Abstract
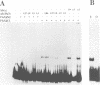
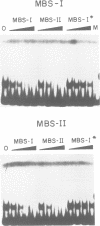
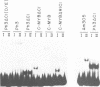
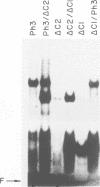
The MYB.Ph3 protein recognized two DNA sequences that resemble the two known types of MYB DNA binding site: consensus I (MBSI), aaaAaaC(G/C)-GTTA, and consensus II (MBSII), aaaAGTTAGTTA. Optimal MBSI was recognized by animal c-MYB and not by Am305 from Antirrhinum, whereas MBSII showed the reverse behaviour. Different constraints on MYB.Ph3 binding to the two classes of sequences were demonstrated. DNA binding studies with mutated MBSI and MBSII and hydroxyl radical footprinting analysis, pointed to the N-terminal MYB repeat (R2) as the most involved in determining the dual DNA binding specificity of MYB.Ph3 and supported the idea that binding to MBSI and MBSII does not involve alternative orientations of the two repeats of MYB.Ph3. Minimal promoters containing either MBSI and MBSII were activated to the same extent by MYB.Ph3 in yeast, indicating that both types of binding site can be functionally equivalent. MYB.Ph3 binding sites are present in the promoter of flavonoid biosynthetic genes, such as the Petunia chsJ gene, which was transcriptionally activated by MYB.Ph3 in tobacco protoplasts. MYB.Ph3 was immunolocalized in the epidermal cell layer of petals, where flavonoid biosynthetic genes are actively expressed. This strongly suggests a role for MYB.Ph3 in the regulation of flavonoid biosynthesis.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- An C., Ichinose Y., Yamada T., Tanaka Y., Shiraishi T., Oku H. Organization of the genes encoding chalcone synthase in Pisum sativum. Plant Mol Biol. 1993 Mar;21(5):789–803. doi: 10.1007/BF00027112. [DOI] [PubMed] [Google Scholar]
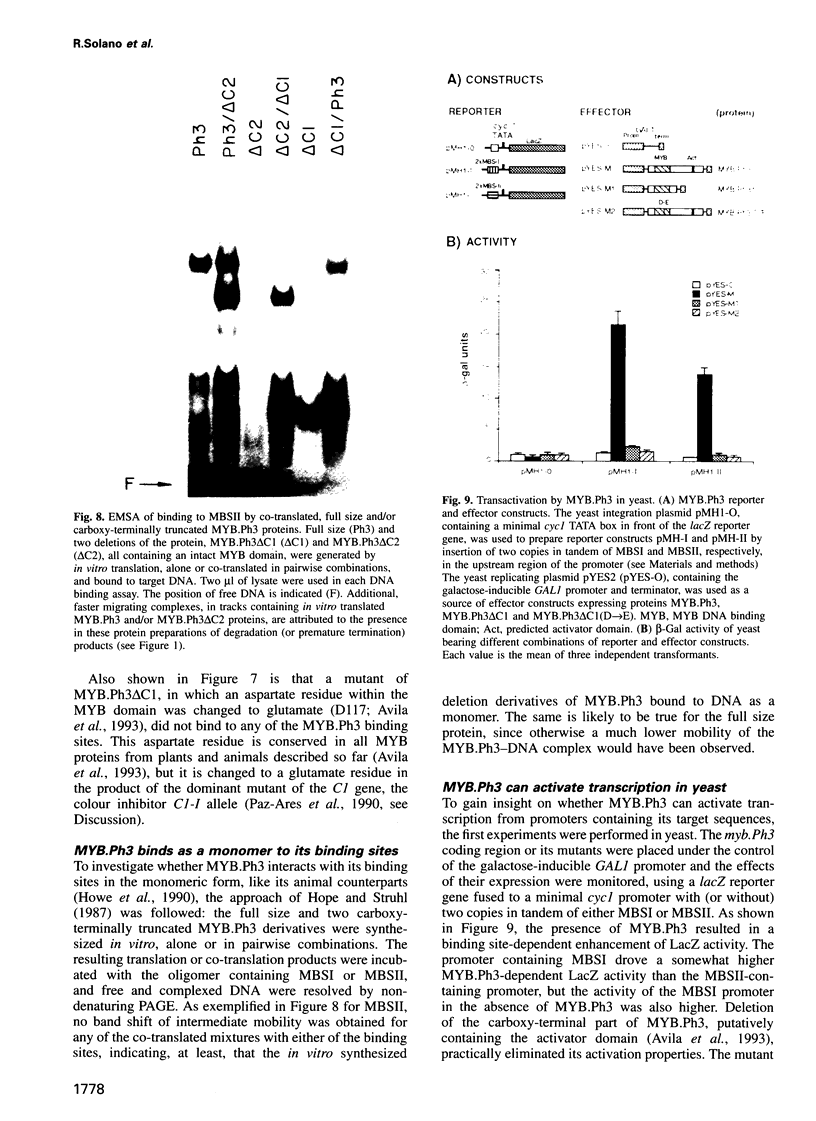
- Avila J., Nieto C., Cañas L., Benito M. J., Paz-Ares J. Petunia hybrida genes related to the maize regulatory C1 gene and to animal myb proto-oncogenes. Plant J. 1993 Apr;3(4):553–562. doi: 10.1046/j.1365-313x.1993.03040553.x. [DOI] [PubMed] [Google Scholar]
- Baranowskij N., Frohberg C., Prat S., Willmitzer L. A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 1994 Nov 15;13(22):5383–5392. doi: 10.1002/j.1460-2075.1994.tb06873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
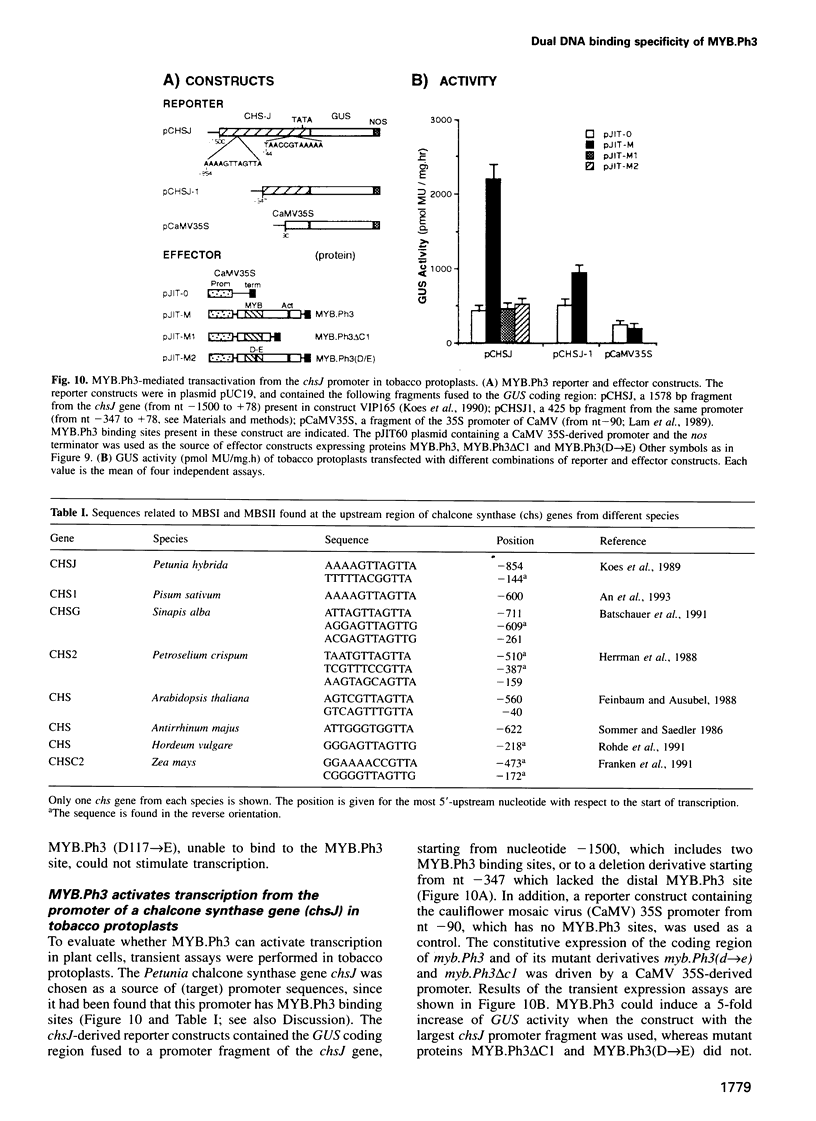
