Abstract
The centromeric histone H3 variant Cse4 in Saccharomyces cerevisiae is polyubiquitylated and degraded in a proteasome-dependent manner. We report here that the proline isomerase Fpr3 regulates Cse4 proteolysis. Structural change in Cse4 by Fpr3 might be important for the interaction between Cse4 and the E3 ubiquitin ligase Psh1.
Keywords: Fpr3, Cse4, Psh1, isomerization, protein degradation
ACCURATE chromosome segregation during mitosis and meiosis is a critical event in the transfer of genetic information to daughter cells. Loss or gain of chromosome is associated with cancer development and genetic disease (Yuen et al. 2005; Holland and Cleveland 2009). The centromere–kinetochore complex is required for faithful segregation. We previously used an in vitro kinetochore assembly system and identified the FK506 binding protein Fpr3 as a novel protein that associates with CEN DNA (Ohkuni and Kitagawa 2011). Fpr3 was also isolated by affinity purification of the Dsn1–Flag-tagged central kinetochore protein (Akiyoshi et al. 2010). These data strongly suggested that Fpr3 has a role in centromere and/or kinetochore function. In this study, we investigated the mitotic function of Fpr3.
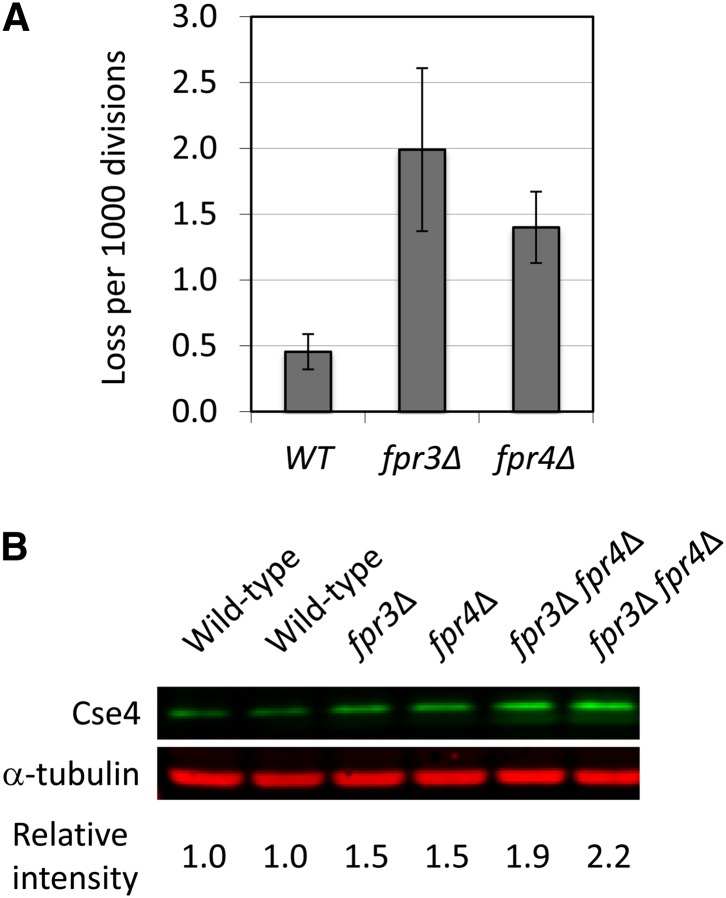
FPR3 and FPR4 encode two related prolyl isomerases that share 46% identity (Arevalo-Rodriguez et al. 2004). Fpr4 regulates histone H3 lysine 36 methylation and gene activity by its proline isomerase activity (Nelson et al. 2006). To characterize the function of Fpr3 and Fpr4, we first generated null mutants of the nonessential FPR3 and FPR4 genes, respectively (Supporting Information, Table S1) (Dolinski et al. 1997; Arevalo-Rodriguez et al. 2004). Single deletion strains (fpr3∆ and fpr4∆) and double deletion strains (fpr3∆ fpr4∆) did not show obvious growth phenotypes such as temperature or benomyl sensitivities (Figure S1). We also examined chromosome stability in fpr3∆ or fpr4∆ strains by a colony color assay (Figure 1A). Both fpr3∆ and fpr4∆ single deletion strains showed a moderate chromosome missegregation phenotype (chromosome fragment loss: 0.2% in fpr3∆, 0.15% in fpr4∆). Because Fpr3 is associated with centromeres (Akiyoshi et al. 2010; Ohkuni and Kitagawa 2011) and because Fpr3 and Fpr4 directly interact with histone H3 (Nelson et al. 2006), we tested a possibility that Fpr3 and/or Fpr4 regulate a centromeric histone H3 variant Cse4. Interestingly, we found that the endogenous protein level of Cse4 was increased in fpr3∆, fpr4∆, and fpr3∆ fpr4∆ cells (Figure 1B). This result suggests that Fpr3 and Fpr4 have a role in regulating the Cse4 protein level in vivo.
