Overexpression of a transcription factor enhances Arabidopsis phosphate uptake by activating transporter expression.
Abstract
The WRKY transcription factor family has more than 70 members in the Arabidopsis (Arabidopsis thaliana) genome, and some of them are involved in plant responses to biotic and abiotic stresses. This study evaluated the role of WRKY45 in regulating phosphate (Pi) uptake in Arabidopsis. WRKY45 was localized in the nucleus and mainly expressed in roots. During Pi starvation, WRKY45 expression was markedly induced, typically in roots. WRKY45 overexpression in Arabidopsis increased Pi content and uptake, while RNA interference suppression of WRKY45 decreased Pi content and uptake. Furthermore, the WRKY45-overexpressing lines were more sensitive to arsenate, the analog of Pi, compared with wild-type seedlings. These results indicate that WRKY45 positively regulates Arabidopsis Pi uptake. Quantitative real-time polymerase chain reaction and β-glucuronidase staining assays showed that PHOSPHATE TRANSPORTER1;1 (PHT1;1) expression was enhanced in the WRKY45-overexpressing lines and slightly repressed in the WRKY45 RNA interference line. Chromatin immunoprecipitation and eclectrophoretic mobility shift assay results indicated that WRKY45 can bind to two W-boxes within the PHT1;1 promoter, confirming the role of WRKY45 in directly up-regulating PHT1;1 expression. The pht1;1 mutant showed decreased Pi content and uptake, and overexpression of PHT1;1 resulted in enhanced Pi content and uptake. Furthermore, the PHT1;1-overexpressing line was much more sensitive to arsenate than WRKY45-overexpressing and wild-type seedlings, indicating that PHT1;1 overexpression can enhance Arabidopsis Pi uptake. Moreover, the enhanced Pi uptake and the increased arsenate sensitivity of the WRKY45-overexpressing line was impaired by pht1;1 (35S:WRKY45-18::pht1;1), demonstrating an epistatic genetic regulation between WRKY45 and PHT1;1. Together, our results demonstrate that WRKY45 is involved in Arabidopsis response to Pi starvation by direct up-regulation of PHT1;1 expression.
Phosphorus is a major essential nutrient for plant growth and development and serves various basic biological functions in plant life cycle (Raghothama, 1999). Phosphate (H2PO4– or, in short, Pi) is the major form that is absorbed and transported into the plant cells (Ullrich-Eberius et al., 1981; Tu et al., 1990). The Pi concentration in the soil, typically 10 μm or less, results in Pi starvation for plant growth and survival, and plants have evolved different strategies to overcome the limited Pi availability. In response to Pi deficiency, plants increase Pi uptake by altering root architecture (López-Bucio et al., 2003; Ticconi and Abel, 2004; Osmont et al., 2007), by altering the expression of Pi-related genes (Bustos et al., 2010), or by changing their metabolic and developmental processes (Raghothama and Karthikeyan, 2005).
Analysis of the Arabidopsis (Arabidopsis thaliana) genome revealed that there are at least nine members of the Pi transporter family (PHOSPHATE TRANSPORTER1 [PHT1] family; Okumura et al., 1998; Mudge et al., 2002). PHT1;1 and PHT1;4, two members of the Arabidopsis PHT1 family, have been demonstrated to be Pi transporters participating in Pi uptake from the soil. Arabidopsis PHT1;1 can complement the yeast (Saccharomyces cerevisiae) mutant pho84 (NS219 [pho3 pho84 ura3]), which lacks the high-affinity Pi transporter Pho84 (Muchhal et al., 1996), and overexpression of the Arabidopsis PHT1;1 gene in tobacco (Nicotiana tabacum) cells increases their Pi uptake capacity (Mitsukawa et al., 1997). PHT1;1 and PHT1;4 are highly expressed in the root epidermis and endoderm and root hairs, where they have been proposed to function in Pi uptake from the soil (Karthikeyan et al., 2002; Mudge et al., 2002), and the double mutant pht1;1Δ4Δ shows a 75% reduction in Pi uptake capacity compared with the wild-type plant (Shin et al., 2004). PHT1;1 can be regulated at transcriptional and/or posttranscriptional levels. The PHT1;1 transcript is the most abundant in the Arabidopsis PHT1 gene family (Mudge et al., 2002), and PHT1;1 expression is clearly induced during Pi starvation (Muchhal et al., 1996; Karthikeyan et al., 2002; Mudge et al., 2002; Shin et al., 2004). Many regulators have been reported to modulate PHT1;1 expression. The transcription of PHT1;1 can be positively regulated by phosphate starvation response1 (PHR1), WRKY75, and sugars and negatively regulated by cytokinin, abscisic acid, myb domain protein62 (MYB62), SPX domain protein3, and actin-related protein6/histone H2A variant Htz1 (Martín et al., 2000; Karthikeyan et al., 2002; Shin et al., 2006; Devaiah et al., 2007a, 2009; Duan et al., 2008; Smith et al., 2010). The phosphate transporter traffic facilitator1 (PHF1) protein, a SEC12-related protein, is necessary for PHT1;1 plasma membrane localization (González et al., 2005). Mutation of PHF1 impairs the localization of PHT1;1 at the plasma membrane, and the phf1 mutant displays a strong reduction in Pi accumulation compared with the wild-type plant (González et al., 2005). PHT1;1 can be phosphorylated (Nühse et al., 2004), and when the Ser-514 residue of PHT1;1 is changed to Asp, the PHT1;1S514D protein accumulates in the endoplasmic reticulum (Bayle et al., 2011).
Transcriptome analysis demonstrated that the expression of many genes, including PHT1 family genes, significantly changes in Oryza sativa (Wasaki et al., 2003) and Arabidopsis (Wu et al., 2003; Misson et al., 2005) in response to Pi starvation, indicating that transcriptional regulation plays important roles in plants’ responses to low-Pi stress. Furthermore, a number of transcription factors have been reported in responses to Pi starvation, such as AtPHR1 (Rubio et al., 2001), rice Pi starvation-induced transcription factor1 (Yi et al., 2005), AtWRKY75 (Devaiah et al., 2007a), zinc finger of Arabidopsis thaliana6 (AtZAT6; Devaiah et al., 2007b), AtMYB62 (Devaiah et al., 2009), and AtWRKY6 (Chen et al., 2009). WRKY proteins are plant-specific transcription factors and are characterized by the presence of one or two highly conserved WRKY domains (Eulgem et al., 2000; Maeo et al., 2001; Zhang and Wang, 2005). The WRKY domain contains the conserved amino acid sequence motif WRKYGQK, followed by a Cys2His2 or Cys2HisCys zinc finger motif, and both conserved motifs of WRKY domain are necessary for the binding affinity of the WRKY protein to the consensus cis-acting element W-box (C/T)TGAC(C/T; Eulgem et al., 2000; Maeo et al., 2001; Zhang and Wang, 2005). Several WRKY proteins, such as AtWRKY75 and AtWRKY6, have been reported to play important roles in plant responses to Pi starvation (Devaiah et al., 2007a; Chen et al., 2009).