- Batschauer A., Ehmann B., Schäfer E. Cloning and characterization of a chalcone synthase gene from mustard and its light-dependent expression. Plant Mol Biol. 1991 Feb;16(2):175–185. doi: 10.1007/BF00020550. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Carra J. H., Schleif R. F. Variation of half-site organization and DNA looping by AraC protein. EMBO J. 1993 Jan;12(1):35–44. doi: 10.1002/j.1460-2075.1993.tb05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone K. C., Burr F. A., Burr B. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9631–9635. doi: 10.1073/pnas.83.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone K. C., Cocciolone S. M., Burr F. A., Burr B. Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell. 1993 Dec;5(12):1795–1805. doi: 10.1105/tpc.5.12.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum R. L., Ausubel F. M. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol. 1988 May;8(5):1985–1992. doi: 10.1128/mcb.8.5.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foos G., Grimm S., Klempnauer K. H. Functional antagonism between members of the myb family: B-myb inhibits v-myb-induced gene activation. EMBO J. 1992 Dec;11(12):4619–4629. doi: 10.1002/j.1460-2075.1992.tb05564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton J., Gibson T. J., Ness S. A., Döderlein G., Graf T. Proposed structure for the DNA-binding domain of the Myb oncoprotein based on model building and mutational analysis. Protein Eng. 1991 Dec;4(8):891–901. doi: 10.1093/protein/4.8.891. [DOI] [PubMed] [Google Scholar]
- Franken P., Schrell S., Peterson P. A., Saedler H., Wienand U. Molecular analysis of protein domain function encoded by the myb-homologous maize genes C1, Zm 1 and Zm 38. Plant J. 1994 Jul;6(1):21–30. doi: 10.1046/j.1365-313x.1994.6010021.x. [DOI] [PubMed] [Google Scholar]
- Goff S. A., Cone K. C., Chandler V. L. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 1992 May;6(5):864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- Goff S. A., Cone K. C., Fromm M. E. Identification of functional domains in the maize transcriptional activator C1: comparison of wild-type and dominant inhibitor proteins. Genes Dev. 1991 Feb;5(2):298–309. doi: 10.1101/gad.5.2.298. [DOI] [PubMed] [Google Scholar]
- Goff S. A., Klein T. M., Roth B. A., Fromm M. E., Cone K. C., Radicella J. P., Chandler V. L. Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. EMBO J. 1990 Aug;9(8):2517–2522. doi: 10.1002/j.1460-2075.1990.tb07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T. Myb: a transcriptional activator linking proliferation and differentiation in hematopoietic cells. Curr Opin Genet Dev. 1992 Apr;2(2):249–255. doi: 10.1016/s0959-437x(05)80281-3. [DOI] [PubMed] [Google Scholar]
