Intracellular transport of processing bodies along actin involves an evolutionarily conserved mechanism of direct interaction between myosins and the processing body core P1.
Abstract
Processing (P)-bodies are cytoplasmic RNA protein aggregates responsible for the storage, degradation, and quality control of translationally repressed messenger RNAs in eukaryotic cells. In mammals, P-body-related RNA and protein exchanges are actomyosin dependent, whereas P-body movement requires intact microtubules. In contrast, in plants, P-body motility is actin based. In this study, we show the direct interaction of the P-body core component DECAPPING PROTEIN1 (DCP1) with the tails of different unconventional myosins in Arabidopsis (Arabidopsis thaliana). By performing coexpression studies with AtDCP1, dominant-negative myosin fragments, as well as functional full-length myosin XI-K, the association of P-bodies and myosins was analyzed in detail. Finally, the combination of mutant analyses and characterization of P-body movement patterns showed that myosin XI-K is essential for fast and directed P-body transport. Together, our data indicate that P-body movement in plants is governed by myosin XI members through direct binding to AtDCP1 rather than through an adapter protein, as known for membrane-coated organelles. Interspecies and intraspecies interaction approaches with mammalian and yeast protein homologs suggest that this mechanism is evolutionarily conserved among eukaryotes.
Newly synthesized mRNAs are processed and transported from the nucleus into the cytoplasm. Their subsequent fate depends on a regulatory balance between cytoplasmic translation and mRNA degradation. Both processes are modulated by mRNA-binding proteins, which influence the translational efficiency, stability, and localization of mRNA molecules. Over the last decade, it became apparent that messenger ribonucleoproteins that are translationally repressed and associated with the cytoplasmic 5′ to 3′ decapping machinery tend to assemble into a newly discovered class of highly dynamic mRNA granules, termed processing (P)-bodies (Sheth and Parker, 2003; Anderson and Kedersha, 2006; Parker and Sheth, 2007; Franks and Lykke-Andersen, 2008). P-bodies were initially identified as discrete cytoplasmic punctae to which the mouse 5′ to 3′ exonuclease localizes (Bashkirov et al., 1997). Since then, they have been established as major cytoplasmic sites for the storage of translationally repressed mRNAs, 5′ to 3′ mRNA degradation, quality control, and microRNA-dependent gene silencing (Dunckley and Parker, 1999; Sheth and Parker, 2003; Cougot et al., 2004; Sen and Blau, 2005).
Although their complete composition is still not known, a set of proteins related to these different functions has been identified and shown to colocalize with each other in these cytoplasmic foci (Ingelfinger et al., 2002; van Dijk et al., 2002; Sheth and Parker, 2003; Eulalio et al., 2007; Parker and Sheth, 2007). P-bodies are commonly described as highly mobile and dynamic structures (Yang et al., 2004; Aizer et al., 2008; Loschi et al., 2009). In Drosophila melanogaster and mammals, they show a microtubule-dependent pattern of motility (Aizer et al., 2008; Loschi et al., 2009), which is inhibited by drugs interfering with the microtubule cytoskeleton (Aizer et al., 2008). Actin, by contrast, is important for the delivery of RNA and P-body components to P-bodies and for their assembly. This is suggested by the colocalization of mammalian myosin Va with the P-body-associated protein eIF4E (for Eukaryotic Translation Initiation Factor4E) and further by the reduction of P-body number and transport of eIF4E to P-bodies when expressing dominant-negative myosin Va fragments (Lindsay and McCaffrey, 2011). An association of myosins with P-bodies was also reported in yeast. In yeast (Saccharomyces cerevisiae), Myo2p was successfully used to immunoprecipitate complex ribonucleoprotein particles, including hundreds of mRNAs and some P-body components, such as DECAPPING PROTEIN1 (Dcp1p), SM-LIKE PROTEIN1, DNA TOPOISOMERASE 2-ASSOCIATED PROTEIN1, DExD/H-BOX HELICASE1, and EXORIBONUCLEASE1 (Chang et al., 2008).
In plants, P-body movement appears to be based on the actin cytoskeleton (Hamada et al., 2012), although myosins have not been implicated in P-body movement. In Arabidopsis (Arabidopsis thaliana), the myosin family contains 17 proteins (Reddy and Day, 2001; Peremyslov et al., 2011), which are grouped into two classes, XI and VIII. Members of the class VIII myosins are associated with endosomal structures, the endoplasmic reticulum, and plasmodesmata (Avisar et al., 2008a; Golomb et al., 2008; Sattarzadeh et al., 2008). The 13 class XI myosins show a domain structure that is similar to those of fungi and animal myosin V proteins. They are composed of an N-terminal motor domain, for both actin association as well as ATP hydrolysis (Kinkema et al., 1994; Tominaga et al., 2003), an internal neck domain, comprising a stretch of six calmodulin-binding IQ motifs (Kinkema and Schiefelbein, 1994), and the C-terminal tail region, which contains a coiled-coil domain of variable extent and a bipartite globular cargo-binding domain (Pashkova et al., 2006; Li and Nebenführ, 2007). The Arabidopsis XI myosins have been implicated in the movement of various membranous organelles and vesicles (for review, see Sparkes, 2010, 2011). Colocalization studies and the analysis of movement in myosin mutants revealed that plant XI myosins are involved in the rapid trafficking of Golgi stacks, mitochondria, and peroxisomes (Avisar et al., 2008b, 2009; Peremyslov et al., 2008, 2010; Prokhnevsky et al., 2008), the long-distance transport of endomembrane vesicles (Peremyslov et al., 2012), plant-specific transport vesicles (Peremyslov et al., 2013), motility of plastids (Natesan et al., 2009; Sattarzadeh et al., 2009), as well as endoplasmic reticulum flow (Peremyslov et al., 2010; Ueda et al., 2010) and remodeling (Sparkes et al., 2009).
In the light of recent data available for the mammalian and yeast systems, myosins appear to be good candidates to also mediate the movement of membraneless RNA granules, such as P-bodies. The molecular basis linking P-bodies to the actin cytoskeleton is not known in any system. In this study, we demonstrate that the P-body core component DECAPPING PROTEIN1 (DCP1) physically interacts with myosins in mammals, yeast, and Arabidopsis and also in interspecies experiments. In support of a direct interaction between myosins and P-bodies, Arabidopsis DCP1 (AtDCP1) colocalizes with the full-size myosin XI-K. Overexpression of dominant-negative myosin tail domains inhibited P-body movement efficiently. The analysis of the respective mutants revealed that in two xi-k alleles, P-body movement was almost completely abolished. This defective motility was efficiently restored in triple myosin knockout plants expressing full-size myosin XI-K. Taken together, these data demonstrate that P-body movement is governed by unconventional myosins and that their anchoring to myosins occurs by direct binding to a core component rather than by connecting to specific adapter proteins, as known for eukaryotic organelle transport.
RESULTS
AtDCP1 Interacts with Myosin XI Tails
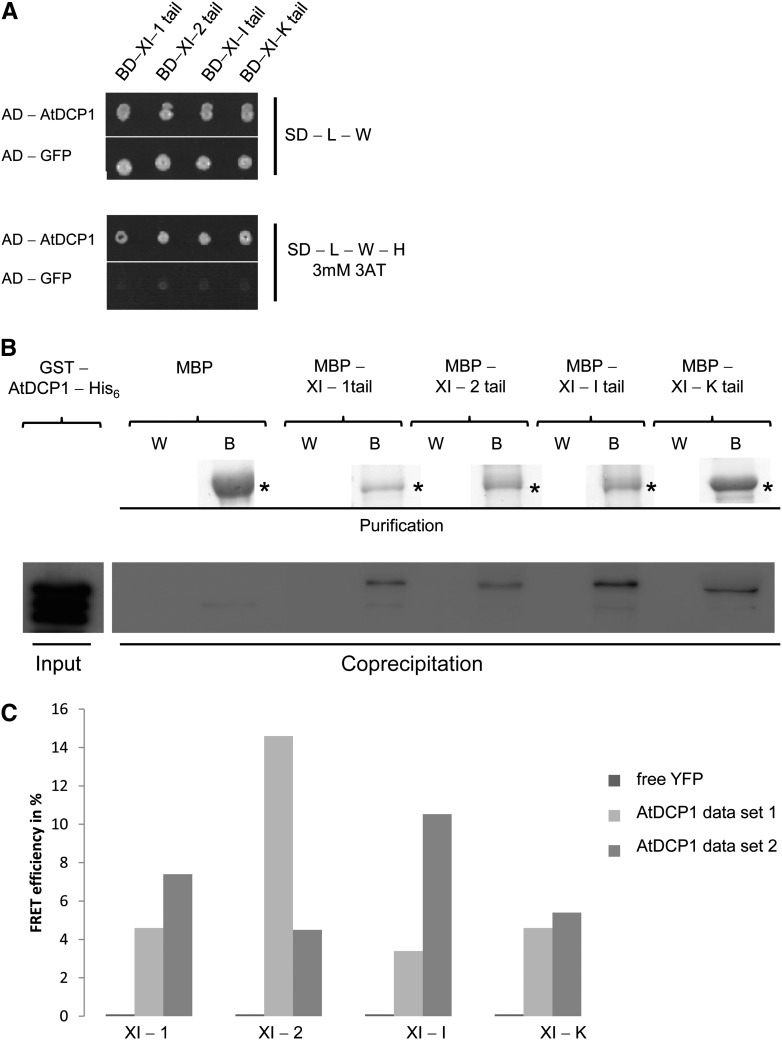
In this study, we focused our analysis on four class XI myosins, XI-1, XI-2, XI-I, and XI-K, which were previously identified to be entirely responsible for the movement of different organelles during sporophyte development in Arabidopsis (Peremyslov et al., 2010). As myosins bind their cargos through their tail domains, these have been widely and successfully used to study myosin-cargo interactions (Pashkova et al., 2005; Li and Nebenführ, 2007; Reisen and Hanson, 2007; Golomb et al., 2008; Avisar et al., 2009; Sattarzadeh et al., 2009; Wang et al., 2012; Peremyslov et al., 2013). Therefore, this strategy was also adopted in our study. The tail domains of XI-1, XI-2, XI-I, and XI-K, lacking their motor and neck domains, were fused to the GAL4 Binding Domain (GAL4-BD), and a GAL4 Activation Domain (GAL4-AD) was fused to AtDCP1. Strikingly, we found an interaction between the prey protein GAL4-AD-AtDCP1 and all four myosin tails investigated as bait constructs (Fig. 1A). As AtDCP1 exhibits strong autoactivation as a GAL4-BD fusion protein, we could not test the reciprocal combinations. This first interaction assay was confirmed by two independent approaches. We performed in vitro coprecipitation assays using bacterially expressed proteins. The Maltose Binding Protein (MBP)-tagged myosin tails were bound to amylose resin and incubated with the cell lysates of Escherichia coli expressing glutathione S-transferase (GST)- and His6-tagged AtDCP1 fusion protein. Coprecipitation of GST-His6-tagged AtDCP1 with the XI-1, XI-2, XI-I, and XI-K tails was confirmed by anti-His antibody staining on a western blot (Fig. 1B; Supplemental Fig. S1A). To provide independent support for these interactions, we used fluorescence resonance energy transfer (FRET) assays that have been successfully applied before for the tail of MyoVb (Roland et al., 2007). As FRET measurements with mobile proteins are error prone, we developed an in vitro FRET assay to measure FRET efficiencies photometrically. AtDCP1-cyan fluorescent protein (CFP) and -yellow fluorescent protein (YFP) fusions of the myosin tails were separately expressed in Arabidopsis cell suspension cultures. Lysates of transfected cells were combined using the YFP and CFP fluorescence intensities to adjust the same ratios between the donor protein AtDCP1-CFP and different YFP-labeled myosin tails as acceptor proteins. FRET was measured photometrically by the acceptor photobleaching method. We could not detect any energy transfer between AtDCP1 and free YFP as a negative control. Although the actual FRET efficiencies between AtDCP1-CFP and the tails of YFP-labeled XI-1, XI-2, XI-I, and XI-K were different in the two independent experiments, both data sets clearly showed interactions between all myosins and AtDCP1 (Fig. 1C). The specificity of our results was furthermore supported by detecting FRET efficiencies not higher than 0.5% between free CFP and YFP fusions of the different myosin tails (Supplemental Fig. S1B). Negative FRET efficiencies of −1.8% and −1.6% between free CFP and YFP fusions in the cases of XI-2 and XI-I tails, respectively, are likely due to protein degradation that we observed over time.