In this study, the function of WRKY45 in Arabidopsis responses to Pi starvation was investigated. We demonstrated that WRKY45 expression was highly induced during Pi deprivation, and overexpression of WRKY45 enhanced Pi uptake and increased arsenate sensitivity. WRKY45 positively regulated PHT1;1 expression by binding to the W-boxes within the PHT1;1 promoter, and overexpression of PHT1;1 improved plant Pi uptake. The pht1;1 mutant suppressed the enhanced Pi uptake and the increased arsenate sensitivity caused by WRKY45 overexpression, demonstrating that PHT1;1 is epistatic to WRKY45. In conclusion, our results demonstrate that WRKY45 is a novel transcription factor that regulates Pi uptake by modulating PHT1;1 expression in Arabidopsis.
RESULTS
WRKY45 Is a Pi Starvation-Responsive Transcription Factor
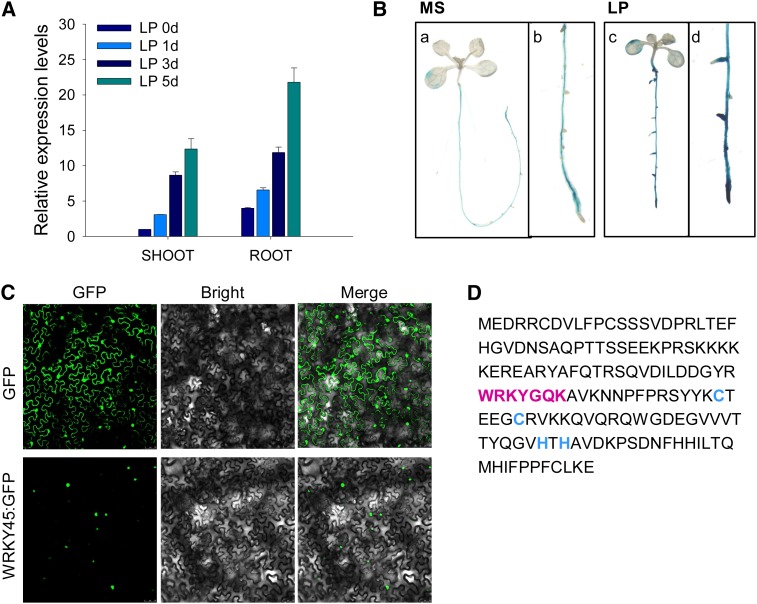
WRKY proteins are plant-specific transcription factors, and two of them, AtWRKY75 (Devaiah et al., 2007a) and AtWRKY6 (Chen et al., 2009), have been reported to participate in Arabidopsis Pi homeostasis. It has been hypothesized that other WRKY proteins may also play roles in plant responses to Pi starvation. To test this hypothesis, we analyzed the expression of WRKY genes in Arabidopsis during Pi starvation. Quantitative real-time (qRT) PCR analysis showed that WRKY45 was mainly expressed in the roots (Fig. 1A), and when wild-type Arabidopsis seedlings were challenged with Pi starvation, the WRKY45 was significantly induced, typically in the roots (Fig. 1A).
Figure 1.
WRKY45 responds to Pi starvation and is localized in the nucleus. A, qRT PCR analysis of WRKY45 expression. Seven-day-old wild-type seedlings were transferred to LP medium, and then the shoots and roots were harvested separately at the indicated time. The data represent the mean values of three replicates ± se. B, GUS staining assay of the ProWRKY45:GUS transgenic line. Seven-day-old ProWRKY45:GUS seedlings were transferred to MS (a and b) or LP (c and d) medium for 5 d and then stained. Details of the roots of the ProWRKY45:GUS transgenic seedlings are shown in b and d. C, Subcellular localization of the WRKY45:GFP fusion protein in the N. benthamiana leaf. The expression of GFP alone was used as the control. D, Deduced amino acid sequence of WRKY45 showing the highly conserved WRKY domain WRKYGQR and the novel C2H2 zinc finger motif in magenta and blue letters, respectively.
To further confirm the expression pattern of WRKY45, homozygous single-copy ProWRKY45:GUS transgenic lines were generated. Seven-day-old ProWRKY45:GUS seedlings were transferred to Murashige and Skoog (MS) medium (Pi-sufficient condition, MS) or MS medium without Pi (Pi-deficient condition, low phosphate medium [LP]) for 5 d and then stained to detect GUS activity. When ProWRKY45:GUS seedlings were grown under Pi-sufficient condition, WRKY45 was mainly expressed in the roots (Fig. 1B, a and b), and when ProWRKY45:GUS seedlings were transferred to Pi-deficient condition, a strong GUS staining appeared in the roots (Fig. 1B, c and d), typically in the root tips (Fig. 1Bd). The qRT PCR and GUS staining results indicate that WRKY45 expression is induced during Pi starvation, mainly in the roots.
To detect subcellular localization of WRKY45 protein, the coding region of WRKY45 was fused with the 3′ end of the GFP reporter gene and expressed under the control of the Super promoter. The GFP gene alone under the control of the Super promoter served as the control. The subcellular localization of WRKY45 was determined in a transient expression system in Nicotiana benthamiana leaves. The WRKY45:GFP fusion protein was exclusively localized in the nucleus (Fig. 1C), indicating that WRKY45 was localized in the nucleus.
WRKY transcription factors typically contain one or more WRKY domains (Eulgem et al., 2000; Maeo et al., 2001; Zhang and Wang, 2005). Amino acid sequence analysis revealed that the deduced amino acid sequence of WRKY45 had a highly conserved WRKYGQK motif and a characteristic Cys2His2 zinc finger motif (Fig. 1D).
Together, these data suggest that WRKY45 is a Pi starvation-responsive transcription factor.
Overexpression of WRKY45 Enhances Arabidopsis Pi Uptake
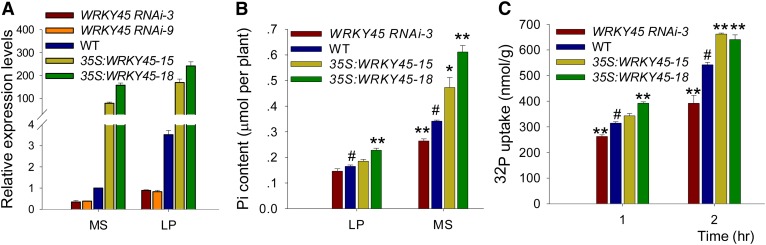
To characterize the role of WRKY45 during Pi starvation, WRKY45-overexpressing (35S:WRKY45) and WRKY45 RNA interference (RNAi) transgenic lines were generated. qRT PCR results showed that under Pi-sufficient (MS) and Pi-deficient (LP) conditions, the WRKY45 expression was significantly elevated in the WRKY45-overexpressing lines and repressed in the WRKY45 RNAi lines compared with that in wild-type seedlings (Fig. 2A). The Pi content of various plants was measured under Pi-sufficient and Pi-deficient conditions. As shown in Figure 2B, the WRKY45-overexpressing lines had higher Pi contents, whereas the WRKY45 RNAi line had a lower Pi content compared with that of wild-type seedlings (Fig. 2B), suggesting that overexpression of WRKY45 enhances Arabidopsis Pi accumulation.
