ABA biosynthetic enzymes expressed in vascular tissues induce stomatal closure, indicating that long-distance ABA signal transfer from the vascular bundle is likely to be mediated by specific transporters.
Abstract
Abscisic acid (ABA) is a phytohormone that responds to environmental stresses, such as water deficiency. Recent studies have shown that ABA biosynthetic enzymes are expressed in the vascular area under both nonstressed and water-stressed growth conditions. However, specific cells in the vasculature involved in ABA biosynthesis have not been identified. Here, we detected the expression of two genes encoding ABA biosynthetic enzymes, ABSCISIC ACID DEFICIENT2 and ABSCISIC ALDEHYDE OXIDASE3, in phloem companion cells in vascular tissues. Furthermore, we identified an ATP-binding cassette transporter, Arabidopsis thaliana ABCG25 (AtABCG25), expressed in the same cells. Additionally, AtABCG25-expressing Spodoptera frugiperda9 culture cells showed an ABA efflux function. Finally, we observed that enhancement of ABA biosynthesis in phloem companion cells induced guard cell responses, even under normal growth conditions. These results show that ABA is synthesized in specific cells and can be transported to target cells in different tissues.
Hormones are chemical substances that exert a biochemical action on target cells at low concentrations. All multicellular organisms, including animals and plants, produce hormones to control their physiological status. In animals, many ordinary hormones are secreted from specific cells (such as endocrine cells) and transported to their target sites in other areas of the body. However, it remains unclear whether the concept of hormones as defined in animals is applicable to plants, because plant hormones are not generally synthesized in specific cells but are broadly produced (Weyers and Paterson, 2001; Gaspar et al., 2003; Forestan and Varotto, 2012).
Abscisic acid (ABA) is a key phytohormone that prevents water loss from the plant body by acting on guard cells, of which stomata (epidermal pores) compose the aerial organs in plants (Hetherington, 2001; Schroeder et al., 2001; Fan et al., 2004; Joshi-Saha et al., 2011). Gene and protein expression analyses using antisense RNA or antibodies specific for ABA biosynthetic enzymes in Arabidopsis (Arabidopsis thaliana) have shown that parenchyma cells in vascular bundles are the abundant expression site of ABA biosynthesis under drought stress and well-watered growth conditions (Cheng et al., 2002; Koiwai et al., 2004; Endo et al., 2008). Because the vasculature is separated from guard cells, it has been suggested that ABA is transported from the site of synthesis to the site of action (Seo and Koshiba, 2011).
We previously found that an ATP-binding cassette (ABC) transporter family, Arabidopsis thaliana ABCG25 (AtABCG25), is expressed mainly in vascular tissues, and it is expected to function as an ABA exporter that transports ABA from inside to outside cells (Kuromori et al., 2010). According to this observation, we have proposed a working model: ABA is exported from ABA-synthesizing cells in vascular tissues by AtABCG25 to reach distant guard cells and induce stomatal closure (Kuromori and Shinozaki, 2010; Umezawa et al., 2010, 2011). However, the parenchyma cells in vascular tissues expressing ABA biosynthetic enzymes or AtABCG25 transporting factor have not been identified.
Here, we explored whether specific cells express ABA biosynthetic enzymes or an ABA transporter and found that their genes were expressed in phloem companion cells of vascular tissues. ABA synthesis in these cells enhances transsignaling to distant guard cells of the epidermis. These results show that ABA is synthesized in specific cells and transported to target cells in another tissue. This result is similar to endocrine hormones in animals and suggests that the ABA transport pathway between tissues in plants may be associated with specific transporters.
RESULTS AND DISCUSSION
In Arabidopsis, the final two steps of ABA biosynthesis are enzymatic processes catalyzed by xanthoxin dehydrogenase, encoded by ABSCISIC ACID DEFICIENT2 (ABA2), and abscisic aldehyde oxidase, encoded by ABSCISIC ALDEHYDE OXIDASE3 (AAO3; Finkelstein and Rock, 2002; Schwartz et al., 2003; Nambara and Marion-Poll, 2005). To determine the location of ABA-biosynthesizing cells, approximately 0.9 kb of the ABA2 promoter region and 1.2 kb of the AAO3 promoter region, which cover the majority of intergenic regions adjacent to the coding genes, were cloned and used to drive expression of the nuclear-localized signal attached to GFP (nGFP) as a promoter-dependent reporter. Each of the recombinant vectors was transformed into Arabidopsis, and the green fluorescence signals were observed in transgenic plants.