Figure 1.
The P-body protein AtDCP1 interacts directly with class XI myosins in Arabidopsis. A, Yeast two-hybrid interaction between AtDCP1 and myosin XI tails. Top, Double-transformed yeast cells on selective dropout (SD) medium lacking Leu (−L) and Trp (−W). Bottom, Interaction between AtDCP1 N-terminally fused to the GAL4-AD and the tails of XI-1, XI-2, XI-I, and XI-K, N-terminally fused to the GAL4-BD, on selective dropout medium lacking Leu, Trp, and His (−H), supplemented with 3 mm 3-aminotriazole (3AT). GFP N-terminally fused to the GAL4-AD was included as a negative control. B, Coprecipitation of AtDCP1 with all myosin XI tails investigated. Input of GST-AtDCP1-His6 (67 kD; 0.18 µg) per reaction was detected by anti-His6 antibody staining. Resin-bound MBP fusions (called B for beads) are shown by Coomassie Blue-stained gels. Coprecipitations of GST-AtDCP1-His6 with the tails of XI-1 (*, approximately 106 kD; 1.6 µg), XI-2 (*, approximately 104 kD; 1.7 µg), XI-I (*, approximately 105 kD; 2.0 µg), and XI-K (*, approximately 100 kD; 2.5 µg) were detected by anti-His6 antibody staining. The wash fraction (W) and bead fraction are presented. No coprecipitation was observed between MBP (*, approximately 42 kD; 2.8 µg) and GST-AtDCP1-His6. C, FRET efficiencies between CFP-tagged AtDCP1 and the YFP-tagged tails of XI-1, XI-2, XI-I, and XI-K in lysates of Arabidopsis cell suspension culture. No FRET could be detected for the negative control AtDCP1-CFP and free YFP. FRET efficiencies from two independent experiments are shown and represent the trend of at least four biological replicates per sample.
The Interaction between Myosins and DCP1 Is Conserved in Mammals and Yeast
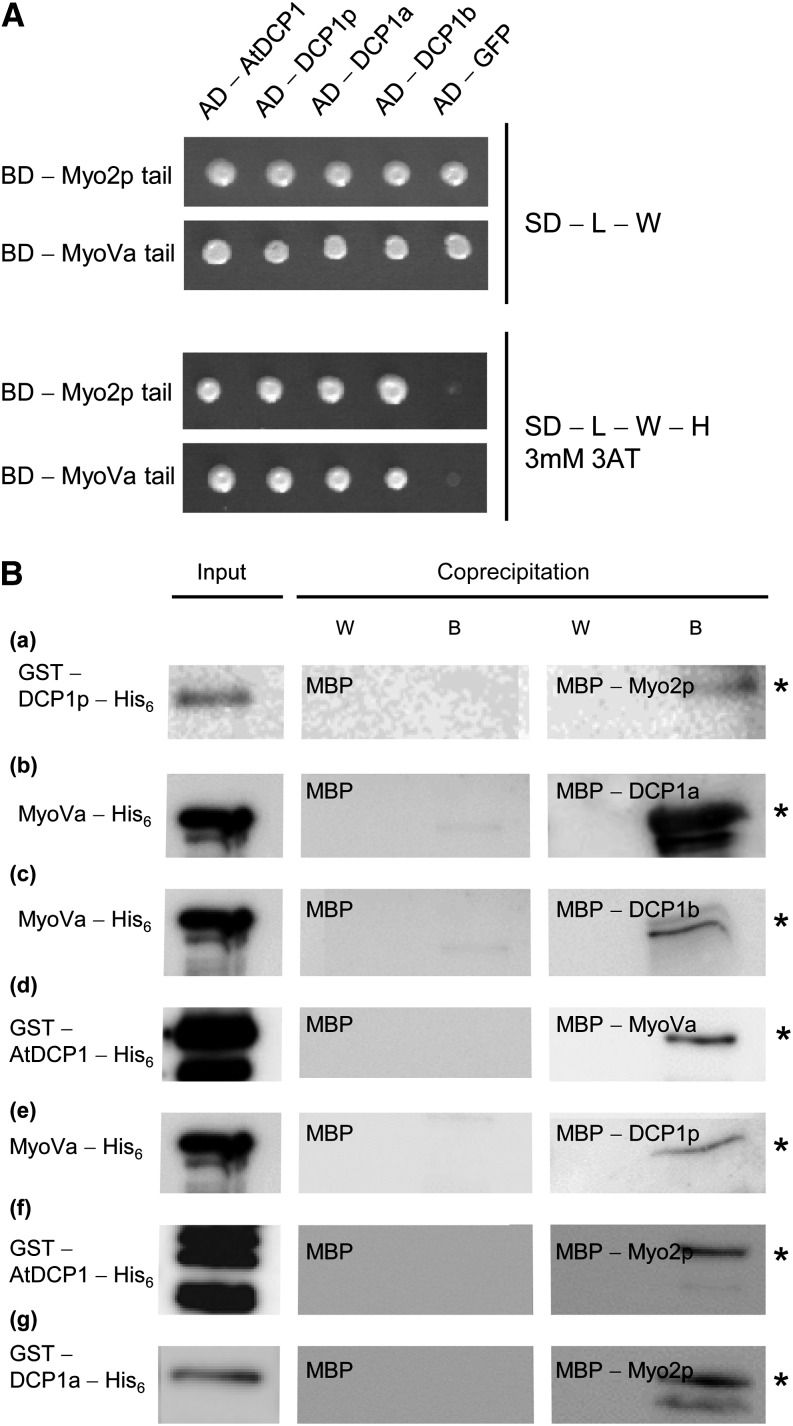
The finding that DCP1 interacts with myosins in Arabidopsis prompted us to test whether this interaction is conserved in other species. Toward this end, we tested the interaction of the yeast Myo2p and the mammalian MyoVa with the respective DCP1 proteins. We found direct interactions between yeast Myo2p and DCP1p and between mouse MyoVa and both human DCP1 homologs in yeast two-hybrid assays and in coprecipitation experiments using bacterially expressed proteins. In addition, we also found interspecies interactions between yeast Myo2p and mouse MyoVa with human, yeast, and Arabidopsis DCP1 (Table I; Fig. 2). Our data indicate that the physical interactions between myosins and DCP1 are evolutionarily conserved.
Table I. Interspecies and intraspecies interactions between DCP1 from Arabidopsis, yeast, and human with yeast and mammalian myosin tails.
Corresponding yeast two-hybrid (Y2H) and coprecipitation (Co-P) data are presented in Figure 2. Positive interactions are indicated by +, and no interaction is indicated by −.
| Interaction Partner/Origin | AtDCP1: Arabidopsis |
DCP1p: Yeast |
DCP1a: Human |
DCP1b: Human |
||||
|---|---|---|---|---|---|---|---|---|
| Y2H | Co-P | Y2H | Co-P | Y2H | Co-P | Y2H | Co-P | |
| Myo2p tail (yeast) | + | + | + | + | + | + | + | − |
| MyoVa tail (Mus musculus) | + | + | + | + | + | + | + | + |
Figure 2.