- Grotewold E., Athma P., Peterson T. Alternatively spliced products of the maize P gene encode proteins with homology to the DNA-binding domain of myb-like transcription factors. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4587–4591. doi: 10.1073/pnas.88.11.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E., Drummond B. J., Bowen B., Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994 Feb 11;76(3):543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
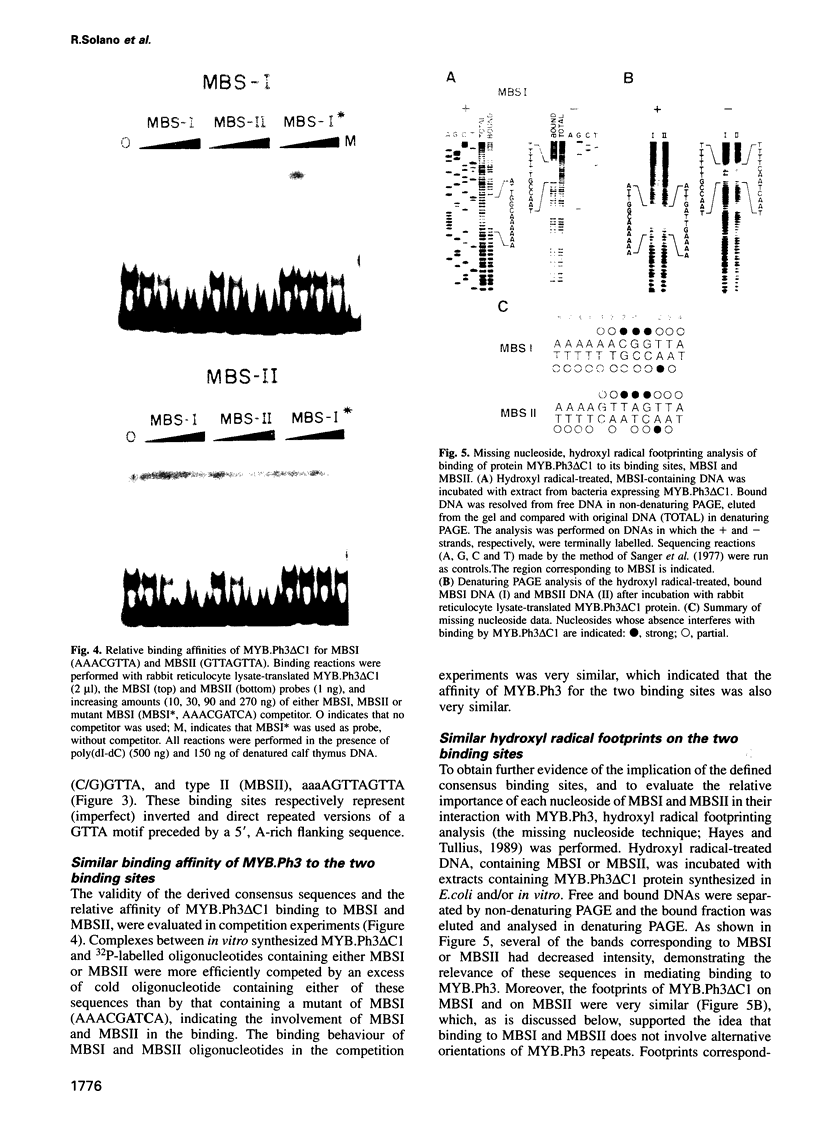
- Hayes J. J., Tullius T. D. The missing nucleoside experiment: a new technique to study recognition of DNA by protein. Biochemistry. 1989 Nov 28;28(24):9521–9527. doi: 10.1021/bi00450a041. [DOI] [PubMed] [Google Scholar]
- Herrmann A., Schulz W., Hahlbrock K. Two alleles of the single-copy chalcone synthase gene in parsley differ by a transposon-like element. Mol Gen Genet. 1988 Apr;212(1):93–98. doi: 10.1007/BF00322449. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987 Sep;6(9):2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K. M., Reakes C. F., Watson R. J. Characterization of the sequence-specific interaction of mouse c-myb protein with DNA. EMBO J. 1990 Jan;9(1):161–169. doi: 10.1002/j.1460-2075.1990.tb08092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D., Culianez-Macia F., Prescott A. G., Roberts K., Martin C. Expression patterns of myb genes from Antirrhinum flowers. Plant Cell. 1991 Feb;3(2):115–125. doi: 10.1105/tpc.3.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamin N., Gabrielsen O. S., Gilles N., Lirsac P. N., Toma F. Secondary structure of the DNA-binding domain of the c-Myb oncoprotein in solution. A multidimensional double and triple heteronuclear NMR study. Eur J Biochem. 1993 Aug 15;216(1):147–154. doi: 10.1111/j.1432-1033.1993.tb18126.x. [DOI] [PubMed] [Google Scholar]
- Katzen A. L., Kornberg T. B., Bishop J. M. Isolation of the proto-oncogene c-myb from D. melanogaster. Cell. 1985 Jun;41(2):449–456. doi: 10.1016/s0092-8674(85)80018-0. [DOI] [PubMed] [Google Scholar]
- Kim J., Tzamarias D., Ellenberger T., Harrison S. C., Struhl K. Adaptability at the protein-DNA interface is an important aspect of sequence recognition by bZIP proteins. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4513–4517. doi: 10.1073/pnas.90.10.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R. E., Spelt C. E., van den Elzen P. J., Mol J. N. Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene. 1989 Sep 30;81(2):245–257. doi: 10.1016/0378-1119(89)90185-6. [DOI] [PubMed] [Google Scholar]