Figure 1.
Fpr3 regulates Cse4 protein level in vivo. (A) The fpr3∆ or fpr4∆ strain displays a moderate chromosome missegregation phenotype. Chromosome loss rate in null mutants was determined by half sector analysis, as previously described (Ohkuni et al. 2008). Wild-type (Y14): 3 half-sectored colonies/6,421 total colonies; fpr3∆ (Y2249, Y2250, and Y2251): 17/8,598; and fpr4∆ (Y2252, Y2253, and Y2254): 14/10,105. P-value (chi-squared test): WT vs. fpr3∆, 0.012; WT vs. fpr4∆, 0.072. (B) Increased protein level of Cse4 in fpr∆ cells. Equal cell numbers of log phase cells, grown in SRaf medium, were visualized by Western blot analysis with anti-Cse4 and anti-α-tubulin antibodies. Cse4 and α-tubulin protein levels were overlayed and quantitated by the Odyssey Imaging System. Cse4 protein levels were normalized by the amount of α-tubulin. Isogenic yeast strains were wild type (YPH499 and YPH500), fpr3∆ (Y2243), fpr4∆ (Y2245), and fpr3∆ fpr4∆ (Y2247 and Y2248).
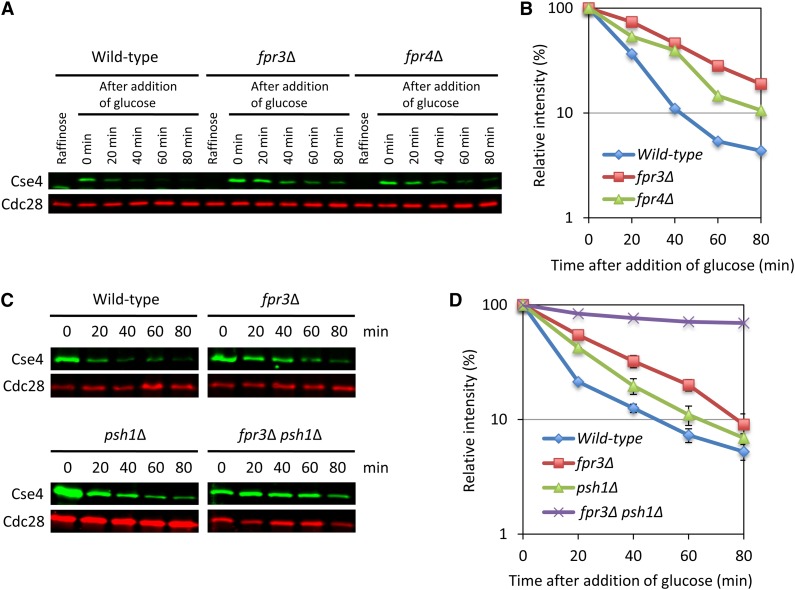
To examine whether Fpr3 and Fpr4 regulate Cse4 proteolysis, we performed a protein stability assay (Figure 2). Cse4 was transiently induced from a GAL1 promoter by the addition of galactose. Then, glucose was added to stop CSE4 transcription, and cells were collected over 80 min. The level of Cse4 was determined by quantitative Western blotting. As expected, deletion of FPR3 or FPR4 moderately stabilized Cse4 protein levels in vivo (Figure 2, A and B). The level of the stabilization in fpr3∆ cells was higher than that in fpr4∆ cells. It has been also previously reported that deletion of PSH1, which is an E3 ubiquitin ligase, moderately stabilized the Cse4 protein level (Hewawasam et al. 2010; Ranjitkar et al. 2010). The level of the stabilization in fpr3∆ cells was higher than that in psh1∆ cells (Figure 2, C and D and Figure S2). Moreover, double deletion mutant, fpr3∆ psh1∆, showed the dramatically increased protein stability of Cse4 (Figure 2, C and D). In all, these results strongly support the idea that Fpr3 and Fpr4 regulate the protein level of Cse4.
