Suppressing the expression of two apyrase genes raises extracellular ATP levels and induces gene expression, growth, and cell wall changes characteristic of stress responses, thus implicating extracellular nucleotides as early signals linking biotic and abiotic stresses to growth inhibition.
Abstract
Plant cells release ATP into their extracellular matrix as they grow, and extracellular ATP (eATP) can modulate the rate of cell growth in diverse tissues. Two closely related apyrases (APYs) in Arabidopsis (Arabidopsis thaliana), APY1 and APY2, function, in part, to control the concentration of eATP. The expression of APY1/APY2 can be inhibited by RNA interference, and this suppression leads to an increase in the concentration of eATP in the extracellular medium and severely reduces growth. To clarify how the suppression of APY1 and APY2 is linked to growth inhibition, the gene expression changes that occur in seedlings when apyrase expression is suppressed were assayed by microarray and quantitative real-time-PCR analyses. The most significant gene expression changes induced by APY suppression were in genes involved in biotic stress responses, which include those genes regulating wall composition and extensibility. These expression changes predicted specific chemical changes in the walls of mutant seedlings, and two of these changes, wall lignification and decreased methyl ester bonds, were verified by direct analyses. Taken together, the results are consistent with the hypothesis that APY1, APY2, and eATP play important roles in the signaling steps that link biotic stresses to plant defense responses and growth changes.
Plant cells release ATP into their extracellular matrix (ECM) or the growth medium when they are wounded (Song et al., 2006), touched, or mechanically stimulated (Jeter et al., 2004; Weerasinghe et al., 2009) and as they grow (Kim et al., 2006; Wu et al., 2007). Dose-response assays indicate that extracellular ATP (eATP) can modulate the rate of cell growth, with low concentrations promoting growth and higher concentrations inhibiting it (Roux and Steinebrunner, 2007; Clark and Roux, 2009). These data implicate a regulatory role for ectoapyrases (ecto-nucleoside triphosphate-diphosphohydrolases) that can hydrolyze extracellular nucleoside triphosphates and thus, limit their accumulation, and in animals, ecto-nucleoside triphosphate-diphosphohydrolases are key regulators of the concentration of extracellular ATP ([eATP]; Knowles, 2011).
Two apyrases (APYs) in Arabidopsis (Arabidopsis thaliana) with high sequence similarity, APY1 and APY2, play a major role in growth regulation (Wu et al., 2007), in part because their expression is required for normal auxin transport (Liu et al., 2012). Recent evidence indicates that they have a Golgi localization (Chiu et al., 2012; Schiller et al., 2012), where they could regulate growth by controlling glycoprotein, glycolipid, and wall polysaccharide synthesis. One source of eATP is secretory vesicles that have a high ATP concentration in their lumen (Rudnick, 2008; Geigenberger et al., 2010). These secretory vesicles are derived from Golgi; therefore, the activity of APY1 and APY2 in Golgi could control the luminal concentration of ATP ([ATP]) in secretory vesicles and thus, indirectly control the [eATP]. It is also possible that some fraction of the cellular APY1 and APY2 population functions as ectoapyrases on the plasma membrane, because externally applied polyclonal antibodies that inhibit the activity of APY1 and APY2 rapidly inhibit the growth of pollen tubes, and the decrease in tube growth is accompanied by a transient increase in the [eATP] (Wu et al., 2007).
The inhibitory effects on both growth and auxin transport that result from suppressing the expression of APY1 and APY2 can be duplicated by raising the concentration of applied nucleotides to 800 μm or higher (Tang et al., 2003; Liu et al., 2012). Moreover, overexpression of APY1 or APY2 can significantly increase the growth rate of diverse tissues (Wu et al., 2007). This result can also be obtained by treating these tissues with low concentrations of nucleotides (Clark et al., 2010a, 2010b). These data, taken together, are consistent with the hypothesis that one function of APY1 and APY2 is to limit the [eATP], regardless of whether this function happens solely in Golgi or both in Golgi and on the plasma membrane.
Both wounding and pathogen attacks would allow cytoplasmic ATP to leak into the ECM of plant cells, and data from several laboratories are consistent with the hypothesis that this eATP could serve as an early signal of biotic stress. This hypothesis would predict that, to the extent that the suppression of APY1 and APY2 leads to an increase in [eATP], it could result in biotic stress-like responses (i.e. induce gene expression and growth inhibitory changes similar to those changes induced by biotic stress). Here, we test this prediction.
