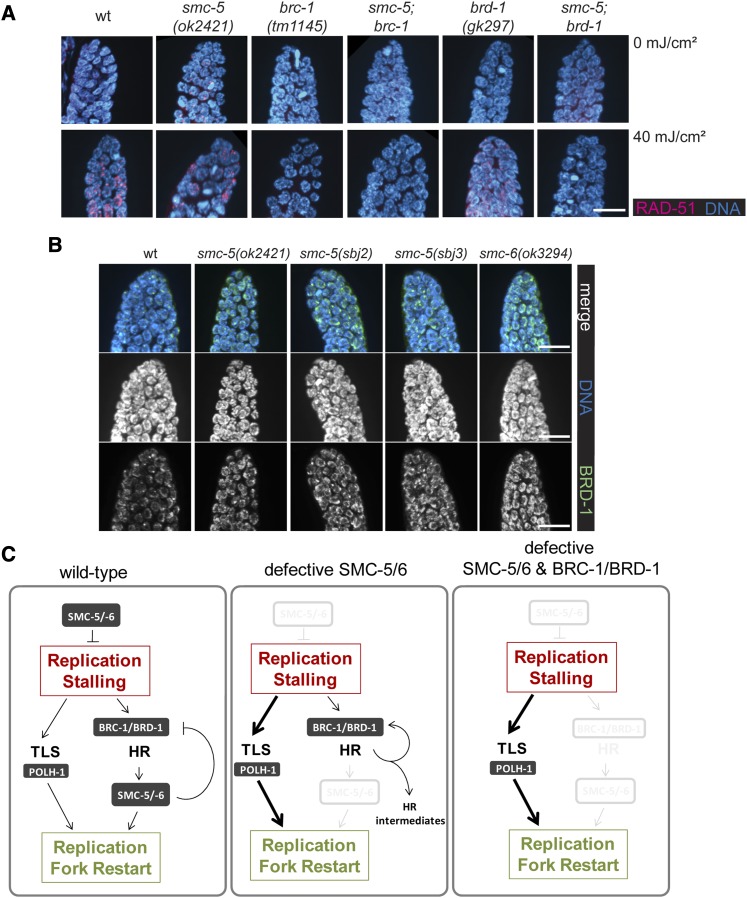
BRC-1/BRD-1 promote genome instability when SMC-5/6 dysfunction leads to replication stalling and aberrant HR. (A) αRAD-51 and DAPI staining of mitotic germ cells. Young adults were dissected 5 hr after either UVB irradiation or mock treatment. Representative images are shown. Bar, 10 µm. (B) αBRD-1 and DAPI staining of mitotic germ cells. Germlines were dissected from young adult worms. Representative images in extended view of stack spanning the whole germline are shown. Bar, 10 µm. (C) Model for DNA repair mechanisms in the presence and absence of SMC-5/6. In proliferating wild-type cells, SMC-5/6 prevents fork stalling. Upon stalling, TLS promotes readthrough, while the BRC-1/BRD-1 complex initiates HR and SMC-5/6 promotes dissociation of BRC-1/BRD-1 from chromatin. Defective SMC-5/6 sensitizes to replication stalling, and TLS becomes the major route for replication fork restart, while BRC-1/BRD-1 initiate HR that, however, cannot be resolved in the absence of SMC-5/6 but instead leads to persistence of HR intermediates. In the absence of BRC-1/BRD-1, the formation of HR intermediates is prevented and TLS can promote lesion bypass.
We observed several specific phenotypes that occur in response to UV irradiation in L1 larvae and adult worms. The UV sensitivity of worms with defective SMC-5/6 complexes is typically indicated by defects in germ-cell proliferation (Figure 1D and Figure 4A); however, when TLS is also impaired, as in smc-5;polh-1 double mutants, UV lesions also compromise somatic growth (Figure 4B and Figure S6). Moreover, smc-5;polh-1 double-mutant worms show germline defects at UV doses that do not impair germline development in smc-5 single-mutant worms, providing evidence that POLH-1 is critically important for bypassing UV lesions when forks stall in the absence of SMC-5 (Figure 4; Figure S6; Figure S7). These observations indicate that smc-5 and polh-1 function in parallel in the germline as well as in somatic development to confer UV resistance.