Interspecies and intraspecies interactions between DCP1 and myosins (related to Table I). A, Yeast two-hybrid interactions. Top, Double-transformed yeast cells on selective dropout (SD) medium lacking Leu (−L) and Trp (−W). Bottom, Interaction between the Arabidopsis AtDCP1, yeast DCP1p, and human DCP1a and DCP1b N-terminally fused to the GAL4-AD and the tails of yeast Myo2p and mouse MyoVa, N-terminally fused to the GAL4-BD, on selective dropout medium lacking Leu, Trp, and His (−H), supplemented with 3 mm 3-aminotriazole (3AT). GFP N-terminally fused to the GAL4-AD was included as a negative control. B, Coprecipitations. a, Input of GST-DCP1p-His6 (left gel; *, approximately 56 kD; 0.18 µg), no coprecipitation of GST-DCP1p-His6 with MBP (7.5 µg) as a negative control (other gels) and coprecipitation with MBP-Myo2p (6.9 µg) detected by anti-His6 antibody staining. b, Input of MyoVa-His6 (left gel; *, approximately 84 kD; 2.9 µg), no coprecipitation of MyoVa-His6 with MBP (7.5 µg) as a negative control (other gels) and coprecipitation with MBP-DCP1a (4.2 µg) detected by anti-His6 antibody staining. c, Input of MyoVa-His6 (left gel; *, approximately 84 kD; 2.9 µg), no coprecipitation of MyoVa-His6 with MBP (7.5 µg) as a negative control (other gels) and coprecipitation with MBP-DCP1b (4.4 µg) detected by anti-His6 antibody staining. d, Input of GST-AtDCP1-His6 (left gel; *, approximately 68 kD; 2.3 µg), no coprecipitation of GST-AtDCP1-His6 with MBP (7.5 µg) as a negative control (other gels) and coprecipitation with MBP-MyoVa (4.4 µg) detected by anti-His6 antibody staining. e, Input of MyoVa-His6 (left gel; *, approximately 84 kD; 2.9 µg), no coprecipitation of MyoVa-His6 with MBP (7.5 µg) as a negative control (other gels) and coprecipitation with MBP-DCP1p (4.3 µg) detected by anti-His6 antibody staining. f, Input of GST-AtDCP1-His6 (left gel; *, approximately 68 kD; 2.3 µg), no coprecipitation of GST-AtDCP1-His6 with MBP (7.5 µg) as a negative control (other gels) and coprecipitation with MBP-Myo2p (6.9 µg) detected by anti-His6 antibody staining. g, Input of GST-DCP1a-His6 (left gel; *, approximately 93 kD; 2.9 µL), no coprecipitation of GST-DCP1a-His6 with MBP (7.5 µg) as a negative control (other gels) and coprecipitation with MBP-Myo2p (6.9 µg) detected by anti-His6 antibody staining. W, Last wash fraction; B, boiled beads.
AtDCP1 Interacts with Class XI Myosin Tails at P-Bodies
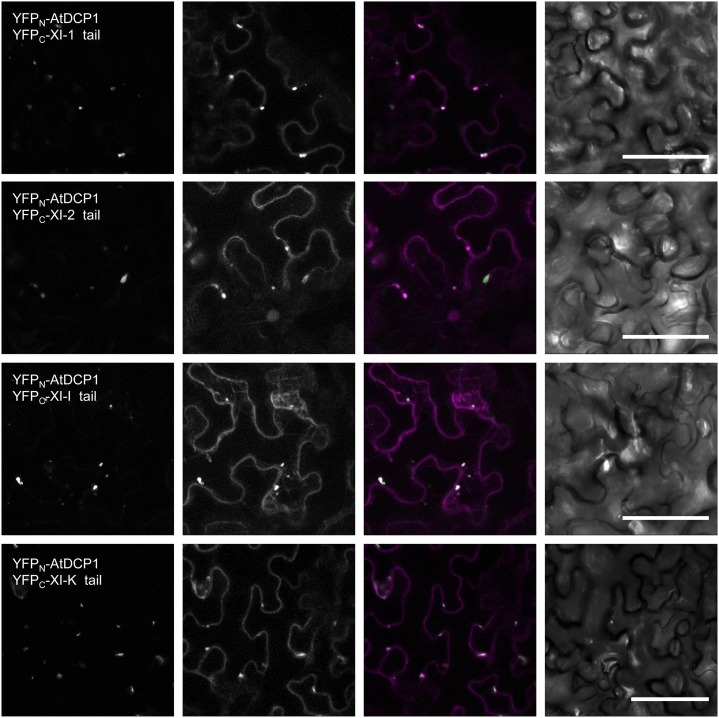
To localize the interaction between AtDCP1 and the different class XI myosin tails in planta, we performed bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana leaf epidermal cells. BiFC interactions were found at mobile dots that colocalized almost completely with AtDCP2, another well-established marker for P-bodies (Xu et al., 2006). This demonstrates that the interactions between AtDCP1 and the tails of XI-1, XI-2, XI-I, and XI-K occur at P-bodies (Fig. 3; Tables II and III). RabD1 was used as a positive control to show BiFC interactions with XI-1 (Supplemental Fig. S2) and as a negative control for AtDCP1, revealing the specificity of AtDCP1’s interactions with the myosin tails. The N-terminal part of YFP was included as a negative control for the myosin interactions (Supplemental Fig. S3; Tables II and III).
Figure 3.
BiFC interaction of AtDCP1 and myosin class XI tails at P-bodies. Interaction is shown for myosin tails and AtDCP1 (column I) at AtDCP2-labeled P-bodies in transiently transformed N. benthamiana leaves 48 h posttransfection. Column II shows AtDCP2-mCherry-labeled P-bodies, column III shows the overlay between the images in column I (green) and column II (magenta), and column IV shows the corresponding transmission images. The interaction between RabD1 and the tails of XI-1 was included as a positive control (Supplemental Fig. S2). Plasmids encoding the N-terminal half of YFP alone were included as negative controls for the myosin tails fused to the C-terminal residues of YFP. RabD1 fused to the C-terminal residues of YFP was used as a negative control for AtDCP1 fused to the N-terminal residues of YFP (Supplemental Fig. S3; Tables II and III). Bars = 50 µm.
Table II. Statistical analysis of BiFC studies, negative controls (related to Fig. 3 and Supplemental Figs. S2 and S3).
| Variables | YFPN-AtDCP1 and YFPC-RabD1 | YFPC-XI-1 Tail and YFPN | YFPC-MYA2 no coil and YFPN | YFPC-XI-I Tail and YFPN | YFPC-XI-K Tail and YFPN |
|---|---|---|---|---|---|
| No. of cells | 60 | 65 | 55 | 65 | 60 |
| Protein accumulations greater than 20-µm diameter (%) | 3 | 0 | 2 | 0 | 0 |
| Cytoplasmic signal (%) | 0 | 2 | 0 | 2 | 0 |
| Cytoplasmic dots (%) | 0 | 0 | 0 | 0 | 0 |
Table III. Statistical analysis of BiFC studies, positive controls and experimental samples (related to Fig. 3 and Supplemental Figs. S2 and S3).
| Variables | YFPN-RabD1 and YFPC-XI-1 Tail | YFPN-AtDCP1 and YFPC-XI-1 Tail | YFPN-AtDCP1 and YFPN-XI-2 Tail | YFPN-AtDCP1 and YFPC-XI-I Tail | YFPN-AtDCP1 and YFPC-XI-K Tail |
|---|---|---|---|---|---|
| No. of cells | 30 | 30 | 30 | 30 | 30 |
| Protein accumulations greater than 20-µm diameter (%) | 0 | 0 | 0 | 0 | 0 |
| Cytoplasmic signal (%) | 0 | 0 | 0 | 0 | 0 |
| Cytoplasmic dots (%) | 30 | 30 | 28 | 29 | 29 |
| Mander’s coefficient between BiFC signals and AtDCP2 labeled P-bodies (%) | ND | 94.2 ± 6.1 | 92.48 ± 7.6 | 95.61 ± 3.4 | 92.93 ± 4.08 |
ND, Not determined.
P-Bodies Show Distinct Motility Patterns
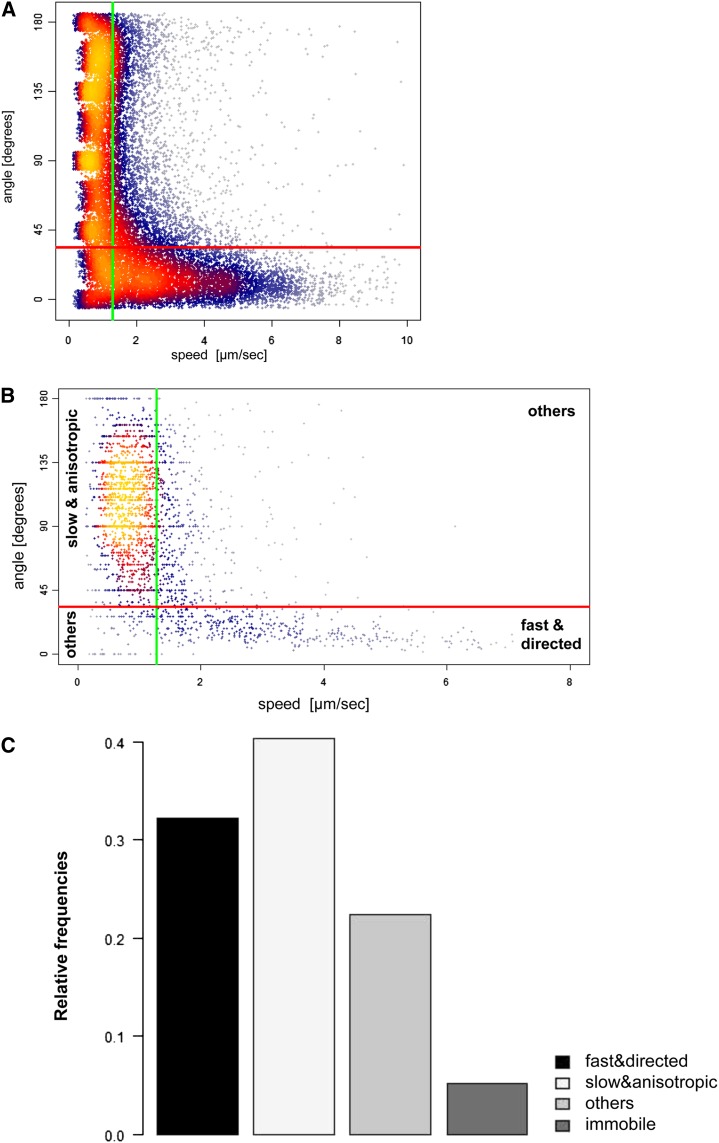
The direct interaction between class XI myosin tails and AtDCP1 at P-bodies strongly suggested a function of myosins in P-body movement. Therefore, we characterized the movement of P-bodies in the elongated Arabidopsis leaf midvein epidermal cells using fluorescent tag fusions of AtDCP1 in transient expression assays (Supplemental Movie S1). The tracking of each P-body yielded a trajectory, a sequence of single steps recorded by speed and angle of movement relative to the preceding movement (for details, see “Materials and Methods”). We focused our analysis on the motility pattern by considering only steps in which movement occurred (Fig. 4A). Steps were classified into fast (respectively slow) and directed (respectively anisotropic) moves. We found a strong association between fast and directed moves and between slow and anisotropic moves (odds ratio of 10, P < 10−10 in a Fisher test). The median values of all steps were used as the thresholds for the classification of the whole trajectories of P-bodies and thereby to define P-body motility classes (Fig. 4B; see “Materials and Methods”). This strategy led to the definition of four classes: immobile, fast and directed, slow and anisotropic, and “other” movers (Fig. 4C). Interestingly, the path of fast and directed P-bodies often showed spontaneous turns of the P-bodies into the opposite direction. About 9% of all steps from this P-body class were backtracking moves, as defined by a change of directionality of more than 150° (Supplemental Fig. S4B).