Although mutants that are null for both APY1 and APY2 (apy1apy2) are male sterile (Steinebrunner et al., 2003), the inducible suppression of APY1 by RNA interference (RNAi) in the background of an apy2 knockout yields viable plants that are dwarf (Wu et al., 2007). In the R2-4A line of mutants, APY2 is knocked out, APY1 suppression is induced by estradiol, and the induced decrease in APY1 transcripts occurs gradually, declining to about 30% of wild-type levels in whole seedlings by 3.5 d, at which point growth inhibitory effects become apparent. By 6 d, the primary root growth of these mutants has virtually ceased, and major phenotypic changes (swollen root tips and disappearance of elongation zone) become especially obvious (Liu et al., 2012). Although apyrase suppression induces expression changes in hundreds of genes, changes in the expression of genes encoding stress-responsive and growth-regulatory wall proteins in Arabidopsis are especially dramatic and highlighted in this report.
RESULTS
Confirmation of Microarray Data by Quantitative Real-Time-PCR
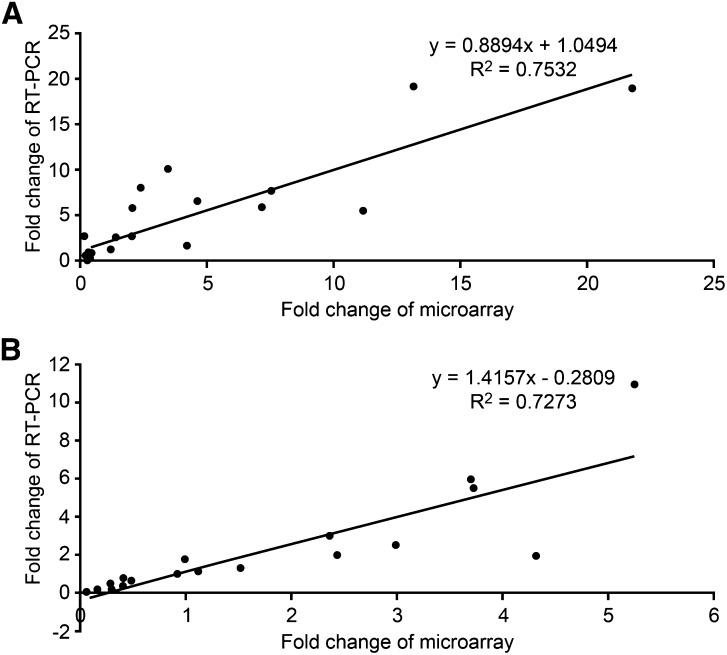
We compared transcript abundance differences between wild-type plants and RNAi mutants grown continuously in the estradiol inducer for 6.0 d in the light using the NimbleGen Arabidopsis 4-Plex microarray with three replicates. Results revealed the identity of genes with expression that significantly changed (i.e. more than 2-fold with false discovery rate of less than 5%) after apyrase suppression. Statistically significant changes in the transcript abundance of several hundred genes, including scores encoding wall/ECM -localized proteins, were observed. Control assays of wild-type plants treated with estradiol and single knockout mutants (apy1 or apy2) were conducted, and only expression changes that were not induced in wild-type plants by estradiol and not induced in single knockout mutants are described in this report. For 18 genes, the changes observed by microarray were confirmed by qRT-PCR (Fig. 1; Table I) and RNA-Sequence (data not shown). We also used quantitative real-time-PCR (qRT-PCR) to show that, for these 18 genes, the transcript abundance changes observed at 6.0 d were also significant at 3.5 d (Table I).
Figure 1.
Correlation analyses between real-time qRT-PCR and microarray results. qRT-PCR was used to validate the two microarray datasets. A, Result of standard linear regression analysis between the fold changes observed in apyrase-suppressed light-grown plants by real-time qRT-PCR (x axis) and microarray analyses (y axis). B, Result of standard linear regression analysis between the fold changes observed in apyrase-suppressed dark-grown plants by real-time qRT-PCR (x axis) and microarray data (y axis). The calculated equation and correlation value (R²) is shown along with the regression line.