BRC-1/BRD-1 mutations suppress genome instability in smc-5/6 mutants
The accumulation of RAD-51 foci in the germlines of smc-5 mutant worms and the synthetic interaction of smc-5 with polh-1 in response to UV are consistent with a model in which HR activity persists or is increased at sites of collapsed replication forks when SMC-5/6 function is disrupted. BRCA1 has been suggested to promote postreplicative repair upon replication fork stalling in mammalian cells (Pathania et al. 2011). In C. elegans, the BRCA1/BARD1 complex, composed of BRC-1 and BRD-1, is required for repair of DSBs following IR treatment (Boulton et al. 2004). To test whether brc-1 and brd-1 genetically interact with smc-5 in response to UV-induced DNA damage, we analyzed the UV sensitivities of smc-5;brc-1 and smc-5;brd-1 double mutants employing deletion alleles of brc-1(tm1145) and brd-1(gk297) as well as using RNA interference (RNAi) against brc-1 and brd-1 in smc-5 and smc-6 mutants. The brc-1 and brd-1 mutant worms showed similar UV resistance as wild-type animals, suggesting that the BRC-1/BRD-1 complex is dispensable for UV repair in otherwise repair-proficient animals (Figure S8). Importantly, the loss of brc-1 and brd-1 by genetic mutations or RNAi strongly suppressed the UV-induced disruption of germline development in smc-5(ok2421) and smc-6(ok3294) mutants (Figure 5A and Figure S9). RNAi against brc-1 and brd-1 can even suppress the severe UV-induced germline defects in the smc-5(ok2421);div-1(or148) double mutant worms (Figure 5A). The loss of brc-1 and brd-1 also alleviated replication-stress-related defects in the smc-5 mutant background caused by HU treatment. The brc-1 and brd-1 mutations suppressed the HU-induced germline development defects in the smc-5(ok2421) mutant background (Figure 5B). Both the HU-enhanced chromatin bridge defect (Figure 5C) and a reduction in mitotic germ cells in the smc-5(ok2421) mutant backgrounds are suppressed by the brc-1(tm1145) mutation (Figure 5, C and D). In contrast, the incorporation of a EdU defect in the smc-5(ok2421) background was largely unaffected by brc-1(tm1145), suggesting that the reduced replicative activity was not alleviated (Figure 5E). These results suggest that the BRC-1/BRD-1 complex does not impact the primary replicative defect of smc-5 mutants but instead alleviates the genome instability resulting from replicative impediments.
HR may function at stalled forks to promote fork progression pass an obstruction, or to repair collapsed replication forks. To examine how BRC-1/BRD-1 affects HR in smc-5 mutant worms following UV damage, we assessed RAD-51 foci formation in adult germlines 5 hr after mock and 40 mJ/cm2 UV treatment. While RAD-51 foci were detected only in wild-type germ cells after UV irradiation, smc-5(ok2421) mutant germ cells exhibited RAD-51 foci already in the absence of UV treatment (Figure 6A; Figure S2; Figure S10). In contrast to both, wild-type and smc-5 mutant germ cells, the brc-1 and brc-1 single-mutant germ cells had substantially reduced RAD-51 foci following UV irradiation (Figure 6A, bottom; Figure S10). Strikingly, smc-5;brc-1 and smc-5;brd-1 double-mutant worms had decreased RAD-51 foci formation compared to smc-5 mutant worms in the absence (Figure 6A, top; Figure S10) and the presence of UV treatment (Figure 6A, bottom; Figure 10). Therefore, the amelioration of smc-5 and smc-6 mutant defects by the loss of brc-1 and brd-1 is likely through the suppression of HR activities.
Mammalian BRCA1 promotes HR at sites of DSBs during S/G2 phase by suppressing the recruitment of error-prone nonhomologous end joining (NHEJ) (Chapman et al. 2013; Escribano-Díaz et al. 2013). In contrast to BRCA1 function early in the DSB repair pathway choice, the Smc5/6 complex is thought to act after HR has been initiated based on evidence in budding and fission yeast (Lehmann et al. 1995; Ampatzidou et al. 2006; Branzei et al. 2006; De Piccoli et al. 2006; Chen et al. 2009; Sollier et al. 2009). If the C. elegans BRC-1/BRD-1 complex biases DSB repair in favor of HR and prevents other DSB repair pathways such as NHEJ, then in smc-5 mutant germ cells, in which HR is impaired, normal BRC-1/BRD-1 function could end up trapping DSB repair in an unproductive defective pathway. To test the consequences of impaired initiation of NHEJ, we employed a mutant strain for hsr-9, the C. elegans 53BP1 homolog (Ryu et al. 2013). In mammals, 53BP1 antagonizes the function of BRCA1 by promoting initiation of NHEJ while inhibiting HR (Zimmermann and De Lange 2013). We found that germline formation after UV treatment in the smc-5(ok2421) mutant background is not enhanced by a loss-of-function mutation in hsr-9 (Figure S11), suggesting that DSB repair, once it is initiated through HR, is not effectively acted upon by NHEJ.