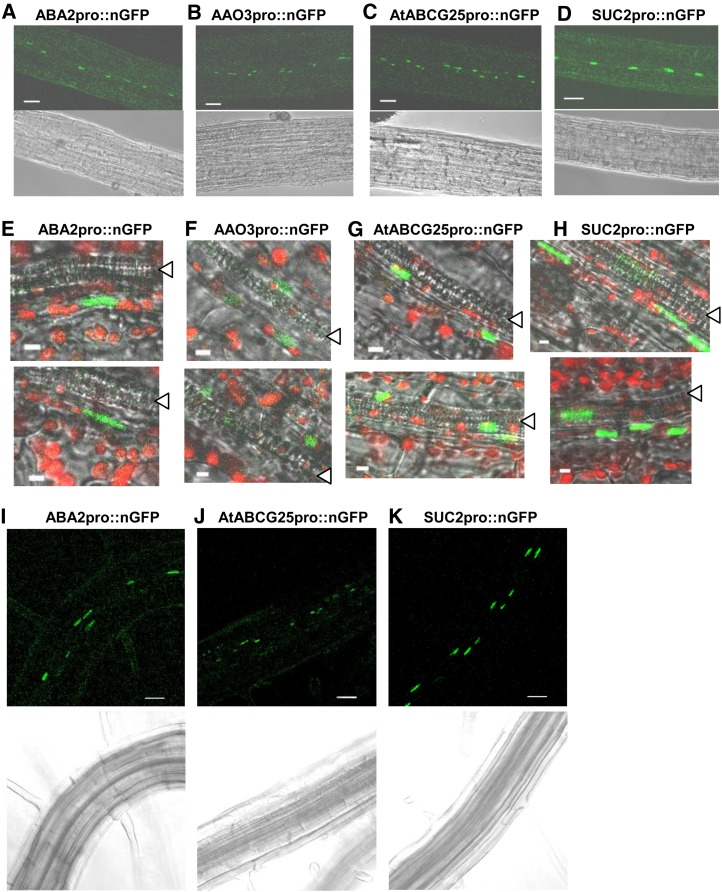
It is difficult to observe fluorescent signals in the leaf vascular site of intact plants because of the strong autofluorescence by chloroplasts. Therefore, we examined various aerial tissues to detect clear signals. We observed an array of nuclear fluorescence of nGFP along the vascular veins in the center of anther filaments in both transgenic plants expressing ABA2 promoter-driven nGFP and those plants expressing AAO3 promoter-driven nGFP using confocal laser scanning microscopy (Fig. 1, A and B). nGFP fluorescence was also observed close to (but not in) xylem vessels in sepals or petals in both transgenic plants (Fig. 1, E and F), although occasionally, it was detected in the sepal epidermis of the AAO3 promoter-driven transgenic plants. These results showed that the expression patterns of the two promoters overlap in the vascular area. Furthermore, the expressions of ABA2 andAAO3 are limited to particular cells and not expressed broadly through every parenchyma cell in the vascular tissue. When observing belowground components, a similar pattern of fluorescence was detected in the root vasculature of ABA2 promoter-driven transformants (Fig. 1I), although there were only faint signals in AAO3 promoter-driven transformants.
Figure 1.