Figure 2.
Overexpression of WRKY45 enhances Arabidopsis Pi accumulation and Pi uptake. A, qRT PCR analysis of WRKY45 expression in the WRKY45 RNAi lines, WRKY45-overexpressing lines, and wild-type plants. Seven-day-old seedlings were transferred to MS or LP medium and then harvested for RNA extraction. The data are the mean values of three replicates ± se. B, Pi content analysis. Seven-day-old seedlings were transferred to MS or LP medium for 10 d and then harvested for Pi content analysis. Data are shown as mean ± se (n = 3). Asterisks indicate statistically significant differences compared with the wild type (Student’s t test, P < 0.05). Wild-type plants were used as a control (#). C, Pi uptake was monitored over a 2-h period in 7-d-old seedlings germinated and grown on MS medium. Data are shown as mean ± se (n = 3). Asterisks indicate statistically significant differences compared with the wild type (Student’s t test, P < 0.05). Wild-type plants were used as a control (#). WT, Wild type.
To analyze Pi uptake rates, 7-d-old seedlings were transferred into a Pi uptake solution containing 50 μm Pi supplemented with 32P orthophosphate, and Pi uptake over a 2-h period was measured. As shown in Figure 2C, the WRKY45-overexpressing lines displayed a substantial increase, whereas the WRKY45 RNAi line showed a reduction in Pi uptake capacity compared with that of the wild-type seedlings. Arsenate is an ion structurally analogous to Pi, which is transported into plant roots via Pi transporters (Asher and Reay, 1979; Shin et al., 2004; Catarecha et al., 2007; Castrillo et al., 2013). Thus, arsenate sensitivity was tested among various plant genotypes. No obvious phenotypic differences were observed among the WRKY45-overexpressing lines, the WRKY45 RNAi line, and wild-type seedlings grown under medium without arsenate (Supplemental Fig. S1A). In the presence of arsenate, compared with wild-type seedlings, the WRKY45-overexpressing lines displayed an arsenate-sensitive phenotype, and the root length of WRKY45-overexpressing lines was significantly shorter than that of wild-type seedlings (Supplemental Fig. S1, A and B), indicating that WRKY45 overexpression enhanced plant arsenate accumulation. No obvious differences were observed between the WRKY45 RNAi line and wild-type seedlings grown on medium with arsenate (Supplemental Fig. S1).
WRKY45 Positively Regulates PHT1;1 Transcription
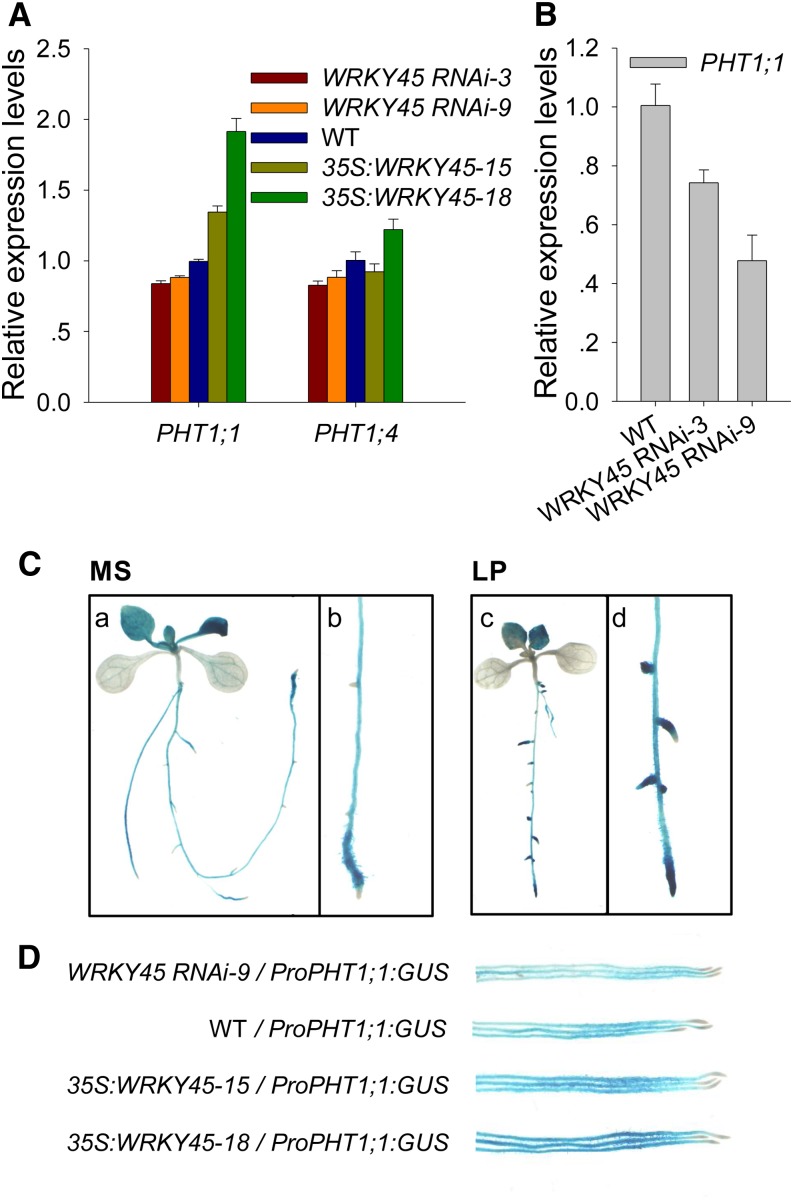
As the WRKY45-overexpressing lines displayed an increase in Pi uptake capacity, we hypothesized that WRKY45 regulates the expression of Pi transporter genes. PHT1;1 and PHT1;4 are the two main Pi transporters for Arabidopsis Pi acquisition from the soil (Shin et al., 2004), which are mainly expressed in the roots (Karthikeyan et al., 2002). Therefore, the expressions of PHT1;1 and PHT1;4 were examined in the roots of the WRKY45-overexpressing lines, the WRKY45 RNAi line, and wild-type plants under Pi-sufficient condition. The transcription of PHT1;1 was elevated in the WRKY45-overexpressing lines and slightly repressed in the WRKY45 RNAi lines compared with that in wild-type roots (Fig. 3A; Supplemental Fig. S2), and there were no obvious differences in PHT1;4 expression among the various plants (Fig. 3A). Then, the expressions of PHT1;1 were detected in the roots of WRKY45 RNAi lines and wild-type seedlings under Pi starvation. The 7-d-old seedlings were transferred to LP medium for 3 d, and then the roots were harvested for RNA extraction. The qRT PCR results showed that the expressions of PHT1;1 in WRKY45 RNAi lines (WRKY45 RNAi-3 and WRKY45 RNAi-9) were obviously lower than that in wild-type plants under Pi starvation (Fig. 3B).
Figure 3.