Since BRCA1 recruitment to DSBs is thought to suppress NHEJ and promote HR, we examined the accumulation of the BRC-1/BRD-1 complex in mitotic germ cells in the smc-5 and smc-6 mutant backgrounds. In C. elegans, immunostaining for BRD-1 has been established to mark the BRC-1/BRD-1 localization (Boulton et al. 2004). Intriguingly, BRD-1 immunostaining showed accumulation of BRD-1 on chromatin in the mitotic germ cells of smc-5 mutant animals (Figure 6B). Taken together, our results suggest that, in the absence of a functional SMC-5/6 complex during replication, the BRC-1/BRD-1 complex accumulates on chromatin and triggers HR as indicated by RAD-51 foci formation. Consequently, BRC-1/BRD-1 activity in smc-5 mutant cells becomes deleterious as it leads to the induction of HR that cannot be resolved in the absence of SMC-5/6, thus leading to genome instability in proliferating cells.
Discussion
Bulky lesions, such as those induced by UV, pose obstacles to the progression of the replication fork (Sale et al. 2012). Replication fork collapse can give rise to mutations and chromosomal aberrations (Petermann and Helleday 2010). In replicating cells, GG-NER is important for surveying the genome for helix-distorting lesions and removes them before they lead to replication fork stalling. Mutations that inactivate GG-NER lead to highly elevated skin cancer susceptibility in xeroderma pigmentosum (XP) patients (Cleaver et al. 2009). Pol η-mediated TLS preserves ongoing DNA replication by incorporating nucleotides at lesions that cannot be accommodated by the replicative DNA polymerase complex (Sale et al. 2012). Mutations in POLH increase UV-dependent mutagenicity of XP variant patients, increasing their susceptibility to skin cancer (Lehmann 2011). Alternatively, the mobilization of HR can facilitate resolution of collapsed replication forks by recombination-dependent repair and template switching (Petermann and Helleday 2010). Recent experiments using a conditional replication fork barrier system in S. pombe revealed that, in contrast to HR repair of IR-induced DSBs, HR-mediated rescue of collapsed replication forks can be mutagenic (Iraqui et al. 2012; Mizuno et al. 2013). In replicating cells, DSBs can form as a secondary consequence of collapsed replication forks at bulky lesions caused by UV-induced DNA damage (Limoli et al. 2002; Garinis et al. 2005). At stalled replication forks, DSBs can also be produced via active incision by the MUS81/EME1 endocucleases (Petermann and Helleday 2010). Neither the recruitment mechanisms of HR, nor the actual resolution of the replication impasse by HR, are completely understood. However, the choice of the repair pathway can have important consequences on the maintenance of genome stability amid replication fork stalling.