Localization of ABA2, AAO3, AtABCG25, or SUC2 promoter-driven nGFP in Arabidopsis vascular tissues. A to D, Confocal images were taken in the filaments of transgenic plants expressing (A) ABA2 (ABA2pro::nGFP), (B) AAO3 (AAO3pro::nGFP), (C) AtABCG25 (AtABCG25pro::nGFP), and (D) SUC2 (SUC2pro::nGFP) promoter-driven nGFP. Fluorescence images (top)and bright-field images (bottom)are shown. Bars in A to D = 40 µm. E to H, Confocal images were taken in the sepals or petals of transgenic plants expressing (E) ABA2 (ABA2pro::nGFP), (F) AAO3 (AAO3pro::nGFP), (G) AtABCG25 (AtABCG25pro::nGFP), and (H) SUC2 (SUC2pro::nGFP) promoter-driven nGFP. nGFP localization is shown in green, and chlorophyll autofluorescence is shown in red. Triangles show xylem vessels. Bars in E to H = 4 µm. I to K, Confocal images were taken in roots of transgenic plants expressing (I) ABA2 (ABA2pro::nGFP), (J) AtABCG25 (AtABCG25pro::nGFP), and (K) SUC2 (SUC2pro::nGFP) promoter-driven nGFP. Confocal images in J were taken after ABA treatment. Fluorescence images (top) and bright-field images (bottom)are shown. Bars in I to K = 50 µm.
To identify cells expressing ABA2 and AAO3 in the vascular area, we first examined a publicly available database of a high-resolution microarray map in Arabidopsis (Brady et al., 2007). Compared with vascular cell types, both ABA2 and AAO3 have expression patterns similar to SUCROSE-PROTON SYMPORTER2 (SUC2), which is commonly used as a gene specifically expressed in phloem companion cells (Stadler and Sauer, 1996; Imlau et al., 1999). Based on this information, it is possible that the specific cells expressing ABA2 and AAO3 are phloem companion cells. Phloem companion cells are located adjacent to phloem sieve tubes, which form a line in the vascular tissues (Oparka and Turgeon, 1999). We generated transgenic plants to observe SUC2 promoter-driven nGFP and determined that the fluorescence in the transformants formed a clear array along the vasculature (Fig. 1, D, H, and K).
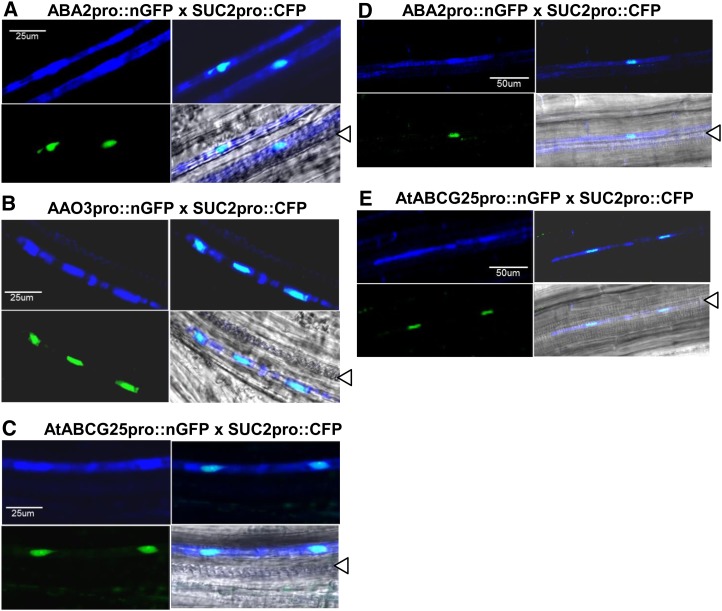
To confirm whether ABA2 or AAO3 promoter-driven fluorescent cells colocalize with SUC2 promoter-driven fluorescent cells, we generated transgenic plants expressing SUC2 promoter-driven cyan fluorescent protein (CFP) and crossed them with the plants expressing ABA2 promoter-driven nGFP or AAO3 promoter-driven nGFP. By visualizing two types of fluorescent proteins, we found that both ABA2 promoter-driven nGFP and AAO3 promoter-driven nGFP colocalized with SUC2 promoter-driven CFP (Fig. 2, A, B, and D; Supplemental Fig. S1). These results indicated that ABA2 and AAO3 are expressed in phloem companion cells.
Figure 2.