WRKY45 promotes PHT1;1 expression. A, qRT PCR analysis of PHT1;1 and PHT1;4 expressions in the roots of the WRKY45-overexpressing lines, WRKY45 RNAi lines, and wild-type plants. The plants were germinated and grown on MS medium for 7 d, and then the roots were harvested for RNA extraction. The data are the mean values of three replicates ± se. B, qRT PCR analysis of PHT1;1 expression in the roots of the WRKY45 RNAi lines and wild-type plants. The 7-d-old seedlings were transferred to LP medium for 3 d and then harvested for RNA extraction. The data are the mean values of three replicates ± se. C, GUS staining of the ProPHT1;1:GUS transgenic line. Seven-day-old ProPHT1;1:GUS seedlings were transferred to MS (a and b) or LP (c and d) medium for 5 d and then stained. Details of the roots of the ProPHT1;1:GUS transgenic line are shown in b and d. D, GUS staining showing the expression patterns of PHT1;1 in the WRKY45 RNAi line, WRKY45-overexpression lines, and wild-type plants. The plants were germinated and grown on MS medium for 7 d and then harvested for GUS staining. WT, Wild type.
The GUS staining results showed that PHT1;1 was mainly expressed in the roots of the single-copy ProPHT1;1:GUS transgenic lines (Fig. 3C). Under Pi starvation, PHT1;1 expression was significantly induced, typically in the root tips (Fig. 3C, c and d), indicating that WRKY45 and PHT1;1 had similar expression patterns. In addition, we crossed the ProPHT1;1:GUS line with the WRKY45-overexpressing lines (35S:WRKY45-15 and 35S:WRKY45-18), the WRKY45 RNAi line (WRKY45 RNAi-9), and wild-type plants and obtained the 35S:WRKY45-15/ProPHT1;1:GUS, 35S:WRKY45-18/ProPHT1;1:GUS, WRKY45 RNAi-9/ProPHT1;1:GUS, and wild-type/ProPHT1;1:GUS plants, respectively. Under Pi-sufficient condition, the root GUS staining results showed that the PHT1;1 expression was promoted in the WRKY45-overexpressing lines (35S:WRKY45-15/ProPHT1;1:GUS and 35S:WRKY45-18/ProPHT1;1:GUS) and slightly repressed in the WRKY45 RNAi line (WRKY45 RNAi-9/ProPHT1;1:GUS) compared with that in the wild type (wild type/ProPHT1;1:GUS; Fig. 3D). These data demonstrate that WRKY45 positively regulates PHT1;1 expression.
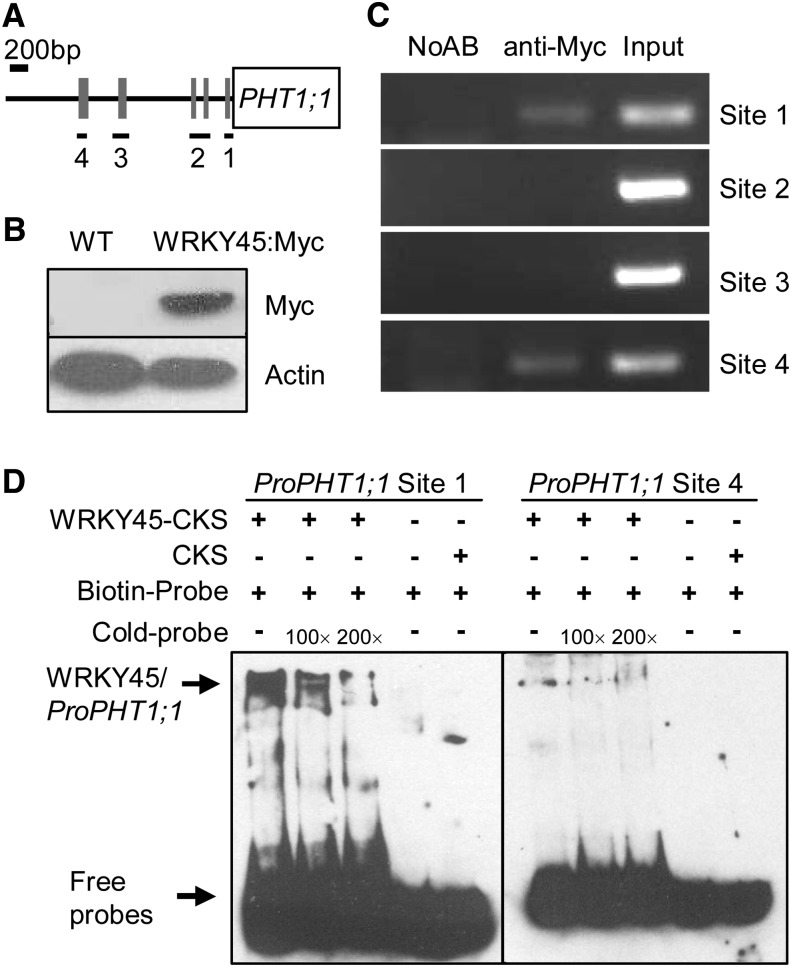
WRKY45 Binds to the PHT1;1 Promoter
WRKY proteins regulate their target genes’ expression by binding to the W-box(es) in their target gene promoters (Rubio et al., 2001; Wu et al., 2003; Misson et al., 2005). There are several W-box motifs within the PHT1;1 promoter (Fig. 4A; Martín et al., 2000). Thus, we further hypothesized that WRKY45 may directly regulate PHT1;1 expression by binding to the W-box(es) of the PHT1;1 promoter. To test this hypothesis, we generated the WRKY45-Myc transgenic lines. The coding region of WRKY45 was fused with the 3′ end of the Myc tag and expressed under the control of the Cauliflower mosaic virus 35S promoter, generating single-copy WRKY45-Myc transgenic lines. Western-blot analysis with anti-Myc showed that the WRKY45-Myc protein was expressed in the WRKY45-Myc line (Fig. 4B). The in vivo interaction between WRKY45 and the W-box motifs within the PHT1;1 promoter was investigated using the chromatin immunoprecipitation (ChIP) method with WRKY45-Myc roots. As shown in Figure 4C, WRKY45 interacted with the PHT1;1 promoter when the primer combinations containing either site 1 or site 4 were applied, while no interaction was observed between WRKY45 and the PHT1;1 promoter containing site 2 or site 3 (Fig. 4C). The eclectrophoretic mobility shift assay (EMSA) was also performed. WRKY45 was expressed in Escherichia coli as a fusion protein with CTP:CMP-3-deoxy-D-mannooctulosonate cytidylyltransferase (CMP-KDO) synthetase (CKS) and affinity purified from the soluble fraction. The WRKY45-CKS fusion protein can bind to the site 1 and site 4 within the PHT1;1 promoter, and the binding was abolished by the addition of increasing amounts of unlabeled competitors with the same sequence (Fig. 4D). By contrast, the CKS protein alone did not show any detectable binding to the PHT1;1 promoter (Fig. 4D). These data demonstrate that WRKY45 can directly bind to two W-boxes within the PHT1;1 promoter.
Figure 4.