We conducted a forward genetics screen to identify genes required for UV resistance in replicating cells, which produced two novel alleles of smc-5. In contrast to GG-NER, the SMC-5/6 complex was dispensable for the removal of UV-induced lesions. Instead, SMC-5/6 dysfunction sensitized cells to perturbed replication. In particular, the synergistic UV sensitivity with polh-1 mutant alleles suggests that SMC-5/6 confers UV resistance by stabilizing replication forks or recovery from fork collapse. In addition, the sensitivity of smc-5 mutants to dysfunctional DIV-1 primase indicates that transient fork stalling in the absence of exogenous DNA damage requires the SMC-5/6 complex for stability and resumption of fork progression (Figure 6C). The RAD-51 foci formation and accumulation of chromatin-associated BRD-1 in smc-5 mutants suggests that stalled replication forks lead to induction of HR, which is then not resolved. POLH-1 counters DSB formation and HR activity via its TLS activity. Particularly in the absence of SMC-5/6, TLS becomes the major route for maintaining genome stability in the presence of UV lesions. Consistently, mutations in smc6 in S. pombe were shown to confer hypersensitivity to UV and IR and were genetically placed in the HR pathway (Lehmann et al. 1995). It is likely that SMC5/6 supports the stabilization of molecular intermediates and proximity during complex HR reactions (Ampatzidou et al. 2006; Branzei et al. 2006; De Piccoli et al. 2006; Sollier et al. 2009). The SMC5/6 complex not only promotes HR but also stabilizes replication forks and facilitates their restart (Irmisch et al. 2009). In S. pombe, the methyl methanesulfonate (MMS) sensitivity of the smc6-74 allele can be suppressed by overexpression of the six-BRCT domain protein Brc1, related to human PTIP, in an HR-dependent manner (Lee et al. 2007). In contrast, brc-1Δ was found to be synthetically lethal with the smc6-74 mutation (Sheedy et al. 2005). These observations are highly consistent with the role of the SMC5/6 complex in HR as mammalian PTIP, in contrast to BRCA1, promotes NHEJ and 53BP1-mediated inhibition of HR (Callén et al. 2013). Rescue of smc6-74 defects requires nucleases related to HR repair and TLS polymerases (Lee et al. 2007). HR activity was also required for alleviation of MMS and HU sensitivity of smc6-9 mutants by a deletion of the gene encoding the FANCM helicase MPH1 (Chen et al. 2009; Chavez et al. 2011). It was suggested that Smc5/6 facilitates resolving of recombination intermediates that are formed through Mph1, Mms2, and the Shu complex during replication in the presence of MMS (Choi et al. 2010).
We demonstrate that inactivation of the BRC-1/BRD-1 complex alleviates RAD-51 foci formation and suppresses genome instability in smc-5 mutants. In contrast, a brc-1 mutation does not alleviate the recombination repair defects of meiotic DSBs in smc-5 mutants (Bickel et al. 2010). The BRC-1/BRD-1 complex plays an important role in HR and, when dysfunctional, evokes hypersensitivity to DSB-inducing IR (Boulton et al. 2004). It is thought that the defects in HR-mediated DSB repair underlie the genome instability and cancer susceptibility caused by BRCA1 mutations (Silver and Livingston 2012). BRCA1 has recently been implicated in promoting postreplicative repair at sites of stalled replication forks (Pathania et al. 2011). Our results suggest that the BRC-1/BRD-1-mediated initiation of HR at stalled replication forks requires the SMC-5/6 complex to resolve the recombination intermediates. SMC-5/6 might in turn regulate the chromatin dissociation of the BRC-1/BRD-1 complex (Figure 6C). In the absence of SMC-5/6, the HR machinery assembles but fails to engage the intra-S-phase checkpoint. Consistent with this, smc5/6 defective budding yeast fail to induce intra-S checkpoints but instead halt the cell cycle after the first mitotic cycle following smc5/6 deprivation (Torres-Rosell et al. 2005). A dominant negative mutation of fission yeast smc6 arrested in response to UV irradiation similar to wild-type cells but displayed mitotic defects due to DNA aberrations, which is evident of a checkpoint maintenance defect (Harvey et al. 2004). Drosophila with a defective Smc5/6 complex are capable of inducing G2/M-phase as well as S-phase checkpoints while being sensitive to DNA damage (Li et al. 2013). In C. elegans smc-5/6 mutants, RAD-51 foci persist, and chromatin bridges are formed during ensuing mitosis. The SMC-5/6 complex thus might play a dual role in maintaining fork stability and in resolving recombination intermediates when BRC-1/BRD-1 initiates HR. Intriguingly, our data suggest that the BRC-1/BRD-1 complex can both promote and antagonize genome stability depending on the genetic background. It is likely that suppression of BRC-1/BRD-1 function facilitates a more effective TLS-mediated rescue of replication fork stalling.