- Koes R. E., Van Blokland R., Quattrocchio F., Van Tunen A. J., Mol JNM. Chalcone Synthase Promoters in Petunia Are Active in Pigmented and Unpigmented Cell Types. Plant Cell. 1990 May;2(5):379–392. doi: 10.1105/tpc.2.5.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa R., Yu V. C., När A., Kyakumoto S., Han Z., Silverman S., Rosenfeld M. G., Glass C. K. Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes Dev. 1993 Jul;7(7B):1423–1435. doi: 10.1101/gad.7.7b.1423. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lam E., Benfey P. N., Gilmartin P. M., Fang R. X., Chua N. H. Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7890–7894. doi: 10.1073/pnas.86.20.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., He X., Gerrero M. R., Mok M., Aggarwal A., Rosenfeld M. G. Spacing and orientation of bipartite DNA-binding motifs as potential functional determinants for POU domain factors. Genes Dev. 1993 Dec;7(12B):2483–2496. doi: 10.1101/gad.7.12b.2483. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. New light on Myc and Myb. Part II. Myb. Genes Dev. 1990 Dec;4(12B):2235–2241. doi: 10.1101/gad.4.12b.2235. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Myrset A. H., Bostad A., Jamin N., Lirsac P. N., Toma F., Gabrielsen O. S. DNA and redox state induced conformational changes in the DNA-binding domain of the Myb oncoprotein. EMBO J. 1993 Dec;12(12):4625–4633. doi: 10.1002/j.1460-2075.1993.tb06151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda K., Glover B. J., Linstead P., Martin C. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature. 1994 Jun 23;369(6482):661–664. doi: 10.1038/369661a0. [DOI] [PubMed] [Google Scholar]
- Ogata K., Hojo H., Aimoto S., Nakai T., Nakamura H., Sarai A., Ishii S., Nishimura Y. Solution structure of a DNA-binding unit of Myb: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6428–6432. doi: 10.1073/pnas.89.14.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K., Morikawa S., Nakamura H., Sekikawa A., Inoue T., Kanai H., Sarai A., Ishii S., Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994 Nov 18;79(4):639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- Oppenheimer D. G., Herman P. L., Sivakumaran S., Esch J., Marks M. D. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991 Nov 1;67(3):483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- Ording E., Kvåvik W., Bostad A., Gabrielsen O. S. Two functionally distinct half sites in the DNA-recognition sequence of the Myb oncoprotein. Eur J Biochem. 1994 May 15;222(1):113–120. doi: 10.1111/j.1432-1033.1994.tb18848.x. [DOI] [PubMed] [Google Scholar]