Figure 2.
Deletion of FPR3 stabilizes Cse4 protein level in vivo. Cse4 was induced from a GAL1 promoter by the addition of galactose for 2 hr. Glucose was added and cells were collected at the time point. Equal cell numbers were visualized by Western blot analysis with anti-Cse4, or anti-Cdc28. We used the Odyssey Imaging System to detect and quantify the signals. (In detail, see File S1) (A and B) Isogenic yeast strains were wild type (Y2255), fpr3∆ (Y2256), and fpr4∆ (Y2257). (C and D) Isogenic yeast strains were wild type (Y2255), fpr3∆ (Y2256), psh1∆ (Y2258), and fpr3∆ psh1∆ (Y2340). Error bars represent SE of two independent experiments.
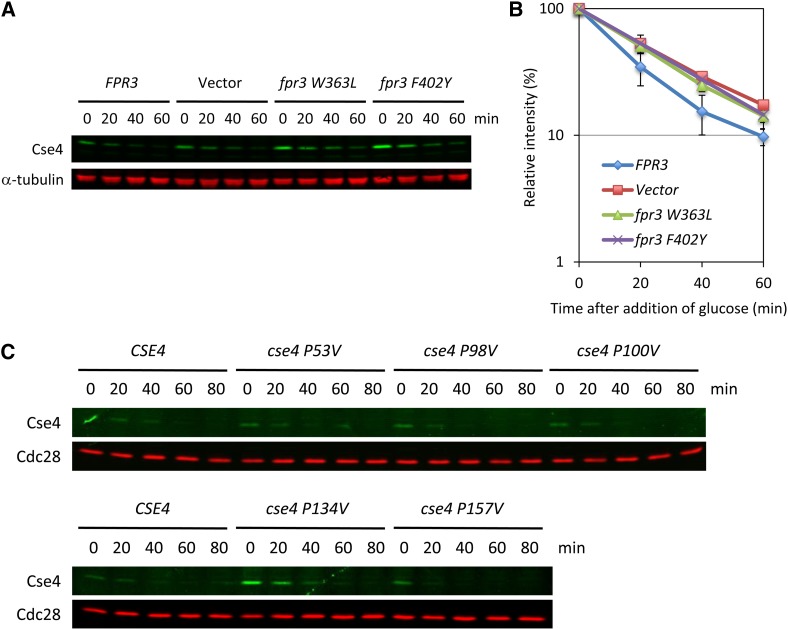
FPR3 encodes a peptidylprolyl cis–trans isomerase (PPIase) (Benton et al. 1994; Shan et al. 1994), which is involved in the meiotic recombination checkpoint pathway (Hochwagen et al. 2005; Macqueen and Roeder 2009). We next tested whether the peptidylprolyl cis–trans isomerase enzymatic activity is important for the Cse4 protein stability. We performed the Cse4 protein stability assay using two Fpr3 catalytic domain point mutants (W363L and F402Y) that have lost PPIase activity (Hochwagen et al. 2005). The two PPIase dead mutations caused a stabilization of Cse4 protein level in vivo, suggesting that the Fpr3 proline isomerase activity is required for Cse4 protein stability (Figure 3, A and B).
Figure 3.