WRKY45 binds to the PHT1;1 promoter. A, Diagram of the PHT1;1 promoter region showing the relative positions of the W-boxes. The W-boxes are marked by gray rectangles, and relative positions and sizes of the different PCR-amplified fragments are indicated by black lines under the W-box(es). B, Western-blot analysis was performed with anti-Myc to detect the WRKY45-Myc protein in the WRKY45-Myc transgenic line and wild-type seedlings. C, ChIP analysis to detect the association between WRKY45 and the W-boxes within the PHT1;1 promoter in the WRKY45-Myc transgenic line. The ChIP signals with (anti-Myc) and without (NoAB) addition of anti-Myc are indicated. D, EMSA assay to analyze the binding of WRKY45 to PHT1;1 promoter. Each biotin-labeled DNA probe was incubated with WRKY45-CKS protein. An excess of unlabeled probes was added to compete with labeled promoter sequences. Biotin-labeled probes incubated with CKS protein served as the negative control. WT, Wild type.
Overexpression of PHT1;1 Enhances Arabidopsis Pi Uptake
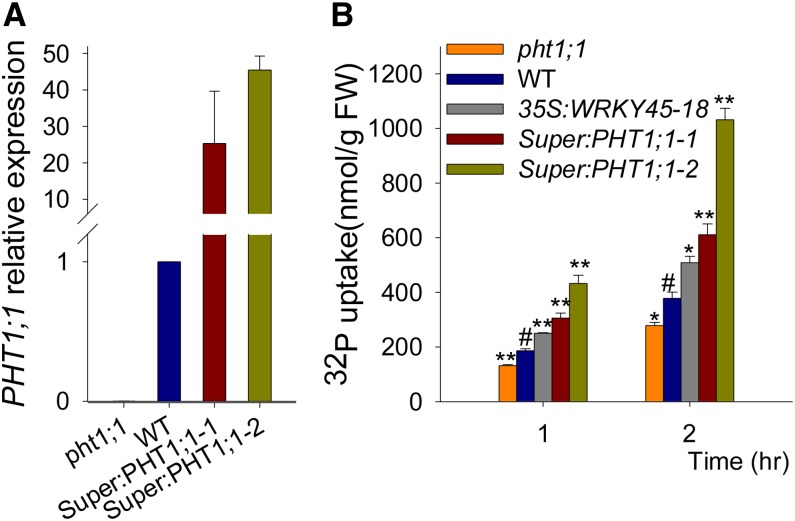
The PHT1;1-overexpressing lines (Super:PHT1;1) were also generated, and the pht1;1 mutant was obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/abrc). qRT PCR analysis showed that the PHT1;1 expression was abolished in the pht1;1 mutant and overexpressed in the PHT1;1-overexpressing lines (Fig. 5A). The Pi uptake capacity of the PHT1;1-overexpressing lines was much higher than that of wild-type plants and even much higher than that of the WRKY45-overexpressing line (Fig. 5B). By contrast, the pht1;1 mutant showed a reduction in Pi uptake compared with that of wild-type seedlings (Fig. 5B). In the presence of arsenate, the PHT1;1-overexpressing line was much more sensitive to arsenate [As(V)] than were wild-type seedlings and the WRKY45-overexpressing line (Supplemental Fig. S3). These data indicate that overexpression of PHT1;1 enhances Arabidopsis Pi uptake.
Figure 5.
Overexpression of PHT1;1 enhances plant Pi uptake. A, qRT PCR analysis of PHT1;1 expression in the pht1;1 mutant, PHT1;1-overexpressing lines, and wild-type seedlings. Seven-day-old seedlings were used for RNA extraction. The data are the mean values of three replicates ± se. B, Pi uptake was monitored over a 2-h period in 7-d-old seedlings germinated and grown on MS medium. Data are shown as mean ± se (n = 3). Asterisks indicate statistically significant differences compared with the wild type (Student’s t test, P < 0.05). Wild-type plants were used as a control (#). WT, Wild type.
Epistatic Relationship between WRKY45 and PHT1;1
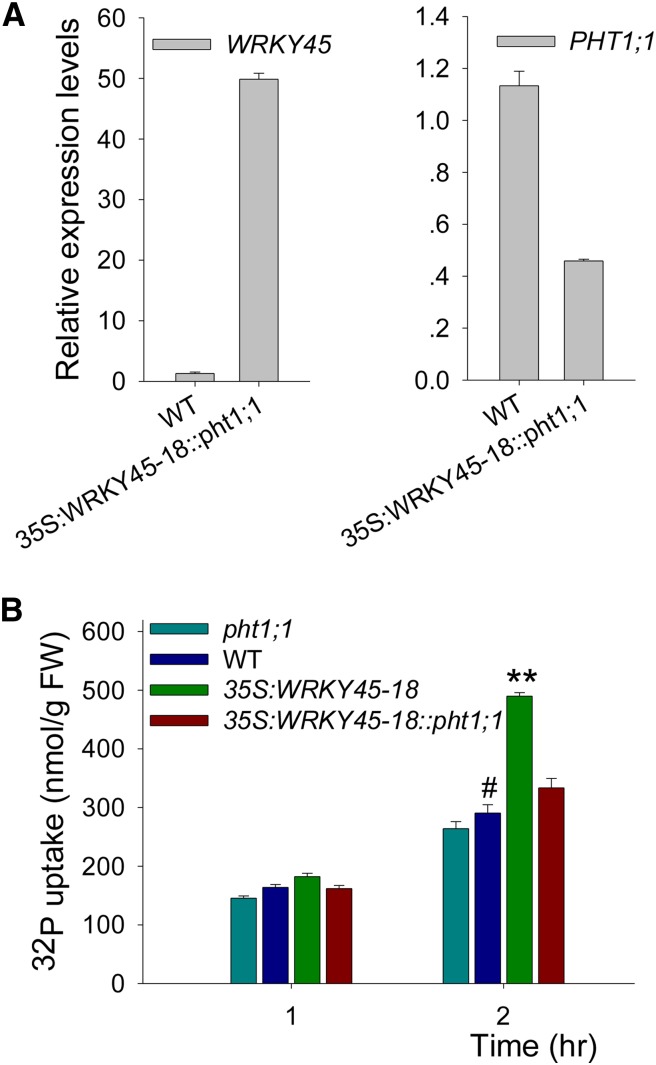
Epistatic relationship between WRKY45 and PHT1;1 was assessed by crossing 35S:WRKY45 and pht1;1 mutant to produce 35S:WRKY45::pht1;1. F2 progeny were genotyped for the presence of both 35S:WRKY45 and pht1;1 mutantation, and the homozygous at both loci was selected for evaluation. In the 35S:WRKY45-18::pht1;1, the WRKY45 was overexpressed and PHT1;1 expression was repressed (Fig. 6A). The Pi uptake of 35S:WRKY45-18::pht1;1 was lower compared with 35S:WRKY45-18 (Fig. 6B), and in the presence of arsenate, pht1;1 suppressed the As(V) sensitivity of 35S:WRKY45 (Supplemental Fig. S4). These results indicate that PHT1;1 is genetically epistatic to WRKY45.
Figure 6.