Figure 4.
P-body movement in Arabidopsis. A, Scatterplot of the speed (x axis) and angle (y axis) values of all single steps. The density of dots is indicated by color, with blue dots representing low densities and yellow dots representing high densities. The vertical green line indicates the median speed value of all single steps (1.3 μm s−1). The horizontal red line indicates the median angle of all single steps (34°). B, Scatterplot of the median speed (x axis) versus the median angle (y axis) of P-body trajectories. The vertical green line indicates the median speed of all single steps (1.3 μm s−1) used to classify P-bodies as fast or slow movers. The horizontal red line indicates the median angle of all single steps (34°) used to classify P-bodies as directed or anisotropic movers. The three movement classes fast and directed, slow and anisotropic, and others are indicated. C, Distribution of P-body motility classes in Col-0.
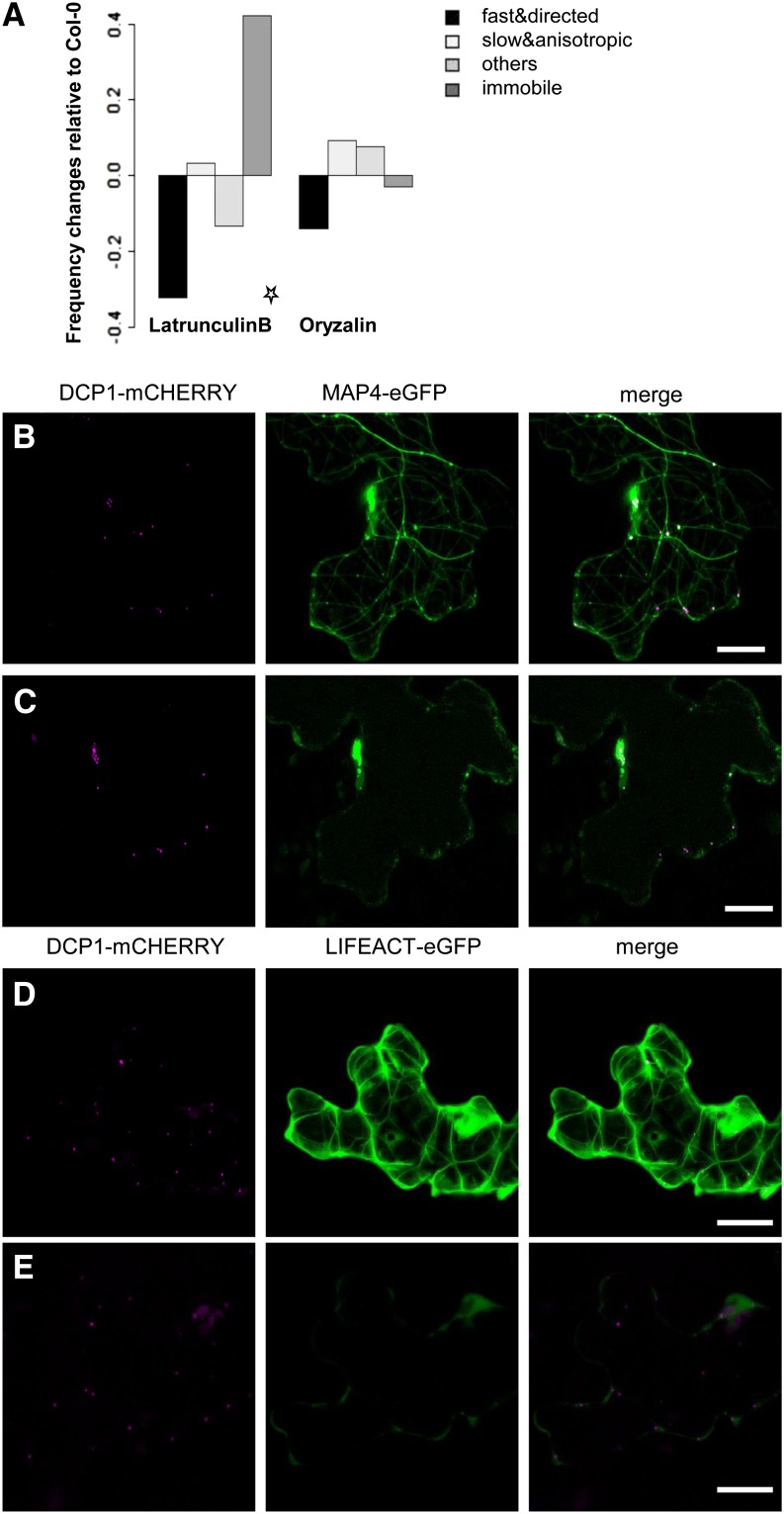
The recent finding that P-body movement in Arabidopsis requires an intact actin cytoskeleton (Hamada et al., 2012) raised the question of whether all types of movement are actin dependent. The role of microtubules in P-body movement was studied by treatment with the microtubule depolymerization agent oryzalin in Columbia-0 (Col-0) leaves cotransfected with AtDCP1-mCherry and MICROTUBULE-ASSOCIATED PROTEIN4 (MAP4)-eGFP (Fig. 5, A–C; Supplemental Fig. S5, A and B; Supplemental Movies S2–S5). In contrast to oryzalin treatments, the application of the actin depolymerization drug latrunculin B inhibited the movement of P-bodies efficiently in cells expressing the actin marker Lifeact-eGFP and AtDCP1-mCherry (Fig. 5, A, D, and E; Supplemental Fig. S5, C and D; Supplemental Movies S6–S9). A χ2 test (significance level 1% after Bonferroni multiple testing correction) revealed no significant differences in the motility classes after oryzalin treatment and significant changes after latrunculin B treatment. Consistent with previous (Hamada et al., 2012) and our own drug inhibitor experiments, we additionally observed close associations between transiently expressed CFP fusions of AtDCP1 and the actin cytoskeleton in living plants expressing the F-actin marker Lifeact-eGFP (Era et al., 2009; Fig. 6A).
Figure 5.
P-body movement is actin dependent. A, Frequency changes of P-body motility classes relative to Col-0 after drug treatment with latrunculin B (left) and oryzalin (right) in wild-type leaf midvein cells expressing the P-body marker AtDCP1-YFP. The star denotes a condition resulting in a significant change in P-body motility class frequency relative to Col-0 (χ2 test, significance level 1% after Bonferroni multiple testing correction). B, Transient coexpression of AtDCP1-mCherry (magenta) and MAP4-eGFP (green) in wild-type Col-0 before treatment with 40 µm oryzalin. C, The same cell as in B after treatment (Supplemental Fig. S5, A and B). D, Transient expression of AtDCP1-mCherry (magenta) in Lifeact-eGFP (green) plants before treatment with 20 µm latrunculin B. E, The same cell as in D after treatment (Supplemental Fig. S5, C and D). Bar = 10 µm.
Figure 6.
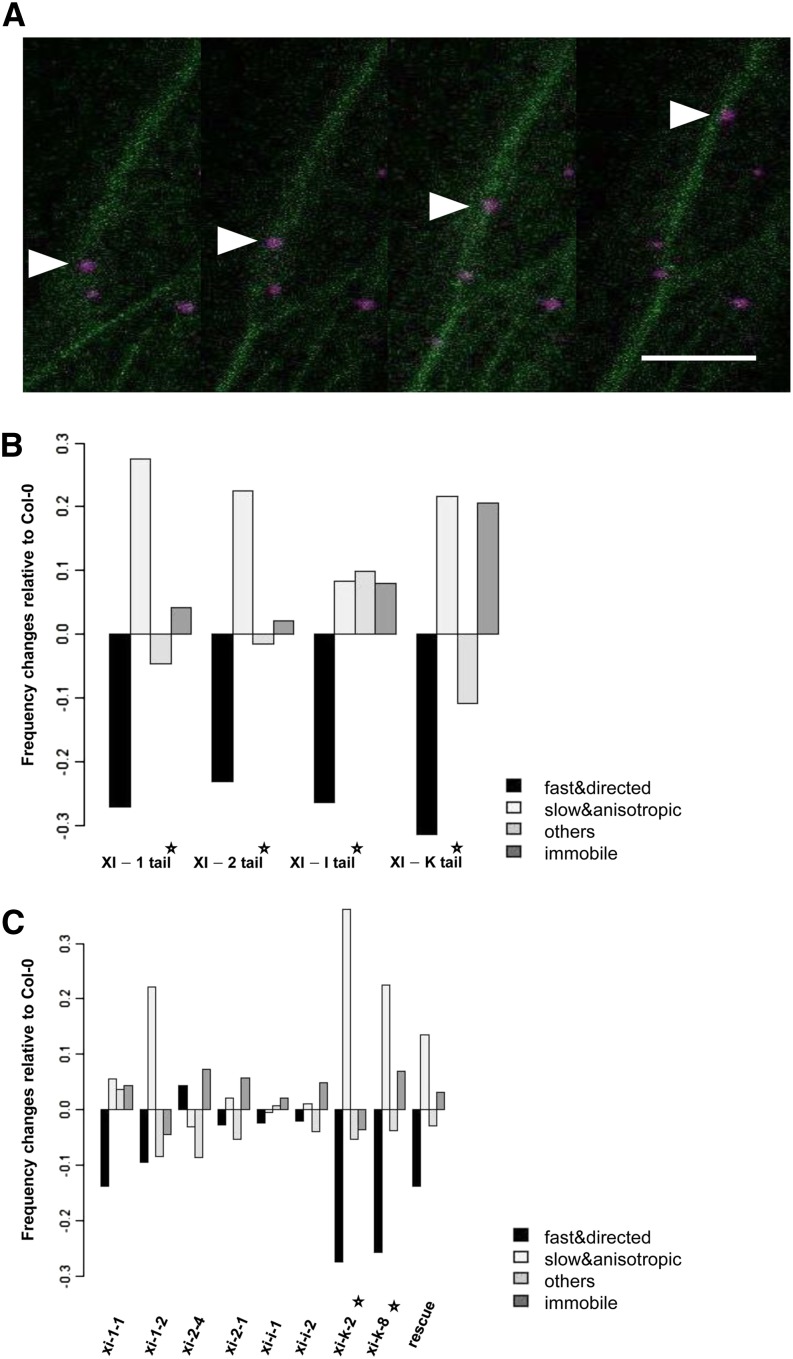
The movement of AtDCP1-CFP requires an intact actomyosin system. A, Movement of one AtDCP1-CFP-labeled P-body (arrowheads) along an actin filament in stably transformed Col-0 plants expressing the F-actin marker Lifeact-eGFP. Time-lapse images were taken at 3-s intervals using laser scanning confocal microscopy. Bar = 12 µm. B, Dominant-negative effect of myosin tail domains on AtDCP1-labeled P-body movement (related to Supplemental Movies S10–S13). Frequency changes of P-body motility classes are shown in cells coexpressing AtDCP1-CFP in combination with a fluorescence-tagged myosin tail relative to AtDCP1-CFP-expressing cells. C, P-body motility class frequency changes in myosin single mutants and the xi-1 xi-2 xi-k triple mutant carrying YFP-tagged full-size myosin XI-K (rescue line) relative to Col-0 (related to Supplemental Fig. S7). Stars denote experimental setups resulting in significant changes in P-body motility class frequencies relative to Col-0 (χ2 test, significance level 1% after Bonferroni multiple testing correction).