Fpr3 isomerization activity is necessary for the Cse4 proteolysis. (A and B) PPIase dead mutants stabilize the Cse4 protein level in vivo. We constructed plasmids harboring PPIase dead mutations (W363L and F402Y) (Table S2). The protein stability assay was performed as described in Figure 2. Isogenic yeast strains were FPR3 (Y2259), Vector (Y2260), fpr3 W363L (Y2261), and fpr3 F402Y (Y2262). Error bars represent SE of two or three independent experiments. (C) Mutation of P134 does stabilize the Cse4 protein level in vivo. There are five proline sites in Cse4 (P53, P98, P100, P134, and P157). We constructed plasmids harboring proline-to-valine mutation (Table S2). The protein stability assay was performed as described in Figure 2. Isogenic yeast strains were Cse4 (Y2255), cse4 P53V (Y2263), cse4 P98V (Y2264), cse4 P100V (Y2265), cse4 P134V (Y2266), and cse4 P157V (Y2267).
We next aimed to identify the target proline related to the Cse4 protein stability. There are five proline sites in Cse4, which are P53, P98, P100, P134, and P157. We generated yeast strains bearing each mutation (proline to valine) in Cse4 (Table S1) and analyzed the stability of Cse4. Only the P134V mutation has a clear effect on Cse4 stabilization (Figure 3C). This result suggests that P134 may be the target of Fpr3 isomerization.
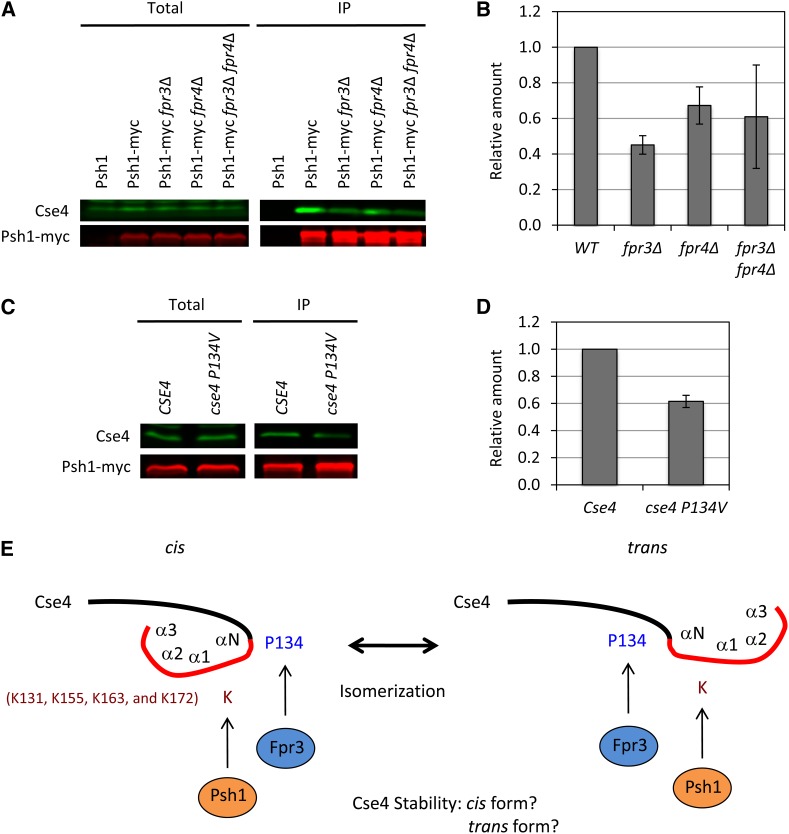
Psh1 ubiquitinates Cse4 at the following lysine residues in its C terminus: K131, K155, K163, and K172 (Hewawasam et al. 2010). As well, Cse4 directly interact with Psh1 via the RING finger domain, which is a hallmark of many E3 ligases (Ranjitkar et al. 2010). Because P134 is located close to those ubiquitylation sites, we tested whether deletion of FPR3 or FPR4 influences the interaction between Cse4 and Psh1. We performed coimmunoprecipitation and Western blotting using Psh1–myc-tagged strains. Psh1–myc-tagged protein physically interacts with Cse4 in wild-type (WT) cells. Interestingly, deletion of FPR3 or FPR4 diminishes the interaction between Cse4 and Psh1–myc (Figure 4, A and B). These data indicate that FPR3 and FPR4 regulate the Cse4–Psh1 interaction.
Figure 4.