- Paz-Ares J., Ghosal D., Saedler H. Molecular analysis of the C1-I allele from Zea mays: a dominant mutant of the regulatory C1 locus. EMBO J. 1990 Feb;9(2):315–321. doi: 10.1002/j.1460-2075.1990.tb08113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J., Ghosal D., Wienand U., Peterson P. A., Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987 Dec 1;6(12):3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro M., García-Olmedo F., Diaz I. Redox modulation of the expression of bacterial genes encoding cysteine-rich proteins in plant protoplasts. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3867–3871. doi: 10.1073/pnas.91.9.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R., Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990 Nov 11;18(21):6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Ramsay R. G., Ishii S., Gonda T. J. Interaction of the Myb protein with specific DNA binding sites. J Biol Chem. 1992 Mar 15;267(8):5656–5662. [PubMed] [Google Scholar]
- Rohde W., Dörr S., Salamini F., Becker D. Structure of a chalcone synthase gene from Hordeum vulgare. Plant Mol Biol. 1991 Jun;16(6):1103–1106. doi: 10.1007/BF00016087. [DOI] [PubMed] [Google Scholar]
- Sablowski R. W., Moyano E., Culianez-Macia F. A., Schuch W., Martin C., Bevan M. A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 1994 Jan 1;13(1):128–137. doi: 10.1002/j.1460-2075.1994.tb06242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakura H., Kanei-Ishii C., Nagase T., Nakagoshi H., Gonda T. J., Ishii S. Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5758–5762. doi: 10.1073/pnas.86.15.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarai A., Uedaira H., Morii H., Yasukawa T., Ogata K., Nishimura Y., Ishii S. Thermal stability of the DNA-binding domain of the Myb oncoprotein. Biochemistry. 1993 Aug 3;32(30):7759–7764. doi: 10.1021/bi00081a022. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tanikawa J., Yasukawa T., Enari M., Ogata K., Nishimura Y., Ishii S., Sarai A. Recognition of specific DNA sequences by the c-myb protooncogene product: role of three repeat units in the DNA-binding domain. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9320–9324. doi: 10.1073/pnas.90.20.9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice-Baldwin K., Fink G. R., Arndt K. T. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science. 1989 Nov 17;246(4932):931–935. doi: 10.1126/science.2683089. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T., Yamaguchi-Shinozaki K., Urao S., Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993 Nov;5(11):1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weston K. Extension of the DNA binding consensus of the chicken c-Myb and v-Myb proteins. Nucleic Acids Res. 1992 Jun 25;20(12):3043–3049. doi: 10.1093/nar/20.12.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]