P-Body Movement Is Inhibited by Coexpression of Myosin Tails
Myosin tail fragments are known to exert a dominant negative effect on the motility of their putative cargo in plant cells (Prokhnevsky et al., 2008; Sparkes et al., 2008; Avisar et al., 2009; Sattarzadeh et al., 2013). To investigate whether they also inhibit the mobility of AtDCP1-labeled P-bodies, we compared the P-body motility class frequency in AtDCP1-expressing cells with P-body movement in cells coexpressing AtDCP1 and one of the myosin tails under consideration (Fig. 6B; Supplemental Movies S10–S13). Overexpression of the tails from XI-1, XI-2, XI-I, and XI-K resulted in a strong reduction of fast and directional moving P-bodies (χ2 test, significance level 1% after Bonferroni multiple testing correction). In the case of XI-1, XI-2, and XI-K, this motility reduction was accompanied by a significant increase of slow and anisotropic P-bodies (χ2 test, significance level 1% after Bonferroni multiple testing correction). The coexpression of the tail from XI-K led to a significant increase of nonmoving P-bodies (χ2 test, significance level 1% after Bonferroni multiple testing correction).
Analysis of P-Body Movement in Myosin Mutants
The reduced P-body mobility due to coexpressed myosin XI truncations pointed to a possible competition between exogenous and endogenous myosins for cargo binding, which would result in an uncoupling of P-bodies from actin filaments. To test this hypothesis, we studied P-body movements in two mutant alleles of each investigated myosin: xi-1-1 and xi-1-2 (for the myosin XI-1 gene), xi-2-1 and xi-2-4 (for the myosin XI-2 gene), xi-i-1 and xi-i-2 (for the myosin XI-I gene), and xi-k-2 and xi-k-8 (for the myosin XI-K gene; Supplemental Table S1). YFP fusions of AtDCP1 were transiently expressed in leaves of homozygous mutants (Fig. 6C; Supplemental Fig. S6). In comparison with the wild type, both xi-k alleles but none of the other myosin mutants studied showed a significant decrease of the fast and directional moving P-bodies (χ2 test, significance level 1% after Bonferroni multiple testing correction). Additionally, the class of slow and anisotropic moving P-bodies was significantly increased (Supplemental Fig. S7, A and B). The mutant analysis suggests that the fast and directed P-body transport is mediated predominantly by myosin XI-K. Given that all four tested myosin tail domains bind to AtDCP1, it is likely that the dominant negative inhibition of P-body movement by the other three myosins is caused by a competition for binding sites. It is possible that they act partially redundantly with XI-K and that their contribution was not seen in the single mutants.
AtDCP1 Colocalizes with the Full-Length Myosin XI-K
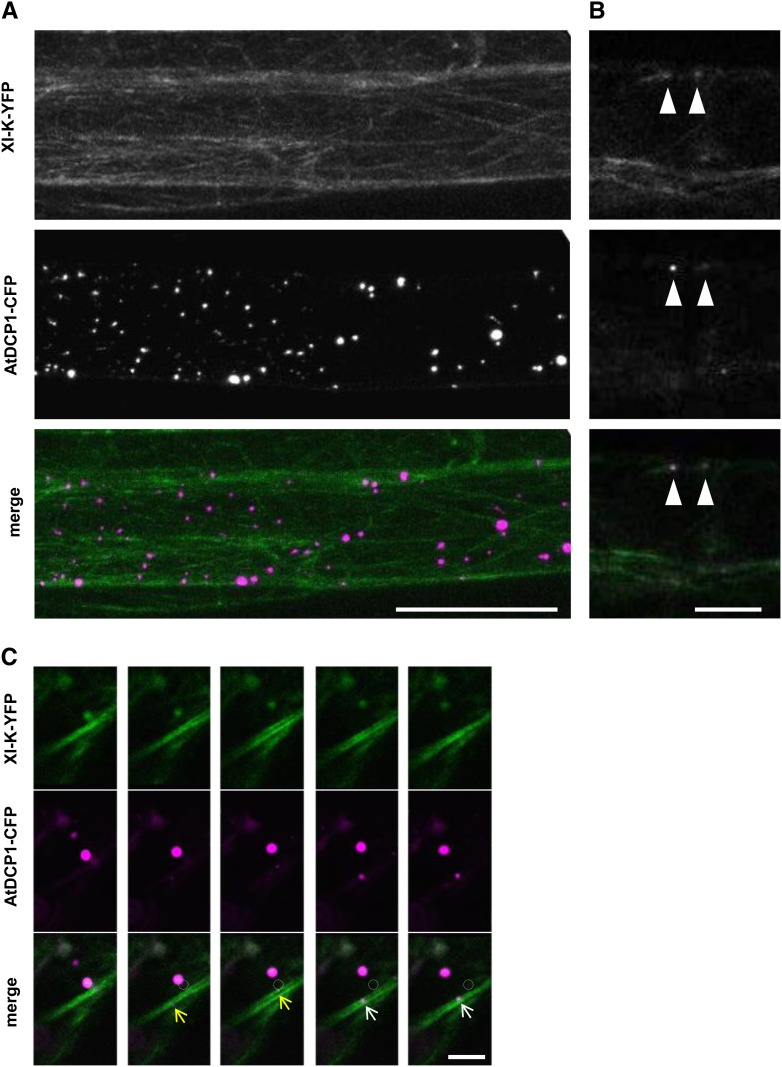
To independently confirm that myosin XI-K is the primary myosin for the fast and directed P-body movement, we analyzed transiently transformed AtDCP1-CFP in leaf epidermal cells of the myosin xi-1 xi-2 xi-k triple mutant carrying the genomic full-length myosin XI-K fused to YFP driven by its native promoter, which was shown to rescue the morphological phenotype of the triple mutant (Peremyslov et al., 2012). As reported before, full-length myosin XI-K broadly labeled the actin cytoskeleton. Occasionally, we observed full-length myosin XI-K-labeled dot-like structures. The analysis of AtDCP1-labeled P-bodies in the xi-1 xi-2 xi-k triple mutant carrying the genomic full-length myosin XI-K-YFP line revealed a rescue of their motility (Fig. 6C; Supplemental Fig. S7, A and B). This indicates that myosin XI-K is essential for fast and directed P-body movement and sufficient to overcome at least the requirement of XI-1 and XI-2 in Arabidopsis (Supplemental Movie S14). In these experiments, AtDCP1-CFP dots exhibited a close association with myosin XI-K-YFP-labeled F-actin-like structures (Fig. 7A). A meaningful colocalization analysis of AtDCP1-CFP and XI-K-YFP was only possible at thin actin strands with clearly visible XI-K-YFP dots. There, we reliably found myosin colabeling with AtDCP1-CFP (Fig. 7B). About 20% to 30% of the AtDCP1-CFP-labeled P-bodies were associated with distinct actin strands and showed no dots for XI-K-YFP. As the XI-K-YFP signal was generally very weak, we cannot decide whether XI-K-YFP dots are present but below the detection limit or AtDCP1-CFP does not exhibit colocalization in these cases.
Figure 7.
Colocalization studies of AtDCP1 and full-size myosin XI-K. A, Transiently expressed AtDCP1-CFP (magenta) in leaf epidermal cells of the xi-1 xi-2 xi-k mutant carrying YFP-tagged full-size myosin XI-K (green). Bar = 50 µm. B, XI-K (green)-labeled dot-like structures that colocalize with AtDCP1-CFP (magenta)-labeled P-bodies (arrowheads) in the xi-1 xi-2 xi-k triple mutant carrying full-size myosin XI-K-YFP. Bar = 10 µm. C, Comovement between transiently expressed AtDCP1-CFP and full-size myosin XI-K-YFP in Arabidopsis leaf epidermis cells (related to Supplemental Movie S15). Comovement of an AtDCP1-CFP (magenta)-labeled P-body and an XI-K-YFP (green)-labeled dot structure in transiently transfected Arabidopsis leaf epidermal cells is shown (circles denote the starting point of movement). The movements of two different P-bodies running along an XI-K-YFP-labeled thick filament are highlighted by yellow and white arrows. Time-lapse images were taken at 2-s intervals. Bar = 10 µm.