Fpr3 regulates the Cse4–Psh1 interaction. (A and B) Interaction between Cse4 and Psh1 was diminished in fpr3∆ cells. The indicated strains were grown to log phase, lysed, and anti-myc immunoprecipitations were performed as previously described (Ohkuni et al. 2008). Total and the immunoprecipitated fraction (IP) were subjected to SDS–PAGE, and Western blots were used to detect Cse4 and myc-tagged Psh1. We used the Odyssey Imaging System to detect and quantify the signals. Isogenic yeast strains were untagged (YPH500), Psh1–myc (Y2280), Psh1–myc fpr3∆ (Y2281), Psh1–myc fpr4∆ (Y2282), and Psh1–myc fpr3∆ fpr4∆ (Y2283). Error bars represent SE of two independent experiments. Significant difference, P = 0.0089 (WT vs. fpr3∆). (C and D) P134V mutation in Cse4 diminishes the Psh1 interaction. Anti-myc immunoprecipitation assay and the quantification were performed as described in Figure 4, A and B. Isogenic yeast strains were Cse4 (Y2284) and cse4 P134V (Y2285). Error bars represent SE of two independent experiments. Significant difference, P = 0.0134. (E) A model for the role of Fpr3 in the Cse4 proteolysis. Psh1 is the E3 ubiquitin ligase that targets Cse4. Four lysine sites (K131, K155, K163, and K172) were ubiquitinated by Psh1. P134 close to the αN-helix (136–147) (Keith et al. 1999) might be the target of Fpr3 isomerization. We propose that the structural change in Cse4 from cis to trans or from trans to cis is important for the Cse4 degradation by Psh1. It is not known which form of Cse4 is ubiquitinated. The N-terminal domain (black) and the histone fold domain (red) of the α-N, α-1, α-2, and α-3 helices are indicated (Keith et al. 1999).
Given that the interaction of Cse4 with Psh1–myc is diminished in fpr3∆ cells, we predicted that mutation of P134V in Cse4 also influences the Cse4–Psh1 interaction. As expected, the interaction between cse4–P134V and Psh1–myc was also diminished (Figure 4, C and D). Taken together, these data suggest that structural change between the cis and trans form of P134 in Cse4 is essential for the Cse4–Psh1 interaction (Figure 4E).
We note that cse4–P134V mutants do not show any benomyl sensitivity or chromosome missegregation phenotype (data not shown). These phenotypes are also consistent with that of psh1∆ cells (Yuen et al. 2007). The phenotypic data also support the idea that Fpr3 regulates the Psh1-dependent Cse4 proteolysis. As Psh1 prevents Cse4 localization at noncentromeric regions (Hewawasam et al. 2010; Ranjitkar et al. 2010), Fpr3 might also prevent Cse4 interaction with noncentromeric regions. However, we found that Cse4 is accumulated at CEN (Figure S3), suggesting that Fpr3 might regulate Cse4 levels at the centromere.
Recently, it has become apparent that the Doa1/Ufd3 (WD repeat protein) is also required for the ubiquitination and proteolysis of Cse4 (Au et al. 2013). Doa1 regulates cellular levels of ubiquitin (Zhao et al. 2009). They propose that Doa1-mediated ubiquitination of Cse4 might be regulated by other ubiquitin ligases than Psh1 (Au et al. 2013). Interestingly, Doa1 is one of the targets of Fpr3 (Collins et al. 2007). We have shown that the stabilization level of Cse4 in fpr3∆ psh1∆ cells is much higher than that in fpr3∆ (Figure 2, C and D), implying that Fpr3 might regulate the proteolysis of Cse4 in both Psh1-dependent and Psh1-independent manners.
Loss of Fpr3 causes chromosome missegregation, but not benomyl sensitivity (Figure 1A and Figure S1). Interestingly, a high copy of FPR3 suppress the Glc7 overexpression lethality and the temperature sensitivity of ipl1-1 mutant (Ghosh and Cannon 2013). Deletion of Fpr3 showed a synthetic sickness phenotype with ipl1-2 mutant (data not shown). The opposing Ipl1 (aurora kinase B) and Glc7 (protein phosphatase 1: PP1) activities ensure a bipolar attachment to the spindle (Lampson and Cheeseman 2011). Thus, Fpr3 might also be involved in another mechanism in the centromere function such as bipolar attachment.