Colocalization of ABA2, AAO3, or AtABCG25 promoter-driven nGFP and SUC2 promoter-driven CFP in Arabidopsis vascular tissues. Confocal images of vascular tissues of (A to C) filaments and (D and E) roots were taken after crossing between the transgenic plants expressing (A and D) ABA2 (ABA2pro::nGFP × SUC2pro::CFP), (B) AAO3 (AAO3pro::nGFP × SUC2pro::CFP), and (C and E) AtABCG25 (AtABCG25pro::nGFP × SUC2pro::CFP) promoter-driven nGFP and transgenic plants expressing SUC2 promoter-driven CFP. Confocal images in E were taken after ABA treatment. The two fluorophores CFP and nGFP were simultaneously visualized; CFP localization is shown in cyan (top left), nGFP localization is shown in green (bottom left), and both were merged (top right). Two fluorescence images and bright-field images were merged (bottom right). Triangles show xylem vessels.
Next, we investigated cells expressing AtABCG25. Approximately 2.0 kb of the AtABCG25 promoter region was used to detect promoter-dependent nGFP expression. A similar pattern of fluorescence for ABA2 and AAO3 was detected in aerial components of AtABCG25 promoter-driven transformants (Fig. 1, C and G). No clear signals were observed in roots, but ABA-treated plants showed similar signals (Fig. 1J). We previously found that AtABCG25 expression was induced by ABA treatment (Kuromori et al., 2010). We then crossed transgenic plants expressing AtABCG25 promoter-driven nGFP with transgenic plants expressing SUC2 promoter-driven CFP. By visualizing the two fluorescent proteins, we found that AtABCG25 promoter-driven nGFP and SUC2 promoter-driven CFP colocalize (Fig. 2, C and E). These results indicated that (in addition to ABA2 and AAO3) AtABCG25 is predominantly expressed in phloem companion cells.
The colocalization of cells expressing ABA2, AAO3, and AtABCG25 supports our working model that AtABCG25 plays the role of an ABA exporter in ABA-biosynthesizing cells (Kuromori and Shinozaki, 2010; Umezawa et al., 2010, 2011). To assess ABA efflux activity of AtABCG25, we expressed AtABCG25 cDNA in cultured Spodoptera frugiperda9 (Sf9) insect cells. After adding ABA isotope to cell cultures, Sf9 insect cells expressing AtABCG25 contained lower radioactive counts of ABA isotopes