The Pi uptake enhancement caused by WRKY45 overexpression is suppressed in the pht1;1 mutant. A, qRT PCR analysis of PHT1;1 or WRKY45 expression in the 35S:WRKY45-18::pht1;1 lines and wild-type seedlings. Seven-day-old seedlings were used for RNA extraction. The data are the mean values of three replicates ± se. B, Pi uptake was monitored over a 2-h period in 7-d-old seedlings germinated and grown on MS medium. Data are shown as mean ± se (n = 3). Asterisks indicate statistically significant differences compared with the wild type (Student’s t test, P < 0.05). Wild-type plants were used as a control (#). WT, Wild type.
WRKY45 Negatively Regulates WRKY75 Expression
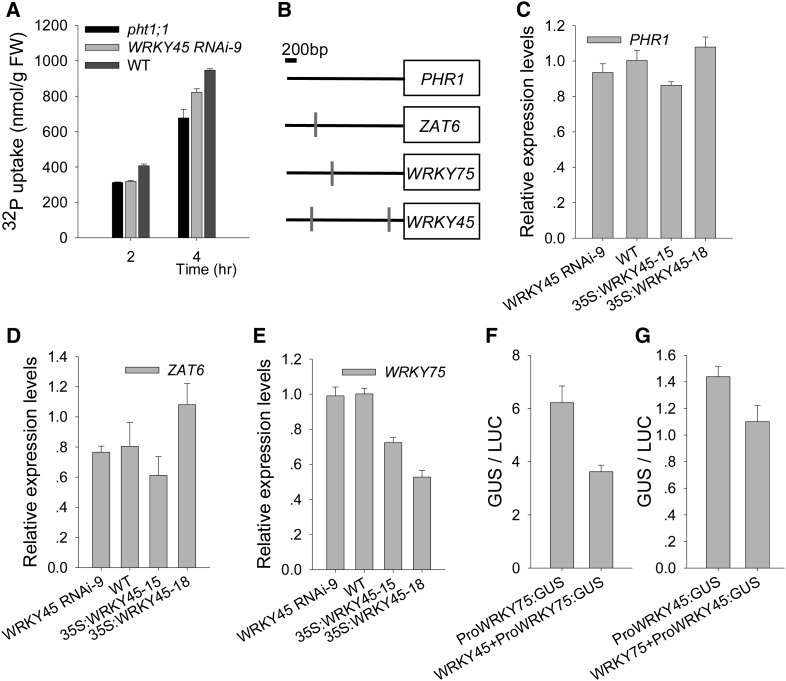
The Pi uptake of pht1;1 mutant was lower than that of the wild-type seedlings, even lower than that of WRKY45 RNAi-9 (Fig. 7A), indicating that in addition to WRKY45, there are other transcription factor(s) regulating the expression of PHT1;1. Previous reports demonstrated that several transcription factors, such as PHR1 (Rubio et al., 2001), WRKY75 (Devaiah et al., 2007a), and ZAT6 (Devaiah et al., 2007b), regulate plant Pi uptake or PHT1 family genes’ expression. PHR1 can bind to P1BS sequence within its target gene promoters to regulate target gene expression (Bustos et al., 2010). Promoter sequence analysis showed that there was no P1BS motif within the WRKY45 promoter (data not shown), and the expression of WRKY45 was not regulated in the phr1 glucocorticoid receptor:PHR1-overexpressing plant (Bustos et al., 2010), indicating that PHR1 cannot regulate the expression of WRKY45. There was no W-box within the PHR1 promoter (Fig. 7B), and the WRKY45 cannot regulate the expression of PHR1 (Fig. 7C). These data indicate that the function of WRKY45 in up-regulating PHT1;1 expression is independent of PHR1.
Figure 7.
Expressions of Pi-related transcription factor genes in WRKY45-overexpressing lines. A, Pi uptake was monitored over a 4-h period in 7-d-old seedlings germinated and grown on MS medium. Data are shown as mean ± se (n = 3). B, Diagram of the promoter regions of PHR1, ZAT6, WRKY75, and WRKY45 showing the relative positions of the W-boxes. The W-boxes are marked by gray rectangles. C to E, qRT PCR analysis of PHR1 (C), ZAT6 (D), and WRKY75 (E) expressions in the WRKY45 RNAi line, WRKY45-overexpressing lines, and wild-type plants. Seven-day-old seedlings were harvested for RNA extraction. The data are the mean values of three replicates ± se. F and G, Transient overexpression of WRKY45 fused to ProWRKY75:GUS (F) and transient overexpression of WRKY75 fused to ProWRKY45:GUS (G) in N. benthamiana leaves. Each data bar represents the means ± se (n = 3). WT, Wild type.
Promoter sequence analysis showed that there was a W-box within the promoters of ZAT6 and WRKY75 separately (Fig. 7B). It is proposed that WRKY45 may regulate the expressions of ZAT6 and/or WRKY75. The qRT PCR assays showed that WRKY45 did not regulate the ZAT6 expression (Fig. 7D), and the expression of WRKY75 was repressed in the WRKY45-overexpressing lines (Fig. 7E). To further test the function of WRKY45 on regulation of WRKY75 expression, transient expression experiments in tobacco leaves were performed. As shown in Figure 7F, WRKY45 inhibited WRKY75 promoter activity. These data demonstrate that WRKY45 negatively regulates the expression of WRKY75.
Promoter sequence analysis results also showed that there were two W-boxes within the WRKY45 promoter (Fig. 7B), and the transient expression assays showed that WRKY75 can repress the activity of the WRKY45 promoter (Fig. 7G), indicating that WRKY75 negatively regulated the WRKY45 expression.
DISCUSSION
WRKY45 Is a Positive Regulator of Plant Pi Uptake
Previous reports have revealed the importance of transcriptional control in plant responses to Pi starvation (Wu et al., 2003; Misson et al., 2005), suggesting that transcription factors play important roles in plant responses to Pi starvation. The molecular mechanisms of gene expression regulation during Pi starvation are poorly understood. Some transcription factors, such as PHR1 (Rubio et al., 2001), WRKY75 (Devaiah et al., 2007a), ZAT6 (Devaiah et al., 2007b), basic helix-loop-helix32 (Chen et al., 2007), MYB62 (Devaiah et al., 2009), and WRKY6 (Chen et al., 2009), have been identified to be involved in the response to Pi starvation. This study demonstrates that the Arabidopsis transcription factor WRKY45 is a positive regulator in plant response to Pi starvation. WRKY45 expression was induced by Pi starvation (Fig. 1), and overexpression of WRKY45 enhanced Arabidopsis Pi uptake and increased arsenate sensitivity (Fig. 2). The genetic and biochemical data support the notion that WRKY45 positively regulates the PHT1;1 expression by binding to the PHT1;1 promoter (Figs. 3 and 4). Overexpression of PHT1;1 enhanced Arabidopsis Pi uptake (Fig. 5) and increased arsenate sensitivity (Supplemental Fig. S3), and genetic data further confirmed that PHT1;1 is genetically epistatic to WRKY45 (Fig. 6; Supplemental Fig. S4). We demonstrated that WRKY45, as a positive regulator of plant responses to Pi starvation, directly regulates PHT1;1 expression to enhance plant Pi uptake.