Table I. qRT-PCR assays confirm microarray data.
qRT-PCR assays of 18 selected genes with transcript abundance that changed significantly in R2-4A seedlings after the suppression of APY1 and APY2 were assayed at 3.5 and 6.0 d. Most gene expression changes persist from 3.5 to 6 d. Numbers in parentheses are the NimbleGen microarray values for fold changes, which were only measured in 6.0-d-old light-grown seedlings. All qRT-PCR numbers represent average fold changes of more than three biological repeats. ANAC, Arabidopis no apical meristem, Arabidopsis transcription activation factor, and cup-shaped cotyledon-domain transcription factor; DREB, Dehydration-responsive element-binding protein; DRM1, Domains rearranged methylase1; ERF/AP2, APETALA2/Ethylene Responsive Factor; MDR1, Multi-drug resistance1; PGP19, P-glycoprotein1; NPA, naphtylphtalamic acid; SKS15, SKU5 similar15; TF, transcription factor.
| Genes | Light | |
|---|---|---|
| 3.5 d | 6 d | |
| AT1G07160 protein phosphatase 2c phosphatase | 1.2 | 0.62 (0.41) |
| AT1G21250 cell wall-associated kinase (WAK1) | 3.16 | 1.52 (4.22) |
| AT1G73330 protease inhibitor | 0.78 | 0.38 (0.25) |
| AT2G18150 Peroxidase (PER 15) ECM | 5.15 | 18.9 (21.86) |
| AT2G31083 Clavata3 | 0.13 | 0.02 (0.39) |
| AT2G33790 Arabinogalactan protein 30 | 0.08 | 0 (0.19) |
| AT2G37440 phosphatidylinositol phosphatase | 0.59 | 0.43 (0.24) |
| AT2G38340 ERF/AP2 TF; DREB subfamily | 2.23 | 2.02 (7.54) |
| AT2G46400 WRKY46 | 4.53 | 5.75 (2.01) |
| AT3G28860 MDR1;PGP19 | 0.9 | 1.22 (1.23) |
| AT4G15290 similar to cellulose synthase | 0.1 | 0.04 (0.29) |
| AT4G17030 expansin related (NPA induced) | 1.8 | 2.53 (2.04) |
| AT4G22950 AGAMOUS-like 19 TF | 0.35 | 2.43 (1.44) |
| AT4G23590 aminotransferase | 2.01 | 0.56 (0.37) |
| AT4G37160 SKS15; pectinesterase like | 0.43 | 0.09 (0.29) |
| AT5G06720 Peroxidase 53 | 5.03 | 7.95 (2.44) |
| AT5G15380 DRM1 | 9.92 | 7.64 (4.45) |
| AT5G64060 ANAC103 | 5.83 | 6.47 (4.66) |
Apyrase Suppression Significantly Alters the Expression of Stress-Related Genes and Genes Encoding Proteins Expressed in the ECM
Genes that were significantly up- or down-regulated (>2-fold; q value < 5%) were assessed by gene ontology (GO) category analyses, and they were assigned a functional categorization by annotation for GO Biological Processes. The significant trends in these results showed that genes related to stress responses were the ones that most changed expression when APY1 and APY2 were suppressed (Supplemental Fig. S1). Included among the GO Biological Processes are almost 24,000 terms listed in Ontology (version 1.3499). Every gene is annotated by at least one specific GO term, and most genes are annotated by more than one GO term. These more specific GO terms allow a statistical evaluation of which categories of functionally related genes changed most significantly when apyrases are suppressed (Table II). Only those GO terms in which the changes had an adjusted P value of less than 0.05 as sorted by Bonferroni-corrected P values and at least five genes in the Overlap count are included in Tables I and II.
Table II. GO terms for genes up-regulated or down-regulated when APY1 and APY2 are suppressed.