In order to substantiate the colocalizations found in these experiments, we aimed to demonstrate not only colocalization but also comovement. Toward this end, we performed transient expression assays. XI-K-YFP showed a similar intracellular localization pattern to that in the transgenic rescue line expressing the full-size genomic XI-K-YFP. We frequently found AtDCP1-labeled P-bodies moving in regions of XI-K-YFP-labeled actin (Fig. 7C, arrows). In cases where we found AtDCP1-labeled dots at distinct thin actin strands, we could observe a clear colocalization and comovement of AtDCP1 and XI-K-YFP, indicating that XI-K-YFP colocalizes with AtDCP1-labeled P-bodies (Fig. 7C; Supplemental Movie S15).
DISCUSSION
Our findings that myosin tail domains physically interact with the respective DCP1 in mammals, yeast, as well as plants, and the further observation that class XI myosins are required for P-body transport in Arabidopsis, shed new light on the molecular mechanism underlying P-body transport. In contrast to organelles, in which membrane-associated adapter proteins provide the link to motor molecules, P-bodies are ill-defined RNA/protein agglomerations that appear to have a set of core proteins in common (Eulalio et al., 2007; Beckham and Parker, 2008). Our data suggest that one of these core components, DCP1, acts as a direct anchor for motor molecules.
What Are the Cargos of Plant Myosins?
The rapid trafficking of various intracellular compartments depends on the function of the sporophytically expressed class XI myosins (Avisar et al., 2008b; Ueda et al., 2010; Peremyslov et al., 2012). In addition, colocalization experiments revealed associations between the respective compartments and class XI myosins. By using marker proteins for Golgi stacks, peroxisomes, mitochondria, as well as other endomembrane compartments, a partial colocalization of XI-1 and XI-2 truncations with peroxisomes could be observed (Hashimoto et al., 2005; Reisen and Hanson, 2007). A partial overlap with Golgi stacks was reported for subdomains of XI-1 (Reisen and Hanson, 2007) and also for the full-length myosin XI-K (Peremyslov et al., 2012). A direct binding of myosins to any of these compartments through adapter proteins was not shown so far. RabD1 and RabC2A have been identified as XI-2 interactors and, therefore, are good candidates for mediating myosin interactions with cellular compartments. RabC2A localizes to peroxisomes and RabD1 to punctate structures associated with Golgi stacks and the trans-Golgi network (Hashimoto et al., 2005; Pinheiro et al., 2009), suggesting that XI-2 mediates the movement of these compartments through binding to the two Rab proteins. Recently, a new class of plant-specific transport vesicles has been identified and was shown to be directly associated with myosin XI-K via their DUF593 domain-containing receptor MyoB1 (Peremyslov et al., 2013). The authors provide a working model that explains the movement of organelles and other compartments by their transient association to actively transported MyoB vesicles or by following the hydrodynamic flow in the cell. Our data indicate that this model cannot be adopted to explain P-body movement. This is suggested by several findings.
First, like the MyoB proteins (Peremyslov et al., 2013), AtDCP1 acts as an anchor protein mediating the direct interaction between membraneless P-bodies and the main sporophytically expressed class XI myosins. The physical interaction between the tails of the investigated myosin class XI members and the P-body core component AtDCP1 was confirmed by three independent approaches and shown to occur at P-bodies using BiFC assays. In addition, we demonstrate that the full-size myosin XI-K can localize to P-bodies. However, it was not possible to functionally test the role of AtDCP1 in P-body movement, as dcp1 mutants are lethal (Xu et al., 2006). Second, we demonstrate that the regular distribution of P-bodies in the cytoplasm of plant cells is achieved by an active process, namely the movement by different myosin class XI members. The movement of P-bodies seems to be specifically regulated, as we identified two different mobility classes (fast and directional; slow and anisotropic) and a stationary fraction. As the fast and directed P-body transport is significantly reduced solely in myosin xi-k mutants, myosin XI-K seems to be essential for P-body movement. Third, 9% of the fast and directionally moving P-bodies spontaneously change their movement to the opposite direction. In particular, the single steps occur preferentially either in a straight forward or straight backward direction (Supplemental Fig. S4B). This is not consistent with a passive movement. Together, these three lines of argument confirm our conclusion of an active myosin XI-K-powered transport of fast and directed moving P-bodies in Arabidopsis. As myosin XI-K was shown to bind predominantly to MyoB receptors (Peremyslov et al., 2013), it is likely that only a minor fraction of the myosin XI-K proteins bind to P-bodies in vivo, possibly in a competitive manner. The idea that XI-K binds to different cargos in a competitive manner is further supported by recent work showing that plant myosin XI-I not only binds vesicle-anchored MyoB receptors but also the outer nuclear membrane receptors WPP DOMAIN INTERACTING TAIL-ANCHORED PROTEIN1 (WIT1) and WIT2 (Tamura et al., 2013). This behavior is like that in yeast, where Myo2p binds competitively to multiple cargoes (Eves et al., 2012).
DCP1 as a P-Body Anchor for Myosins
At first glance, the molecular mechanism underlying P-body movement differs between animals and plants, in that plants use the actin cytoskeleton (Hamada et al., 2012) whereas animals utilize microtubules (Aizer et al., 2008). In animals and yeast, however, the actomyosin system has been implicated in the assembly and disassembly of P-bodies (Chang et al., 2008; Lindsay and McCaffrey, 2011). Human myosin Va colocalizes with P-body markers and is required for P-body integrity. In addition, myosin Va is suggested to couple P-bodies in collaboration with kinesins to microtubules, as is also reported for other cargo (Huang et al., 1999; Ali et al., 2008; Hodges et al., 2009; Lindsay and McCaffrey, 2011). In yeast, Myo2p associates with P-bodies and has also been implicated in P-body integrity (Chang et al., 2008). It is conceivable that the role of myosins in the assembly/disassembly of P-bodies is due to a function in the transport of mRNA/P-body components to and possibly away from P-bodies. Together with our observation that myosins are essential for P-body motility in plants, it is tempting to speculate that the role of myosins in aspects of P-body function is evolutionarily conserved. This view is supported by our finding of an evolutionarily conserved physical interaction between myosin tails and one evolutionarily highly conserved core component of P-bodies. Given that previous studies have identified only nonconserved interactions for plant myosins (Li and Nebenführ, 2008), it is remarkable that myosins and DCP1 from yeast, mammals, and plants interact not only interspecifically but also intraspecifically. In mammals and yeast, this interaction might serve as an identifier tag to facilitate the short-range transport of P-body components to P-bodies, whereas in plants, the same interaction was adapted to mediate the transport of P-bodies over long distances along actin.
MATERIALS AND METHODS
Plant Material
The Arabidopsis (Arabidopsis thaliana) myosin mutants used in this study are listed in Supplemental Table S1 (Ojangu et al., 2007; Peremyslov et al., 2008; Ueda et al., 2010). Positions of all transfer DNA (T-DNA) insertions were confirmed by sequencing the flanking genomic regions (Supplemental Fig. S6A). Total RNA of homozygous mutants was isolated (Qiagen RNeasy Mini Kit) and DNaseI treated (Thermo Scientific), complementary DNA (cDNA) was synthesized (SuperScript III; Invitrogen), and loss of full-length transcript expression was confirmed by qualitative reverse transcription-PCR with three different primer pairs, amplifying cDNA fragments upstream of, downstream of, and spanning the T-DNA insertion sites (Supplemental Fig. S6).
Plasmids
DNA sequences corresponding to the C-terminal tail regions of myosin XI-1 (AT1G17580), XI-2 (AT5G43900), XI-I (AT4G33200), and XI-K, as well as the genomic sequence of the full-size XI-K (AT5G20490), were described previously (Reisen and Hanson, 2007; Peremyslov et al., 2012). Coding sequences of AtDCP1 (AT1G08370), AtDCP2 (AT5G13570), and XI-2 tail including the coiled-coil region were amplified from Arabidopsis Col-0 cDNA; DCP1p (Q12517) and Myo2p (P19524) from yeast strain AH109; and DCP1a (Q9NPI6) and DCP1b (Q8IZD4) from HEK298 cDNA, cloned into pDONR201 (Invitrogen), and sequenced. Murine MyoVa tail encoding the last C-terminal 701 amino acids of the melanocyte-specific myosin Va (NP_034994) was amplified with primers containing attB sites on melanocyte MyosinVa long tail in mCherry-C1, which has been described before (Chen et al., 2012), recombined in pDONR201, and sequenced. RabD1 (AT3G11730) was amplified with primers containing attB sites on pUNI clone U50900, recombined in pDONR207, and sequenced. A complete overview of primer pairs used for cloning is given in Supplemental Table S2. The following Gateway destination vectors were used for protein expression in planta: pENSG/pEXSG-YFP (Feys et al., 2005) for N-terminal/C-terminal fusions with YFP, pENSG/pEXSG-CFP (Feys et al., 2005) for N-terminal/C-terminal fusions with CFP, pAMARENA (Marc Jakoby; GenBank identifier FR695418) for N-terminal fusions with mCherry, pCL112/pCL113 (J.F. Uhrig, unpublished data) for N-terminal fusion with YFPN/YFPC, and pSPYNE/pSPYCE (Walter et al., 2004) for C-terminal fusions with YFPN/YFPC; for protein expression in yeast: pAS/pACT (Clontech) for N-terminal fusions of the GAL4 binding and activation domains; for protein expression in Escherichia coli: pGEX-2TM-GW (kindly received from Imre Sommsich and Bekir Ülker) for creating fusion proteins with an N-terminal GST tag and a C-terminal His6 tag, pDEST17 (Invitrogen) for N-terminal fusions with a His6-tag, and pETG-40A (EMBL) for N-terminal fusions with an MBP tag.
Yeast Two-Hybrid Assays
The yeast two-hybrid assays, using the yeast strain AH109, were done as described previously (Gietz et al., 1995). Interactions were analyzed by plating double-transformed yeast cells on synthetic dropout interaction medium lacking Leu, Trp, and His supplemented with 3, 5, and 10 mm 3-amino-1,2,3-triazole.