remaining in cells than control cells containing the empty vector (Fig. 3A). This result indicated that ABA excretion was more efficient in cells expressing AtABCG25 than in control cells. This count reduction was not detected when other isotope-labeled phytophormones, indole-3-acetic acid (auxin) or jasmonic acid, were added to cell cultures (Fig. 3, B and C), indicative of the specificity of the target molecule. These results support the conclusion that AtABCG25 is an ABA exporter with efflux activity from inside to outside cells.
Figure 3.
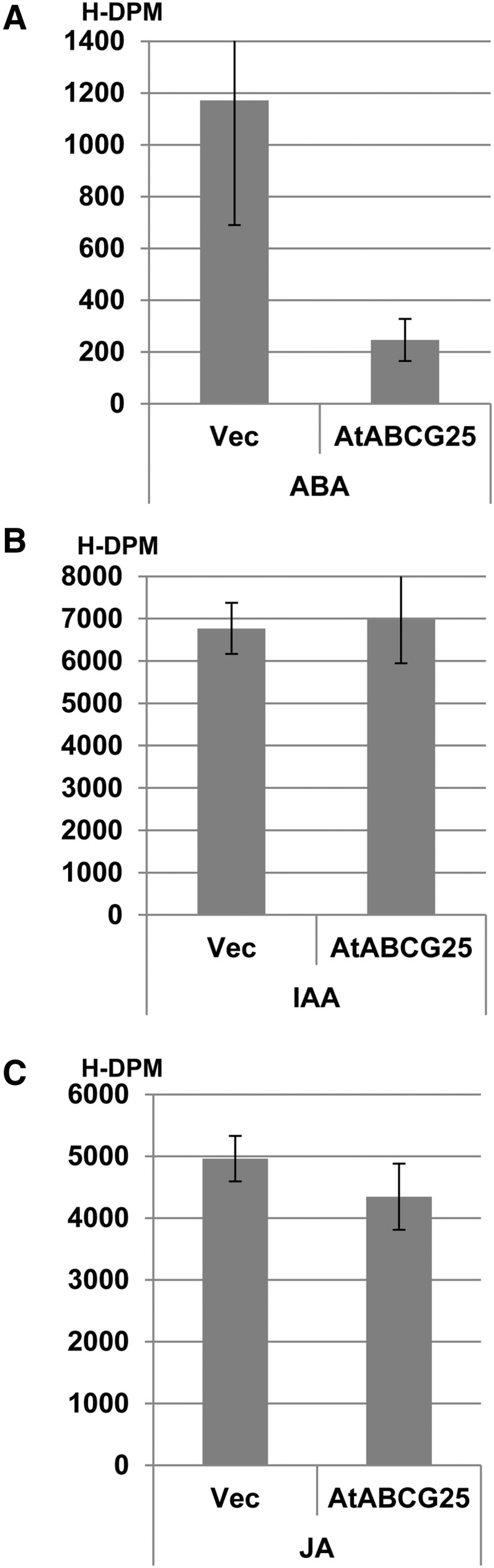
Transport assay of isotope-labeled phytophormones by AtABCG25-expressing Sf9 culture cells; (A) ABA ([3H]ABA), (B) indole-3-acetic acid ([3H]IAA), or (C) jasmonic acid ([3H]JA) efflux activity was measured in Sf9 culture cells expressing AtABCG25 and empty vector (Vec). Each bar represents the mean ± sd (n = 4). H-DPM, 3H-disintegrations per minute.
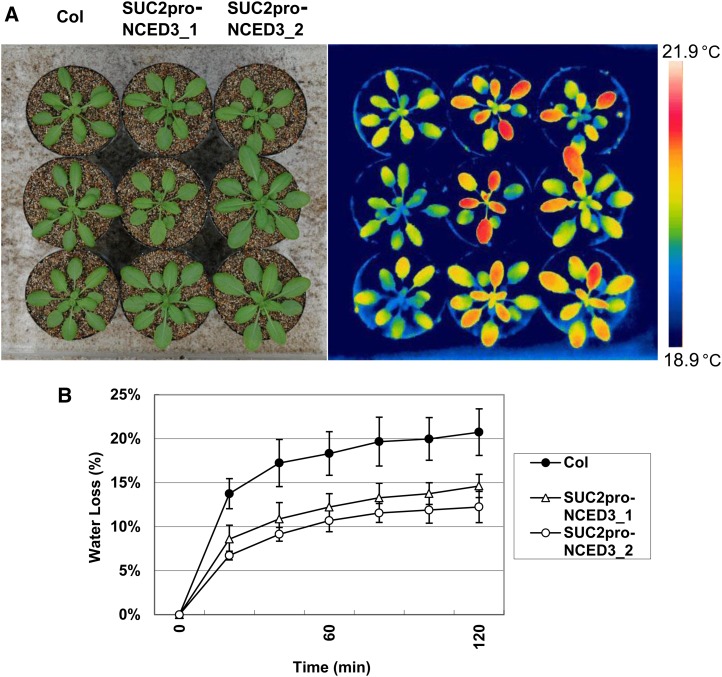
ABA induces stomatal closure through its action on guard cells, which are located in the epidermis and separated from vascular tissue (Hetherington, 2001; Schroeder et al., 2001; Fan et al., 2004; Joshi-Saha et al., 2011). To investigate the intertissue signal transfer of ABA, we induced ABA biosynthesis in phloem companion cells. We generated transgenic plants expressing SUC2 promoter-driven NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3), because it enhances ABA biosynthesis (Iuchi et al., 2001). Transgenic plants expressing SUC2 promoter-driven NCED3 did not show any visible phenotypes (Fig. 4A). However, when plants were observed using a thermographic camera, we found that the leaf temperature of transgenic plants was significantly higher than the leaf temperature in wild-type plants under well-watered growth conditions (Fig. 4A). Additionally, water loss from detached leaves of the transgenic plants was reduced compared with water loss from detached leaves of wild-type plants (Fig. 4B). These results suggest that transgenic plants expressing SUC2 promoter-driven NCED3 contain limited stomata open on the leaves and that ABA biosynthesis induced in phloem companion cells influences guard cells in the absence of environmental stress.