Several transcription factors, such as PHR1 (Rubio et al., 2001), WRKY75 (Devaiah et al., 2007a), and ZAT6 (Devaiah et al., 2007b), have been reported to regulate plant Pi uptake or PHT1 family gene expression. The expression of PHR1 is not induced during Pi deprivation, and PHR1 is a small ubiquitin-like modifier (SUMO)ylation target of SAP and Miz protein (SIZ1; SUMO E3 ligase; Rubio et al., 2001; Miura et al., 2005). PHR1 cannot regulate the expression of WRKY45 (Bustos et al., 2010), and the expression of PHR1 was not regulated by WRKY45 (Fig. 7C), indicating that the regulating of WRKY45 on PHT1;1 is independent of PHR1.
Previous reports demonstrated that some WRKY proteins have redundant function, as Arabidopsis WRKY40, WRKY18, and WRKY60 play partial redundant functions in plant responses to pathogens (Xu et al., 2006) and abscisic acid signal (Shang et al., 2010), and Arabidopsis WRKY6 and WRKY42 both activate the expression of senescence-induced receptor-like kinase (Robatzek and Somssich, 2002). Robatzek and Somssich (2002) also found that Arabidopsis WRKY6 negatively regulated the expression of WRKY42. Similar to previous reports, WRKY75 (Devaiah et al., 2007a) and WRKY45 (Fig. 3) both positively regulate PHT1;1 expression, and WRKY45 and WRKY75 can repress each other’s expression (Fig. 7, E–G).
PHR1 (Rubio et al., 2001), WRKY75 (Devaiah et al., 2007a), ZAT6 (Devaiah et al., 2007b), and MYB62 (Devaiah et al., 2009) can regulate PHT1;1 and PHT1;4 expression. By contrast, WRKY45 directly up-regulated PHT1;1 expression (Figs. 3 and 4), while PHT1;4 expression was not regulated in WRKY45-overexpressing lines (Fig. 3A), suggesting that WRKY45 specifically regulates PHT1;1 expression.
The Transcription of PHT1;1 Is Specifically Regulated
Pi is an essential nutrient for plants, and the low bioavailability of Pi in the soil is a major factor limiting growth, development, and productivity of plants. The Pi transporters transport Pi from external media containing very low levels of Pi into the cytoplasm (Nilsson et al., 2007). Two Arabidopsis Pi transporters, PHT1;1 and PHT1;4, have been demonstrated to play a significant role in Pi acquisition from both poor- and rich-Pi environments (Shin et al., 2004). The expressions of PHT1;1 and PHT1;4 are induced preferentially in the roots during Pi starvation (Shin et al., 2004), consistent with their role in Pi acquisition. In addition to these two members of the PHT1 family, the expressions of other family members are also induced during Pi starvation (Muchhal et al., 1996; Karthikeyan et al., 2002). Thus, it has been concluded that the transcription of Pi transporter genes is activated upon Pi starvation, although the mechanisms are still not fully understood.
The PHT1;1 gene is the Pi transporter gene with the most abundant expression level in the Arabidopsis PHT1 gene family (Mudge et al., 2002). Several transcription factors have been reported to influence PHT1;1 expression. In the WRKY75 RNAi line, the expression of PHT1;1 was repressed, and the Pi uptake capacity was reduced during Pi deprivation (Devaiah et al., 2007a), indicating that WRKY75 may positively regulate PHT1;1 expression directly or indirectly. PHR1 and PHR1-Like1 (PHL1) are two important MYB-related transcription factors. In the phr1 mutant or the phr1phl1 double mutant, the transcript level of PHT1;1 was low (Rubio et al., 2001), and overexpression of PHR1 in the wild-type plant resulted in increased Pi content (Nilsson et al., 2007), suggesting that PHR1 and PHL1 can regulate PHT1;1 expression. In this study, we demonstrated that WRKY45 can bind to the two W-box motifs within the PHT1;1 promoter to directly up-regulate PHT1;1 expression. These data indicate that the transcription of PHT1;1 is positively regulated by several transcription factors.
In addition to positive regulation, PHT1;1 expression is also negatively regulated. Upon Pi starvation, the expression of PHT1;1 in ZAT6-overexpressing lines is lower than that in wild-type plants (Devaiah et al., 2007b), indicating that PHT1;1 expression can be negatively regulated even during Pi starvation. The PHT1;1 expression is also down-regulated in the MYB62-overexpressing lines (Devaiah et al., 2009).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used in this study. The Arabidopsis transfer DNA insertion line (Salk_088586c) of PHT1;1 was obtained from the Arabidopsis Biological Resource Center (Ohio State University) and named pht1;1 mutant.
The Arabidopsis seeds were surface sterilized and incubated at 4°C in darkness for 3 d. Then, the seeds were plated on MS medium containing 1.25 mm Pi, 3% (w/v) Suc, and 0.8% (w/v) agar and grown at 22°C with constant illumination at 80 μmol m–2 s–1, unless otherwise indicated.
For Pi starvation treatment, 7-d-old seedlings were transferred to LP medium. The LP medium was made by modifying the MS medium to contain 10 μm Pi (supplied with KH2PO4), and the agar was replaced by agarose (Promega) to avoid phosphorous contamination.
For arsenate treatment, the sterilized seeds were plated on MS medium with 500 μm Pi and 150 μm As(V).
Quantification of Total Pi
Seven-day-old Arabidopsis seedlings were transferred to MS medium for 7 to 10 d, and then the seedlings were harvested for Pi content measurements. The total Pi content in the samples was quantified as described previously (Clough and Bent, 1998; Wasaki et al., 2003).
Pi Uptake Assay
The WRKY45-overexpressing lines, WRKY45 RNAi lines, PHT1;1-overexpressing lines, pht1;1 mutant, and wild-type plants were germinated and grown on MS medium for 7 d, and then the seedlings were used for Pi uptake assays. A group of 15 seedlings was used as one biological sample. Pi uptake assay was modified from Devaiah et al. (2007a), and radioactivity was measured with a scintillation counter (Beckman Coulter).
Plasmid Construction and Plant Transformation
To construct 35S:WRKY45 and Super:PHT1;1, WRKY45 and PHT1;1 complementary DNA fragments were amplified by PCR using gene-specific primers (Supplemental Table S1) and cloned into pBI121 or pCAMBIA1300 vector. To construct the WRKY45 RNAi line, the WRKY45 fragment was amplified by PCR using the primers listed in Supplemental Table S1 and cloned into pBI121 vector. To construct ProWRKY45:GUS and ProPHT1;1:GUS, the WRKY45 and PHT1;1 promoter fragments were amplified by PCR using the primers listed in Supplemental Table S1 and cloned into pCAMBIA1381 vector. All constructs were introduced into an Agrobacterium tumefaciens strain (GV3101) and transformed into plants via floral dip transformation (Clough and Bent, 1998).
qRT PCR
qRT PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, Life Technologies) following the manufacturer’s protocol. Three real-time PCR reactions were repeated independently using Actin2/8 gene as an internal control. The primers used are listed in Supplemental Table S1.