In summary, we characterized the mitotic function of Fpr3, which was identified by our in vitro kinetochore assembly system. Fpr3 has a role in Cse4 protein stability. Fpr3 PPIase dead mutants stabilized Cse4 protein, suggesting that the isomerization activity of Fpr3 is necessary for the Cse4 proteolysis. The interaction between Cse4 and Psh1 was diminished in the fpr3∆ mutant. Furthermore, a mutation on P134, a target of Fpr3 isomerization, reduced the Cse4–Psh1 interaction. Thus, we propose that the structural change between the cis and trans form in Cse4 is important for the interaction with E3 ubiquitin ligase Psh1 (Figure 4E).
Supplementary Material
Acknowledgments
We thank Vivien Measday and members of the Kitagawa lab for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health grant GM68418.
Footnotes
Communicating editor: N. Hunter
Literature Cited
- Akiyoshi B., Sarangapani K. K., Powers A. F., Nelson C. R., Reichow S. L., et al. , 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468: 576–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo-Rodriguez M., Wu X., Hanes S. D., Heitman J., 2004. Prolyl isomerases in yeast. Front. Biosci. 9: 2420–2446. [DOI] [PubMed] [Google Scholar]
- Au W. C., Dawson A. R., Rawson D. W., Taylor S. B., Baker R. E., et al. , 2013. A novel role of the N terminus of budding yeast histone h3 variant cse4 in ubiquitin-mediated proteolysis. Genetics 194: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton B. M., Zang J. H., Thorner J., 1994. A novel FK506- and rapamycin-binding protein (FPR3 gene product) in the yeast Saccharomyces cerevisiae is a proline rotamase localized to the nucleolus. J. Cell Biol. 127: 623–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., et al. , 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810. [DOI] [PubMed] [Google Scholar]
- Dolinski K., Muir S., Cardenas M., Heitman J., 1997. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94: 13093–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Cannon J. F., 2013. Analysis of protein phosphatase-1 and aurora protein kinase suppressors reveals new aspects of regulatory protein function in Saccharomyces cerevisiae. PLoS ONE 8: e69133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G., Shivaraju M., Mattingly M., Venkatesh S., Martin-Brown S., et al. , 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell 40: 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A., Tham W. H., Brar G. A., Amon A., 2005. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell 122: 861–873. [DOI] [PubMed] [Google Scholar]
- Holland A. J., Cleveland D. W., 2009. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10: 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith K. C., Baker R. E., Chen Y., Harris K., Stoler S., et al. , 1999. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell. Biol. 19: 6130–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M. A., Cheeseman I. M., 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macqueen A. J., Roeder G. S., 2009. Fpr3 and Zip3 ensure that initiation of meiotic recombination precedes chromosome synapsis in budding yeast. Curr. Biol. 19: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. J., Santos-Rosa H., Kouzarides T., 2006. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126: 905–916. [DOI] [PubMed] [Google Scholar]
- Ohkuni K., Abdulle R., Tong A. H., Boone C., Kitagawa K., 2008. Ybp2 associates with the central kinetochore of Saccharomyces cerevisiae and mediates proper mitotic progression. PLoS One 3: e1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K., Kitagawa K., 2011. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr. Biol. 21: 1695–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P., Press M. O., Yi X., Baker R., MacCoss M. J., et al. , 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 40: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X., Xue Z., Melese T., 1994. Yeast NPI46 encodes a novel prolyl cis-trans isomerase that is located in the nucleolus. J. Cell Biol. 126: 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K. W., Montpetit B., Hieter P., 2005. The kinetochore and cancer: What’s the connection? Curr. Opin. Cell Biol. 17: 576–582. [DOI] [PubMed] [Google Scholar]
- Yuen K. W., Warren C. D., Chen O., Kwok T., Hieter P., et al. , 2007. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc. Natl. Acad. Sci. USA 104: 3925–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Li G., Schindelin H., Lennarz W. J., 2009. An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation. Proc. Natl. Acad. Sci. USA 106: 16197–16202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.