FRET
Dark-grown Arabidopsis cell suspension culture (Landsberg erecta) was transformed as described previously (Berger et al., 2007). After 4 d of incubation, 500-µL cultures were harvested, resuspended in 150 μL of protein extraction buffer (50 mm NaH2PO4/Na2HPO4, pH 7.2, 1 mm EDTA, pH 8, and 0.1% [v/v] Triton X-100), and shaken with glass beads (1.8–2.2 mm; Roth) for 15 min at 1,400 rpm, 8°C in a thermoblock (Eppendorf). After centrifugation (10 min, 15,000 rpm, 4°C), total protein concentrations of the supernatants were determined using the BCA Protein Assay Kit (Thermo Scientific). Fluorescence intensities were measured at the respective wavelengths. Cell lysates were combined with exactly the same ratios between the CFP-tagged donor and the different YFP-tagged acceptor proteins and incubated for 3 h at 4°C on ice. FRET was analyzed in samples containing (1) combined lysates of cells expressing the CFP-tagged donor in combination with free YFP as a negative control, (2) the CFP-tagged donor in combination with a YFP-tagged acceptor, (3) the lysates of untransformed cells as background corrective, and (4) combined lysates of cells expressing free CFP and the YFP-tagged acceptor proteins. FRET efficiency was determined by subtracting the emission spectra of background-corrected CFP fusions (405 nm), pre acceptor bleaching (514 nm), from background-corrected emission spectra of CFP fusions post acceptor bleaching. Emission spectra were detected using the Tecan Infinite 200 Titerplate reader and Tecan i-Control 1.4.5.0 software. FRET could be clearly detected between DCP1-CFP and the different YFP-labeled myosin tails in at least four independent experiments, but strong variations between FRET efficiencies preclude the combination of different data sets. Therefore, we provide data for two representative data sets.
Bacterial Protein Expression and Coprecipitation
MBP-tagged fusion proteins, with MBP as a negative control, as well as His6- or GST/His6-tagged proteins were expressed in E. coli [BL21 (DE3) RIL]. Bacteria were grown in Terrific Broth medium (37°C to an optical density at 600 nm of 0.8), induced by isopropyl-β-d-1-thiogalactopyranoside, and incubated (20°C, 6 h). Cells were harvested, resuspended in HEPES buffer (500 mm HEPES and 500 mm NaCl, pH 8), and sonicated. MBP fusion proteins were purified from cleared bacterial lysates using amylose resin according to the manufacturer’s instructions (New England Biolabs). The indicated protein amounts were determined using calibration series of defined protein amounts on Coomassie Blue-stained SDS gels and subsequent ImageJ analysis of protein band intensities. For coprecipitations, purified MBP fusions immobilized on amylose resin were incubated with lysates of BL21 (DE3) RIL cells expressing GST/His6-tagged or His6-tagged proteins. To perform each coprecipitation with the same amount of available prey protein, the cleared lysates were distributed in equal quantities (500 µL; Figs. 1B and 2B) per bait. The incubation was performed under constant shaking at 4°C for 45 min. Immobilized fractions were washed three times with HEPES buffer. Bound proteins were eluted by boiling in SDS loading buffer and analyzed by Coomassie Blue staining (bait proteins) and immunoblotting using a primary monoclonal anti-His6 antibody from mouse (Qiagen; 1:2,000 dilution) and secondary anti-mouse antibody coupled to horseradish peroxidase (Sigma; 1:10,000 dilution; prey proteins). Detections of target proteins were performed with a Fujifilm LAS3000 using 300 µL of SuperSignal West Femto Substrate (Thermo Scientific) per western blot. Exposure times for each data set presented were the same. The specificities of the primary and secondary antibodies were confirmed on western blots with a negative control (MBP fusion protein; Supplemental Fig. S1A).
Transient Expression in Plants
Transient transformation of Arabidopsis leaves was performed by biolistic transformation (Mathur et al., 2003) and analyzed after 12 to 16 h by confocal laser scanning microscopy. Transformation of Nicotiana benthamiana was carried out by leaf infiltration with Agrobacterium tumefaciens (GV3101 pMP90RK) according to Yang et al. (2000).
Drug Treatment
Oryzalin and latrunculin B treatments were performed on transiently transformed leaves of 14- to 16-d-old Arabidopsis plants, which were cut and incubated at room temperature for 20 min in 20 μm latrunculin B or for up to 90 min in 40 μm oryzalin.
Confocal Laser Scanning Microscopy
Confocal laser scanning microscopy was performed on the Leica DMRE fluorescence microscope equipped with a TCS-SP2 imaging system (Leica Microsystems). Images were collected with a Leica HCxAPO L40x0.8 water-immersion objective. CFP was excited at 405 nm, and emission was detected between 460 and 480 nm. GFP was excited at 488 nm, and emission was detected between 500 and 525 nm. YFP was excited at 514 nm, and emission was detected between 530 and 570 nm. mCherry was excited at 561 nm, and emission was detected between 600 and 635 nm. For BiFC analysis, cells were counted as transfected dependent on the presence of the cotransfected marker protein. To exclude that variation of fluorescence intensities led to erroneous data interpretations and to ensure a comparison between weak and strongly expressing cells, we always used the same laser intensities and exposure times. For colocalization approaches, images were collected by sequential scanning between frames to avoid cross talk between different fluorescently tagged proteins. For comovement studies, images were collected by parallel scanning between lines using an LSM700 fluorescence microscope equipped with an X40EC-Plan Neofluar objective (1.3; oil). Quantification of colocalizations was performed by calculating Mander’s coefficients using the ImageJ JaCoP plugin (Bolte and Cordelières, 2006). Thresholds for background correction were set independently for individual cells.
Analysis of P-Body Movement
P-body movement was analyzed in transiently transformed midvein epidermal cells of rosette leaves of 14- to 18-d-old Arabidopsis plants. Series of 50 consecutive images were obtained by scanning a single plane every 1.6 s by confocal laser scanning microscopy for general P-body motility analyses and every 3 s for drug treatments due to covisualization of the cytoskeleton. P-body trajectories were determined by manual tracking using the ImageJ Manual Tracker plugin (written by Fabrice Cordelires, Institut Curie, Orsay, France).
For each experimental setup, we analyzed time-lapse movies of an average of eight cells. Each experimental set is represented by an average of 162 P-bodies that were tracked for a maximum of 50 frames (Supplemental Fig. S4A). Each P-body is characterized by its trajectory (xt,yt), t = 1,…,T. We first identified immobile P-bodies that did not move (change their coordinates) more than two times. For the analysis of the mobile P-bodies, we removed all time points of a trajectory for which the respective P-body did not move in the preceding interval. The pruned trajectories were converted into a sequence of steps consisting of angles and speeds by calculating the angle between two consecutive moves and the mean step distance of the previous move and the current move divided by the time interval between two frames.
The median speed and median angle values from all single steps were plotted (Fig. 4A). The medians of the single-step speeds and angles were determined and used as thresholds to classify the P-body trajectories that were plotted in Figure 4B. P-bodies were classified into fast (respectively slow) movers if the median speed of its trajectory was larger (respectively smaller) than the median speed of 1.3 µm s−1 (Fig. 4B, green vertical line) of all single-step speeds measured. Analogously, P-bodies were classified as directed (respectively anisotropic) if the median angle of its trajectory was smaller (respectively larger) than the median angle of 34° (Fig. 4B, red horizontal line) of all single-step angles measured.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XI-1 (AT1G17580), XI-2 (AT5G43900), XI-I (AT4G33200), XI-K (AT5G20490), AtDCP1 (AT1G08370), AtDCP2(AT5G13570), RabD1 (AT3G11730) DCP1p (Q12517), Myo2p (P19524),DCP1a (Q9NPI6), DCP1b (Q8IZD4), and MyoVa (NP_034994).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Additional control experiments (related to Figs. 1 and 2).
Supplemental Figure S2. Confocal images of N. benthamiana epidermal leaf cells expressing BIFC-positive controls (related to Fig. 3; Tables II and III).
Supplemental Figure S3. Confocal images of N. benthamiana epidermal leaf cells expressing BIFC-negative controls (related to Fig. 3; Tables II and III).
Supplemental Figure S4. Scatterplot of P-body movement.
Supplemental Figure S5. Tracks of AtDCP1-labeled P-bodies under different conditions in transiently transformed Arabidopsis cells at three different time points (related to Fig. 5).
Supplemental Figure S6. Arabidopsis T-DNA insertion lines (related to Fig. 6C).
Supplemental Figure S7. Median speed and median angle of P-body trajectories.
Supplemental Table S1. Arabidopsis myosin XI mutants used in this study.
Supplemental Table S2. Sequences of primer pairs used for cloning.
Supplemental Movie S1. P-body movement in an Arabidopsis leaf epidermis cell transiently expressing YFP-tagged AtDCP1.
Supplemental Movie S2. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell before oryzalin treatment.
Supplemental Movie S3. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell before oryzalin treatment (same cell presented in Supplemental Movie S2 without manual tracking).
Supplemental Movie S4. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell after oryzalin treatment.
Supplemental Movie S5. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell after oryzalin treatment (same cell presented in Supplemental Movie S4 without manual tracking).
Supplemental Movie S6. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell before latrunculin B treatment.
Supplemental Movie S7. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell before latrunculin B treatment (same cell presented in Supplemental Movie S6 without manual tracking).
Supplemental Movie S8. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell after latrunculin B treatment.
Supplemental Movie S9. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell after latrunculin B treatment (same cell presented in Supplemental Movie S8 without manual tracking).
Supplemental Movie S10. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell coexpressing the tail of XI-1.
Supplemental Movie S11. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell coexpressing the tail of XI-2.
Supplemental Movie S12. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell coexpressing the tail of XI-I.
Supplemental Movie S13. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell coexpressing the tail of XI-K.
Supplemental Movie S14. Movement of AtDCP1-labeled P-bodies in an Arabidopsis leaf epidermis cell in the myosin xi-1 xi-2 xi-k triple mutant rescued with the genomic full-length myosin XI-K fused to YFP (Peremyslov et al., 2012).