Subcellular Localization Assay
For the subcellular localization assay, WRKY45 fused to GFP and GFP alone were cloned into pSuper1300:GFP vector, resulting in WRKY45:GFP and GFP constructs. The plasmids were then transformed into A. tumefaciens (GV3101). The transient expression assays were conducted as described previously (Chen et al., 2009). GFP fluorescence in the transformed leaves was imaged using a confocal laser scanning microscope (LSM510, Carl Zeiss).
ChIP Assay
The WRKY45-Myc transgenic line was generated first by cloning the coding sequence of WRKY45-Myc into pBI121 vector, resulting in the 35S:WRKY45-Myc construct. Then, the 35S:WRKY45-Myc plasmid was transformed into wild-type plants by the floral dip method (Clough and Bent, 1998). The WRKY45-Myc transgenic line used in the ChIP assay was a single-copy homozygous line.
The 7-d-old WRKY45-Myc seedlings were transferred to LP medium for 10 d, and then the roots were harvested for ChIP experiments with anti-Myc, which were conducted as described previously (Saleh et al., 2008; Chen et al., 2009). The primer combinations amplifying fragments within the PHT1;1 promoter are listed in Supplemental Table S1.
EMSA Assay
The EMSA was conducted using LightShift Chemiluminescent EMSA Kit (Pierce) according to the manufacturer’s protocol. The recombinant WRKY45-CKS protein was purified from Escherichia coli. The fragments of the PHT1;1 promoter were obtained by PCR using biotin-labeled or -unlabeled primers (Supplemental Table S1). Biotin-unlabeled fragments of the same sequences were used as competitors.
Transient Expression Assays in Nicotiana benthamiana
The transient GUS expression assays were performed as described elsewhere (Chen et al., 2009). The constructs (ProWRKY45:GUS and ProWRKY75:GUS) alone and in various combinations were transformed into A. tumefaciens strain GV3101. The infiltrated plants were kept under a 14-h-light/10-h-dark photoperiod at 23°C for 72 h to express GUS and Luciferase (LUC) proteins. The GUS and LUC activities of the infiltrated leaves were quantitatively determined, and the GUS/LUC ratio was used to quantify the promoter activity.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AtWRKY45 (At3g01970), PHT1;1 (At5g43350), PHT1;4 (At2g38940), PHR1 (At4g28610), WRKY75 (At5g13080), and ZAT6 (At5g04340).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Arsenate tolerance phenotype of WRKY45-overexpressing lines, WRKY45 RNAi lines, and wild-type seedlings.
Supplemental Figure S2. RNA gel-blot analysis of PHT1;1 expression in the roots of the WRKY45-overexpressing lines, WRKY45 RNAi line, and wild-type plants.
Supplemental Figure S3. Arsenate tolerance phenotype of PHT1;1-overexpressing line and wild-type seedlings.
Supplemental Figure S4. Arsenate tolerance phenotype of 35S:WRKY45-18, pht1;1 mutant, 35S:WRKY45-18::pht1;1, and wild-type seedlings.
Supplemental Table S1. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Zhi-Zhong Gong (College of Biological Sciences, China Agricultural University) for providing the pSuper1300:GFP vector.
Glossary
- Pi
phosphate
- RNAi
RNA interference
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- qRT
quantitative real-time
- MS
Murashige and Skoog
- LP
low phosphate medium
- WT
wild-type
- As(V)
arsenate
Footnotes
This work was supported by the National Science Foundation of China Projects (grant nos. 30970220 and 31170248 to Y.-F.C.), the Chinese National Key Basic Research Project (grant no. 2011CB100305 to Y.-F.C.), and the Program of Introducing Talents of Discipline to Universities (grant no. B06003 to W.-H.W.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Asher CJ, Reay PF. (1979) Arsenic uptake by barley seedlings. Aust J Plant Physiol 6: 459–466 [Google Scholar]
- Bayle V, Arrighi JF, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L. (2011) Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo G, Sánchez-Bermejo E, de Lorenzo L, Crevillén P, Fraile-Escanciano A, Tc M, Mouriz A, Catarecha P, Sobrino-Plata J, Olsson S, et al. (2013) WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. Plant Cell 25: 2944–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarecha P, Segura MD, Franco-Zorrilla JM, García-Ponce B, Lanza M, Solano R, Paz-Ares J, Leyva A. (2007) A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 19: 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH. (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Nimmo GA, Jenkins GI, Nimmo HG. (2007) BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem J 405: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. (2007a) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. (2009) Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant 2: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. (2007b) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. (2008) Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- González E, Solano R, Rubio V, Leyva A, Paz-Ares J. (2005) PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Mukatira UT, D’Urzo MP, Damsz B, Raghothama KG. (2002) Regulated expression of Arabidopsis phosphate transporters. Plant Physiol 130: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 [DOI] [PubMed] [Google Scholar]
- Maeo K, Hayashi S, Kojima-Suzuki H, Morikami A, Nakamura K. (2001) Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci Biotechnol Biochem 65: 2428–2436 [DOI] [PubMed] [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Peña A, Leyva A, Paz-Ares J. (2000) Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J 24: 559–567 [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsukawa N, Okumura S, Shirano Y, Sato S, Kato T, Harashima S, Shibata D. (1997) Overexpression of an Arabidopsis thaliana high-affinity phosphate transporter gene in tobacco cultured cells enhances cell growth under phosphate-limited conditions. Proc Natl Acad Sci USA 94: 7098–7102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG. (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Stensballe A, Jensen ON, Peck SC. (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura S, Mitsukawa N, Shirano Y, Shibata D. (1998) Phosphate transporter gene family of Arabidopsis thaliana. DNA Res 5: 261–269 [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. (2007) Hidden branches: developments in root system architecture. Annu Rev Plant Biol 58: 93–113 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Raghothama KG, Karthikeyan AS. (2005) Phosphate acquisition. Plant Soil 274: 37–49 [Google Scholar]
- Robatzek S, Somssich IE. (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z. (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, et al. (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Chen R, Harrison MJ. (2006) Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J 45: 712–726 [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Smith AP, Jain A, Deal RB, Nagarajan VK, Poling MD, Raghothama KG, Meagher RB. (2010) Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol 152: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticconi CA, Abel S. (2004) Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- Tu SI, Cavanaugh JR, Boswell RT. (1990) Phosphate uptake by excised maize root tips studied by in vivo 31P nuclear magnetic resonance spectroscopy. Plant Physiol 93: 778–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich-Eberius CI, Novacky A, Fischer E, Lüttge U. (1981) Relationship between energy-dependent phosphate uptake and the electrical membrane potential in Lemna gibba G1. Plant Physiol 67: 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasaki J, Yonetani R, Kuroda S, Shinano T, Yazaki J, Fujii F, Shimbo K, Yamamoto K, Sasaki T. (2003) Transcripomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ 26: 1515–1523 [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW. (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. (2005) OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol 138: 2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L. (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.