CPD lesions are the major cause of UV-induced carcinogenesis (Jans et al. 2005). As Pol η preferably inserts nontemplated deoxy-ATP opposite UV-induced thymidine dimers, its activity likely leads only to limited mutagenicity (Sale et al. 2012). In contrast, HR recruitment might lead to more deleterious chromosomal rearrangements at collapsed replication forks (Iraqui et al. 2012; Mizuno et al. 2013). However, TLS by Pol η at noncognate lesions or by other TLS polymerases may be more mutagenic when replication is blocked by bulky lesions caused by genotoxins other than UV irradiation. Particularly relevant for the development of breast cancer, endogeneous metabolism of estradiol forms quinone derivatives that can induce bulky adducts (Yager and Davidson 2006). It is conceivable that BRCA1-mediated resolution of stalled replication forks at such adducts might to some extent counteract TLS-mediated mutagenicity. It will be highly interesting to explore how the antagonizing functions of BRCA1/BARD1 in the maintenance of genome stability affect replication stress-induced genome instability during both cancer development and therapeutic responses in BRCA1-mutated tumor cells. Intriguingly, the suppression of genome instability in the context of replication stalling amid dysfunctional SMC-5/6 suggests that the maintenance of familial BRCA1 carrier mutations might confer selective advantages under certain conditions of genotoxic challenges.
Supplementary Material
Acknowledgments
We thank S. Boulton for providing BRD-1 antibodies; M. Mueller for mn156 sequence analysis; I. Faulkner, K. Kirkconnell, and W. Bian for assistance in counting EdU-positive cells, measuring nuclei diameter, and intestinal chromatin bridges; and J. Bianco and A. Williams for discussions on the manuscript. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources. S.W. received the Dieter Platt and the International Graduate School in Development Health and Disease graduate fellowships, and M.A.E. received a European Molecular Biology Organization long-term fellowship. R.C.C. is supported by NIH grant GM093173. B.S. acknowledges funding from the Deutsche Forschungsgemeinschaft [Cologne Cluster of Excellence in Cellular Stress Responses in Aging-Associated Diseases, Sonderforschungsbereich (SFB) 829, Klinische Forschergruppe (KFO) 286], the European Research Council (ERC) (starting grant 260383), European Commision FP7-PEOPLE-2012-ITN Marie Curie [FP7 Initial Training Network (ITN) CodeAge 316354, aDDRess 316390, MARRIAGE 316964, and European Reintegration Grant (ERG) 239330], the German–Israeli Foundation (2213-1935.13/2008 and 1104-68.11/2010), the Deutsche Krebshilfe (109453), and the Bundesministerium für Bildung und Forschung (BMBF) (SyBaCol).
Footnotes
Communicating editor: M. P. Colaiácovo
Literature Cited
- Adamo A., Montemauri P., Silva N., Ward J. D., Boulton S. J., et al. , 2008. BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep. 9: 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S., Hodgkin J., 2000. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403: 159–164. [DOI] [PubMed] [Google Scholar]
- Alpi A., Pasierbek P., Gartner A., Loidl J., 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16. [DOI] [PubMed] [Google Scholar]
- Ampatzidou E., Irmisch A., O’Connell M. J., Murray J. M., 2006. Smc5/6 is required for repair at collapsed replication forks. Mol. Cell. Biol. 26: 9387–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono N., Sutani T., Tomonaga T., Mochida S., Yanagida M., 2002. Cnd2 has dual roles in mitotic condensation and interphase. Nature 417: 197–202. [DOI] [PubMed] [Google Scholar]
- Bickel J. S., Chen L., Hayward J., Yeap S. L., Alkers A. E., et al. , 2010. Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability. PLoS Genet. 6: e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S. J., Martin J. S., Polanowska J., Hill D. E., Gartner A., et al. , 2004. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr. Biol. 14: 33–39. [DOI] [PubMed] [Google Scholar]
- Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., et al. , 2006. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callén E., Di Virgilio M., Kruhlak M. J., Nieto-Soler M., Wong N., et al. , 2013. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153: 1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. R., Barral P., Vannier J.-B., Borel V., Steger M., et al. , 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49: 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A., Agrawal V., Johnson F. B., 2011. Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes (Smc) 5/6 complex. J. Biol. Chem. 286: 5119–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-H., Choi K., Szakal B., Arenz J., Duan X., et al. , 2009. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc. Natl. Acad. Sci. USA 106: 21252–21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Szakal B., Chen Y.-H., Branzei D., Zhao X., 2010. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol. Biol. Cell 21: 2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver J. E., Lam E. T., Revet I., 2009. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat. Rev. Genet. 10: 756–768. [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Leonhard K. A., Byrd D. T., Kimble J., 2006. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell 17: 3051–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bont R., van Larebeke N., 2004. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 19: 169–185. [DOI] [PubMed] [Google Scholar]