Figure 4.
Transpiration phenotypes of transgenic plants expressing SUC2 promoter-driven NCED3. A, Thermal images of the transgenic plants expressing SUC2 promoter-driven NCED3. Rosette leaves of 5-week-old wild-type plants (Col) and two independent transgenic plants expressing SUC2 promoter-driven NCED3 (SUC2pro-NCED3_1 and SUC2pro-NCED3_2) were imaged by a visible light camera (left) and an infrared thermography device (right). B, Transpiration ratio of the transgenic plants expressing SUC2 promoter-driven NCED3. The water loss of the detached rosette leaves of 5-week-old plants was determined as a percentage of the initial fresh weight. Values are shown as means ± sds of three independent plants.
Specific organs or cells that supply traditional phytohormones have not been identified. For example, auxin, the best characterized phytohormone, is synthesized in many plant tissues through different pathways (Normanly, 2010; Zhao, 2010; Forestan and Varotto, 2012). In this study, we have indicated that phloem companion cells express ABA biosynthetic enzymes and an ABA transporter. Phloem companion cells are developmentally derived from the unequal division of phloem mother cells and closely associated with phloem sieve tube cells differentiated from phloem mother cells to develop the sieve tube element (Oparka and Turgeon, 1999; Beck, 2010). Interestingly, the sieve tube–companion cell complexes are largely symplastically isolated from other parenchyma cells (Beck, 2010); thus, ABA exporters are required to secrete ABA from the site of biosynthesis.
In addition to vascular ABA synthesis, guard cell-autonomous ABA synthesis has been reported recently (Bauer et al., 2013). Guard cell-autonomous ABA synthesis could allow the plant to respond rapidly to changing environmental conditions to maintain water status homeostasis. According to this report, probably both vascular ABA and guard cell-autonomous ABA may be related to stomatal regulation dependently or independently; however, the initial induction and the spatiotemporal process of drought response at guard cells are not completely understood (Okamoto et al., 2009). Additionally, guard cells are also arranging the physiological appearances by diurnal regulation (Chen et al., 2012; Hills et al., 2012). The balance of the ABA synthesis in vasculature and guard cells under stress or nonstress conditions is to be investigated on the basis to trigger and/or maintain guard cell responses.
ABA transport assays performed directly using insect cell culture were consistent with our previous vesicle transport experiments, in which the efflux activity was detectable as ABA uptake of the regenerated membrane vesicles, including inside-out transport (Kuromori et al., 2010). In both cases, we detected AtABCG25 export activity in a heterologous system of Sf9 insect culture cells. This finding suggested that ABA export activity of AtABCG25 could be activated under no or very little posttranslational regulation. Based on the increased stomatal closure after enhancing ABA biosynthesis in phloem companion cells, we showed a route of ABA signaling from phloem companion cells at the vasculature to epidermal guard cells, which is likely mediated by ABA exporters. In addition, two ABA importers, AtABCG40 and ABA-IMPORTING TRANSPORTER1, have been reported, indicating a complex system of intercellular ABA transport in plants (Kang et al., 2010; Kanno et al., 2012).
Floral stimulus proteins, such as FLOWERING LOCUS T in Arabidopsis, are known to be produced at phloem companion cells and undergo long-distance movement into the shoot apex to initiate flowering (Corbesier et al., 2007; Mathieu et al., 2007). Thus, phloem companion cells may secrete different types of remote signals, likely corresponding to endocrine cells in animals. Because they supply multiple remote signals, phloem companion cells may play a role in integrating environmental recognition into the remote signaling output.