Supplemental Movie S15. Comovement of AtDCP1-mCherry and genomic full-length XI-K-YFP in a cotransfected Arabidopsis leaf epidermis cell.
Supplementary Material
Acknowledgments
We thank Valera V. Peremyslov and Valerian V. Dolja for providing full-length gXI-K-YFP and stably transformed xi-k xi-1 xi-2 XI-K-YFP plants, Amirali Sattarzadeh and Maureen Hanson for the generous gift of pENTR clones containing the myosin tails, Jennifer Lippincott-Schwartz for providing the tail of mammalian myosin Va, Joachim Uhrig for the pCL112/pCL113 vectors, Philipp Thomas for providing Liveact-eGFP plants and the corresponding plasmid, and Arijt Das for discussions on P-body movement analysis.
Glossary
- P
processing
- FRET
fluorescence resonance energy transfer
- BiFC
bimolecular fluorescence complementation
- Col-0
Columbia-0
- T-DNA
transfer DNA
- cDNA
complementary DNA
Footnotes
The online version of this article contains Web-only data.
References
- Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. (2008) The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell 19: 4154–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MY, Lu H, Bookwalter CS, Warshaw DM, Trybus KM. (2008) Myosin V and kinesin act as tethers to enhance each others’ processivity. Proc Natl Acad Sci USA 105: 4691–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. (2006) RNA granules. J Cell Biol 172: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Abu-Abied M, Belausov E, Sadot E, Hawes C, Sparkes IA. (2009) A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiol 150: 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Dolja VV. (2008a) Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. J Virol 82: 2836–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV. (2008b) Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol 146: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. (1997) A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol 136: 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham CJ, Parker R. (2008) P bodies, stress granules, and viral life cycles. Cell Host Microbe 3: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Stracke R, Yatusevich R, Weisshaar B, Flügge UI, Gigolashvili T. (2007) A simplified method for the analysis of transcription factor-promoter interactions that allows high-throughput data generation. Plant J 50: 911–916 [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP. (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224: 213–232 [DOI] [PubMed] [Google Scholar]
- Chang W, Zaarour RF, Reck-Peterson S, Rinn J, Singer RH, Snyder M, Novick P, Mooseker MS. (2008) Myo2p, a class V myosin in budding yeast, associates with a large ribonucleic acid-protein complex that contains mRNAs and subunits of the RNA-processing body. RNA 14: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang Y, Zhang J, Deng Y, Jiang L, Song E, Wu XS, Hammer JA, Xu T, Lippincott-Schwartz J. (2012) Rab10 and myosin-Va mediate insulin-stimulated GLUT4 storage vesicle translocation in adipocytes. J Cell Biol 198: 545–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Séraphin B. (2004) Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Parker R. (1999) The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J 18: 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Era A, Tominaga M, Ebine K, Awai C, Saito C, Ishizaki K, Yamato KT, Kohchi T, Nakano A, Ueda T. (2009) Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol 50: 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. (2007) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Eves PT, Jin Y, Brunner M, Weisman LS. (2012) Overlap of cargo binding sites on myosin V coordinates the inheritance of diverse cargoes. J Cell Biol 198: 69–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE. (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. (2008) The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360 [DOI] [PubMed] [Google Scholar]
- Golomb L, Abu-Abied M, Belausov E, Sadot E. (2008) Different subcellular localizations and functions of Arabidopsis myosin VIII. BMC Plant Biol 8: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Tominaga M, Fukaya T, Nakamura M, Nakano A, Watanabe Y, Hashimoto T, Baskin TI. (2012) RNA processing bodies, peroxisomes, Golgi bodies, mitochondria, and endoplasmic reticulum tubule junctions frequently pause at cortical microtubules. Plant Cell Physiol 53: 699–708 [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Igarashi H, Mano S, Nishimura M, Shimmen T, Yokota E. (2005) Peroxisomal localization of a myosin XI isoform in Arabidopsis thaliana. Plant Cell Physiol 46: 782–789 [DOI] [PubMed] [Google Scholar]
- Hodges AR, Bookwalter CS, Krementsova EB, Trybus KM. (2009) A nonprocessive class V myosin drives cargo processively when a kinesin-related protein is a passenger. Curr Biol 19: 2121–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JD, Brady ST, Richards BW, Stenolen D, Resau JH, Copeland NG, Jenkins NA. (1999) Direct interaction of microtubule- and actin-based transport motors. Nature 397: 267–270 [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T. (2002) The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501 [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Schiefelbein J. (1994) A myosin from a higher plant has structural similarities to class V myosins. J Mol Biol 239: 591–597 [DOI] [PubMed] [Google Scholar]
- Kinkema M, Wang H, Schiefelbein J. (1994) Molecular analysis of the myosin gene family in Arabidopsis thaliana. Plant Mol Biol 26: 1139–1153 [DOI] [PubMed] [Google Scholar]
- Li JF, Nebenführ A. (2007) Organelle targeting of myosin XI is mediated by two globular tail subdomains with separate cargo binding sites. J Biol Chem 282: 20593–20602 [DOI] [PubMed] [Google Scholar]
- Li JF, Nebenführ A. (2008) The tail that wags the dog: the globular tail domain defines the function of myosin V/XI. Traffic 9: 290–298 [DOI] [PubMed] [Google Scholar]
- Lindsay AJ, McCaffrey MW. (2011) Myosin Va is required for P body but not stress granule formation. J Biol Chem 286: 11519–11528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M, Leishman CC, Berardone N, Boccaccio GL. (2009) Dynein and kinesin regulate stress-granule and P-body dynamics. J Cell Sci 122: 3973–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kirik V, Kernebeck B, Srinivas BP, Hülskamp M. (2003) Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130: 3137–3146 [DOI] [PubMed] [Google Scholar]
- Natesan SK, Sullivan JA, Gray JC. (2009) Myosin XI is required for actin-associated movement of plastid stromules. Mol Plant 2: 1262–1272 [DOI] [PubMed] [Google Scholar]
- Ojangu EL, Järve K, Paves H, Truve E. (2007) Arabidopsis thaliana myosin XIK is involved in root hair as well as trichome morphogenesis on stems and leaves. Protoplasma 230: 193–202 [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. (2007) P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Pashkova N, Catlett NL, Novak JL, Wu G, Lu R, Cohen RE, Weisman LS. (2005) Myosin V attachment to cargo requires the tight association of two functional subdomains. J Cell Biol 168: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Jin Y, Ramaswamy S, Weisman LS. (2006) Structural basis for myosin V discrimination between distinct cargoes. EMBO J 25: 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Klocko AL, Fowler JE, Dolja VV. (2012) Arabidopsis myosin XI-K localizes to the motile endomembrane vesicles associated with F-actin. Front Plant Sci 3: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Mockler TC, Filichkin SA, Fox SE, Jaiswal P, Makarova KS, Koonin EV, Dolja VV. (2011) Expression, splicing, and evolution of the myosin gene family in plants. Plant Physiol 155: 1191–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Morgun EA, Kurth EG, Makarova KS, Koonin EV, Dolja VV. (2013) Identification of myosin XI receptors in Arabidopsis defines a distinct class of transport vesicles. Plant Cell 25: 3022–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, Avisar D, Dolja VV. (2008) Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol 146: 1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, Dolja VV. (2010) Class XI myosins are required for development, cell expansion, and F-actin organization in Arabidopsis. Plant Cell 22: 1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro H, Samalova M, Geldner N, Chory J, Martinez A, Moore I. (2009) Genetic evidence that the higher plant Rab-D1 and Rab-D2 GTPases exhibit distinct but overlapping interactions in the early secretory pathway. J Cell Sci 122: 3749–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky AI, Peremyslov VV, Dolja VV. (2008) Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc Natl Acad Sci USA 105: 19744–19749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Day IS. (2001) Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen D, Hanson MR. (2007) Association of six YFP-myosin XI-tail fusions with mobile plant cell organelles. BMC Plant Biol 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. (2007) Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell 18: 2828–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattarzadeh A, Franzen R, Schmelzer E. (2008) The Arabidopsis class VIII myosin ATM2 is involved in endocytosis. Cell Motil Cytoskeleton 65: 457–468 [DOI] [PubMed] [Google Scholar]
- Sattarzadeh A, Krahmer J, Germain AD, Hanson MR. (2009) A myosin XI tail domain homologous to the yeast myosin vacuole-binding domain interacts with plastids and stromules in Nicotiana benthamiana. Mol Plant 2: 1351–1358 [DOI] [PubMed] [Google Scholar]
- Sattarzadeh A, Schmelzer E, Hanson MR. (2013) Arabidopsis myosin XI sub-domains homologous to the yeast myo2p organelle inheritance sub-domain target subcellular structures in plant cells. Front Plant Sci 4: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Blau HM. (2005) Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol 7: 633–636 [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I. (2011) Recent advances in understanding plant myosin function: life in the fast lane. Mol Plant 4: 805–812 [DOI] [PubMed] [Google Scholar]
- Sparkes I, Runions J, Hawes C, Griffing L. (2009) Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell 21: 3937–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA. (2010) Motoring around the plant cell: insights from plant myosins. Biochem Soc Trans 38: 833–838 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Teanby NA, Hawes C. (2008) Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. J Exp Bot 59: 2499–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I. (2013) Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr Biol 23: 1776–1781 [DOI] [PubMed] [Google Scholar]
- Tominaga M, Kojima H, Yokota E, Orii H, Nakamori R, Katayama E, Anson M, Shimmen T, Oiwa K. (2003) Higher plant myosin XI moves processively on actin with 35 nm steps at high velocity. EMBO J 22: 1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Yokota E, Kutsuna N, Shimada T, Tamura K, Shimmen T, Hasezawa S, Dolja VV, Hara-Nishimura I. (2010) Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc Natl Acad Sci USA 107: 6894–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Séraphin B. (2002) Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang G, Wang F, Wang G, Wang F, Zhang X, Zhong M, Zhang J, Lin D, Tang Y, Xu Z, et al (2012) Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell 24: 3447–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang JY, Niu QW, Chua NH. (2006) Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18: 3386–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M. (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22: 543–551 [DOI] [PubMed] [Google Scholar]
- Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EK. (2004) GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci 117: 5567–5578 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.