- De Piccoli G., Cortes-Ledesma F., Ira G., Torres-Rosell J., Uhle S., et al. , 2006. Smc5–Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat. Cell Biol. 8: 1032–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett M., Westlund B., Schedl T., 2009. METT-10, a putative methyltransferase, inhibits germ cell proliferative fate in Caenorhabditis elegans. Genetics 183: 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley M. L., Riddle D. L., 2001. LG II balancer chromosomes in Caenorhabditis elegans: mT1(II;III) and the mIn1 set of dominantly and recessively marked inversions. Mol. Genet. Genomics 266: 385–395. [DOI] [PubMed] [Google Scholar]
- Encalada S. E., Martin P. R., Phillips J. B., Lyczak R., Hamill D. R., et al. , 2000. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228: 225–238. [DOI] [PubMed] [Google Scholar]
- Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J. T. F., et al. , 2013. A cell cycle-dependent regulatory circuit composed of 53BP1–RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49: 872–883. [DOI] [PubMed] [Google Scholar]
- Fousteri M. I., Lehmann A. R., 2000. A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 19: 1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse T., Boulton S. J., 2005. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 24: 4345–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinis G. A., Mitchell J. R., Moorhouse M. J., Hanada K., de Waard H., et al. , 2005. Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. EMBO J. 24: 3952–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A., MacQueen A. J., Villeneuve A. M., 2004. Methods for analyzing checkpoint responses in Caenorhabditis elegans. Methods Mol. Biol. 280: 257–274. [DOI] [PubMed] [Google Scholar]
- Hartman P. S., Herman R. K., 1982. Radiation-sensitive mutants of Caenorhabditis elegans. Genetics 102: 159–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey S. H., Sheedy D. M., Cuddihy A. R., O’Connell M. J., 2004. Coordination of DNA damage responses via the Smc5/Smc6 complex. Mol. Cell. Biol. 24: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale J. T., Ball A. R., Schmiesing J. A., Kim J.-S., Kong X., et al. , 2006. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol. Cell 21: 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., 2006. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7: 311–322. [DOI] [PubMed] [Google Scholar]
- Holway A. H., Kim S.-H., La Volpe A., Michael W. M., 2006. Checkpoint silencing during the DNA damage response in Caenorhabditis elegans embryos. J. Cell Biol. 172: 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui I., Chekkal Y., Jmari N., Pietrobon V., Fréon K., et al. , 2012. Recovery of arrested replication forks by homologous recombination is error-prone. PLoS Genet. 8: e1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch A., Ampatzidou E., Mizuno K., O’Connell M. J., Murray J. M., 2009. Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J. 28: 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. P., Bartek J., 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J., Schul W., Sert Y. G., Rijksen Y., Rebel H., et al. , 2005. Powerful skin cancer protection by a CPD-photolyase transgene. Curr. Biol. 15: 105–115. [DOI] [PubMed] [Google Scholar]
- Kegel A., Betts-Lindroos H., Kanno T., Jeppsson K., Ström L., et al. , 2011. Chromosome length influences replication-induced topological stress. Nature 471: 392–396. [DOI] [PubMed] [Google Scholar]
- Kim J.-S., Krasieva T. B., LaMorte V., Taylor A. M. R., Yokomori K., 2002. Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 277: 45149–45153. [DOI] [PubMed] [Google Scholar]
- Lans H., Marteijn J. A., Schumacher B., Hoeijmakers J. H. J., Jansen G., et al. , 2010. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet. 6: e1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. M., Nizza S., Hayes T., Bass K. L., Irmisch A., et al. , 2007. Brc1-mediated rescue of Smc5/6 deficiency: requirement for multiple nucleases and a novel Rad18 function. Genetics 175: 1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-J., Gartner A., Hyun M., Ahn B., Koo H.-S., 2010. The Caenorhabditis elegans Werner syndrome protein functions upstream of ATR and ATM in response to DNA replication inhibition and double-strand DNA breaks. PLoS Genet. 6: e1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A. R., 2011. DNA polymerases and repair synthesis in NER in human cells. DNA Repair (Amst.) 10: 730–733. [DOI] [PubMed] [Google Scholar]