MATERIALS AND METHODS
Vector Construction
A 0.9-kb ABA2 promoter region, a 1.2-kb AAO3 promoter region, and a 1.2-kb SUC2 promoter region were amplified using KOD plus polymerase (Toyobo) with the primer sets ABA2pro_fw (5′-CACCTATCATCAATTCATCATGTAAACAATAA-3′) and ABA2pro_rev (5′-AATAGCTTTAGCTCCTTAGATCTTCTTT-3′), AAO3pro_fw (5′-CACCTTGAAAGTGATAAACAACTTACATAGTG-3′) and AAO3pro_rev (5′-CAGAATTTTTCCAATTATAAGGTTAGAT-3′), and SUC2pro_fw2 (5′-CACCTTCATATTAATTTCACACACCAAGTTAC-3′) and SUC2pro_rev3 (5′-ATTTGACAAACCAAGAAAGTAAGA-3′), respectively. Each promoter region was cloned into the pENTR/D-TOPO vector (Invitrogen). The AtABCG25 promoter clone was described previously (Kuromori et al., 2010). Each clone was integrated into the nuclear-localized signal-attaching GFP vector pBGGN (Inplanta Inovations Inc.). Additionally, the SUC2 promoter clone was integrated into the CFP vector pHGC (Kubo et al., 2005). An open reading frame clone of the NCED3 (At3g14440) gene was amplified from the Arabidopsis (Arabidopsis thaliana) Columbia genome temperate with the primer set NCED3_TOPO_F2 (5′-CACCATGGCTTCTTTCACGGCAA-3′) and NCED3_TOPO_RS (5′-TCACACGACCTGCTTCGCCAAATCAT-3′) and cloned into the pENTR/D-TOPO vector (Invitrogen). To generate a plasmid of SUC2 promoter-driven NCED3, NCED3 open reading frame blunt fragments digested with NotI and AscI were inserted into the AscI site blunted just after the SUC2 promoter region in the SUC2 promoter–nGFP vector.
Plant Growth and Observations
Each constructed vector was electroporated into Agrobacterium spp. GV3101 for introduction into Arabidopsis by floral dipping of an Agrobacterium-mediated transformation system. In the T2 transgenic plants, fluorescent observation was performed using a confocal laser scanning microscope (Carl Zeiss) in accordance with the manufacturer’s instructions. We observed similar fluorescent patterns in more than four independent transgenic lines of each vector. Furthermore, after cross pollination between T2 plants, plants of the next generation were used for subsequent confocal observation of the dual fluorescent proteins. For ABA treatment, seedlings of 1-week-old transgenic plants were soaked in 10 μm ABA solution for 20 h.
Plants were germinated and grown on 0.5× Murashige and Skoog medium containing 1% (w/v) Suc and 0.8% (w/v) agar in a growth chamber or soil under well-watered conditions at 22°C ± 2°C and 60–70% relative humidity under a 16-h-light/8-h-dark cycle. Thermal images were captured using an infrared thermography device (FLIR). After taking thermal images, each transgenic plant expressing SUC2 promoter-driven NCED3 was confirmed to contain vector insertion based on PCR genotyping with the primer set 5′-AAGTGTCTTTGAGAATCGAACG-3′ and 5′-TGGAGTCATACAGGACCCTATC-3′.
Transport Assay Using Sf9 Insect Culture Cells
AtABCG25 was expressed in Sf9 cells using a baculovirus expression system (Invitrogen) as described previously (Kuromori et al., 2010). Sf9 cells (1 × 106 cells mL−1) were infected with the P3 virus (1/100 v/v) and cultured in liquid culture medium, Sf-900 III SFM (Gibco) with 4% (v/v) fetal bovine serum (Gibco), 100 units mL−1 penicillin, and 100 μg mL−1 streptomycin in a shaking incubator at 100 rpm and 28°C for 48 h. Cells were collected by centrifugation at 1,000 rpm for 5 min, washed with culture medium two times, and finally, resuspended at a final concentration of 25 mg mL−1. Isotope solution of [3H]ABA (GE Healthcare), [3H]indolylacetic acid (Perkin Elmer), or [3H]jasmonic acid (American Radiolabeled Chemicals, Inc.) was added (1/1,000 v/v), and the solution was incubated at room temperature for 16 min. In total, 100 μL each sample was passed through a 0.45-μm membrane filter (Millipore), and the filter was washed with 2 mL liquid culture medium. The radioactivity retained on the filter was determined using a liquid scintillation counter (ALOKA).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure S1. Confocal images of vascular tissues of leaves.
Supplementary Material
Acknowledgments
We thank RIKEN Center for Sustainable Resource Science for transgenic and sequencing support and Drs. Misato Ohtani and Taku Demura for use of the vectors.
Glossary
- ABA
abscisic acid
Footnotes
This work was supported by The Ministry of Education, Culture, Sports, Science, and Technology Japan (grant no. 24570063 to T.K.) and The Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (to K.S.).
The online version of this article contains Web-only data.
References
- Bauer H, Ache P, Lautner S, Fromm J, Hartung W, Al-Rasheid KA, Sonnewald S, Sonnewald U, Kneitz S, Lachmann N, et al. (2013) The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr Biol 23: 53–57 [DOI] [PubMed] [Google Scholar]
- Beck CB. (2010) The Phloem. An Introduction to Plant Structure and Development, Ed 2 Cambridge University Press, Cambridge, United Kingdom, pp 222–246 [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Chen ZH, Hills A, Bätz U, Amtmann A, Lew VL, Blatt MR. (2012) Systems dynamic modeling of the stomatal guard cell predicts emergent behaviors in transport, signaling, and volume control. Plant Physiol 159: 1235–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Endo A, Sawada Y, Takahashi H, Okamoto M, Ikegami K, Koiwai H, Seo M, Toyomasu T, Mitsuhashi W, Shinozaki K, et al. (2008) Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol 147: 1984–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LM, Zhao Z, Assmann SM. (2004) Guard cells: a dynamic signaling model. Curr Opin Plant Biol 7: 537–546 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Rock CD. (2002) Abscisic acid biosynthesis and response. Arabidopsis Book 1: e0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C, Varotto S. (2012) The role of PIN auxin efflux carriers in polar auxin transport and accumulation and their effect on shaping maize development. Mol Plant 5: 787–798 [DOI] [PubMed] [Google Scholar]
- Gaspar T, Kevers C, Faivre-Rampant O, Crèvecoeur M, Penel CL, Greppin H, Dommes J. (2003) Changing concepts in plant hormone action. In Vitro Cell Dev Biol Plant 39: 85–106 [Google Scholar]
- Hetherington AM. (2001) Guard cell signaling. Cell 107: 711–714 [DOI] [PubMed] [Google Scholar]
- Hills A, Chen ZH, Amtmann A, Blatt MR, Lew VL. (2012) OnGuard, a computational platform for quantitative kinetic modeling of guard cell physiology. Plant Physiol 159: 1026–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N. (1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Joshi-Saha A, Valon C, Leung J. (2011) A brand new START: abscisic acid perception and transduction in the guard cell. Sci Signal 4: re4. [DOI] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y. (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107: 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M. (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109: 9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwai H, Nakaminami K, Seo M, Mitsuhashi W, Toyomasu T, Koshiba T. (2004) Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol 134: 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K. (2010) ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA 107: 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Shinozaki K. (2010) ABA transport factors found in Arabidopsis ABC transporters. Plant Signal Behav 5: 1124–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Normanly J. (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol 2: a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Tanaka Y, Abrams SR, Kamiya Y, Seki M, Nambara E. (2009) High humidity induces abscisic acid 8′-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis. Plant Physiol 149: 825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka KJ, Turgeon R. (1999) Sieve elements and companion cells-traffic control centers of the phloem. Plant Cell 11: 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JA. (2003) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol 131: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Koshiba T. (2011) Transport of ABA from the site of biosynthesis to the site of action. J Plant Res 124: 501–507 [DOI] [PubMed] [Google Scholar]
- Stadler R, Sauer N. (1996) The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot Acta 109: 299–306 [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51: 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Hirayama T, Kuromori T, Shinozaki K. (2011) The regulatory networks of plant responses to abscisic acid. Adv Bot Res 57: 201–248 [Google Scholar]
- Weyers JDB, Paterson NW. (2001) Plant hormones and the control of physiological processes. New Phytol 152: 375–407 [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.