- Lehmann A. R., Walicka M., Griffiths D. J., Murray J. M., Watts F. Z., et al. , 1995. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 15: 7067–7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhuo R., Tiong S., Di Cara F., King-Jones K., et al. , 2013. The Smc5/Smc6/MAGE complex confers resistance to caffeine and genotoxic stress in Drosophila melanogaster. PLoS ONE 8: e59866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli C. L., Giedzinski E., Bonner W. M., Cleaver J. E., 2002. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma-H2AX formation, and Mre11 relocalization. Proc. Natl. Acad. Sci. USA 99: 233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D., Korta D. Z., Capua Y., Hubbard E. J. A., 2010. Insulin signaling promotes germline proliferation in C. elegans. Development 137: 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K., Miyabe I., Schalbetter S. A., Carr A. M., Murray J. M., 2013. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature 493: 246–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Haering C. H., 2005. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74: 595–648. [DOI] [PubMed] [Google Scholar]
- Pathania S., Nguyen J., Hill S. J., Scully R., Adelmant G. O., et al. , 2011. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol. Cell 44: 235–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E., Helleday T., 2010. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 11: 683–687. [DOI] [PubMed] [Google Scholar]
- Roerink S. F., Koole W., Stapel L. C., Romeijn R. J., Tijsterman M., 2012. A broad requirement for TLS polymerases η and κ, and interacting sumoylation and nuclear pore proteins, in lesion bypass during C. elegans embryogenesis. PLoS Genet. 8: e1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J.-S., Kang S. J., Koo H.-S., 2013. The 53BP1 homolog in C. elegans influences DNA repair and promotes apoptosis in response to ionizing radiation. PLoS ONE 8: e64028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale J. E., Lehmann A. R., Woodgate R., 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13: 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa Maria S. R., Gangavarapu V., Johnson R. E., Prakash L., Prakash S., 2007. Requirement of Nse1, a subunit of the Smc5-Smc6 complex, for Rad52-dependent postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 8409–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy D. M., Dimitrova D., Rankin J. K., Bass K. L., Lee K. M., et al. , 2005. Brc1-mediated DNA repair and damage tolerance. Genetics 171: 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D. P., Livingston D. M., 2012. Mechanisms of BRCA1 tumor suppression. Cancer Discov. 2: 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J., Driscoll R., Castellucci F., Foiani M., Jackson S. P., et al. , 2009. The Saccharomyces cerevisiae Esc2 and Smc5–6 proteins promote sister chromatid junction-mediated intra-S repair. Mol. Biol. Cell 20: 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiou L., Doukoumetzidis K., Sendoel A., Hengartner M. O., 2007. The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ. 14: 1129–1138. [DOI] [PubMed] [Google Scholar]
- Ström L., Lindroos H. B., Shirahige K., Sjögren C., 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16: 1003–1015. [DOI] [PubMed] [Google Scholar]
- Taylor E. M., Moghraby J. S., Lees J. H., Smit B., Moens P. B., et al. , 2001. Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Spr18 complex. Mol. Biol. Cell 12: 1583–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J., Machín F., Farmer S., Jarmuz A., Eydmann T., et al. , 2005. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 7: 412–419. [DOI] [PubMed] [Google Scholar]
- Tsang C. K., Wei Y., Zheng X. F. S., 2007. Compacting DNA during the interphase: condensin maintains rDNA integrity. Cell Cycle 6: 2213–2218. [DOI] [PubMed] [Google Scholar]
- Unal E., Arbel-Eden A., Sattler U., Shroff R., Lichten M., et al. , 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16: 991–1002. [DOI] [PubMed] [Google Scholar]
- Verkade H. M., Bugg S. J., Lindsay H. D., Carr A. M., O’Connell M. J., 1999. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 10: 2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. D., Barber L. J., Petalcorin M. I., Yanowitz J., Boulton S. J., 2007. Replication blocking lesions present a unique substrate for homologous recombination. EMBO J. 26: 3384–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. L., Liang Y., Li K., Chen J., 2008. Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J. Biol. Chem. 283: 29586–29592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Yu H., 2012. The Smc complexes in DNA damage response. Cell Biosci. 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager J. D., Davidson N. E., 2006. Estrogen carcinogenesis in breast cancer. N. Engl. J. Med. 354: 270–282. [DOI] [PubMed] [Google Scholar]
- Zimmermann M., de Lange T., 2013. 53BP1: pro choice in DNA repair. Trends Cell Biol. 24: 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.