| GO Term | Description | Overlap | GO | P Value |
|---|---|---|---|---|
| Counta | Countb | |||
| Description of 93 up-regulated genes | ||||
| GO:0009627 | Systemic acquired resistance | 9 | 243 | 1.32E-06 |
| GO:0050832 | Defense response to fungus | 10 | 329 | 1.81E-06 |
| GO:0009862 | Systemic acquired resistance | 9 | 266 | 2.83E-06 |
| GO:0042742 | Defense response to bacterium | 10 | 362 | 4.35E-06 |
| GO:0009697 | Salicylic acid biosynthetic process | 8 | 209 | 4.66E-06 |
| GO:0010310 | Regulation of H2O2 metabolic process | 7 | 185 | 2.31E-05 |
| GO:0009595 | Detection of biotic stimulus | 5 | 102 | 0.00013 |
| GO:0043069 | Negative regulation, programmed cell death | 6 | 166 | 0.00013 |
| GO:0000165 | MAP kinase cascade | 6 | 207 | 0.00046 |
| GO:0010363 | Regulation of hypersensitive response | 7 | 368 | 0.00183 |
| GO:0031348 | Negative regulation of defense response | 6 | 268 | 0.00185 |
| Description of 215 down-regulated genes | ||||
| GO:0010106 | Cellular response to iron starvation | 10 | 116 | 2.82E-07 |
| GO:0048765 | Root hair cell differentiation | 10 | 143 | 2.06E-06 |
| GO:0006826 | Iron ion transport | 9 | 117 | 3.39E-06 |
| GO:0018925 | Oxygen binding | 11 | 232 | 2.67E-05 |
| GO:0000041 | Transition metal ion transport | 8 | 114 | 2.71E-05 |
| GO:0010054 | Trichoblast differentiation | 5 | 47 | 0.00017 |
| GO:0015706 | Nitrate transport | 9 | 206 | 0.00035 |
| GO:0010167 | Response to nitrate | 8 | 195 | 0.00129 |
| GO:0008194 | UDP-glycosyltransferase activity | 5 | 100 | 0.00583 |
| GO:0016126 | Sterol biosynthetic process | 6 | 164 | 0.01149 |
Number of genes that are annotated by a particular GO term and present within the set.
Total number of genes annotated in the particular GO term.
The GO Biological Processes analysis revealed that, among the up-regulated genes, 11 different GO terms had statistically significant overlap counts, with nine of them clearly related to genes that are up-regulated in response to biotic stress (Table II). Among the genes down-regulated, 10 different GO terms had statistically significant overlap counts, and three of the top five categories related to metal ion transport (Table II), a key function found to be down-regulated in response to biotic stress in plants (Franza and Expert, 2013).
Apyrase Suppression Induces the Up-Regulation of Type III Wall Peroxidases
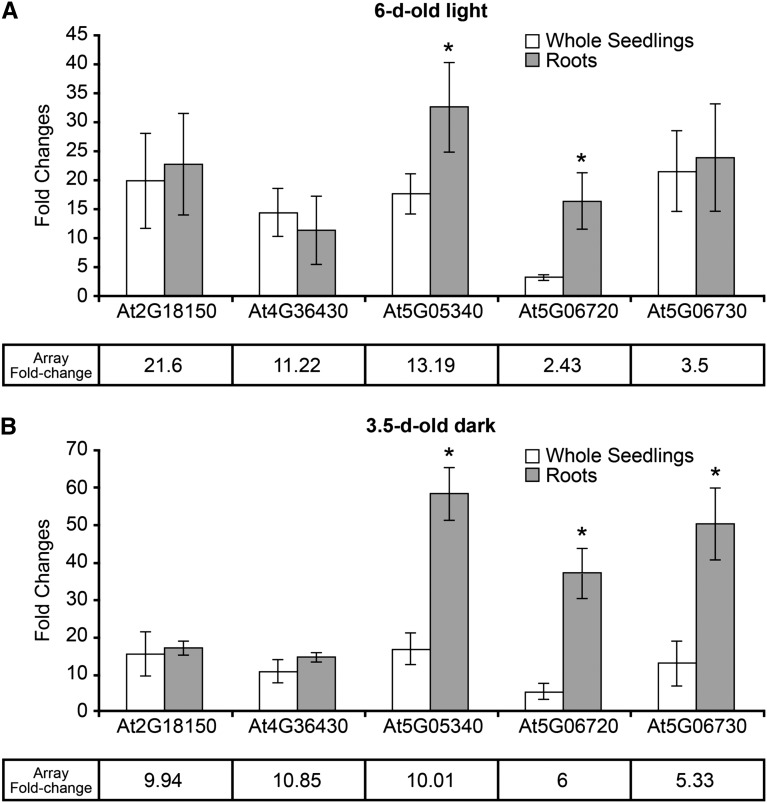
Five type III wall peroxidases that are coexpressed and up-regulated in roots undergoing abiotic stress (http://csbdb.mpimp-golm.mpg.de/csbdb/dbcor/ath/ath_tsgq.html) are all significantly up-regulated when APY1 and APY2 are suppressed in R2-4A mutants, which was judged by qRT-PCR (Fig. 2). All are also strongly up-regulated as judged by microarray analysis, but two of them (AT5G05340 and AT5G06730) have q values higher than 5%. Because the microarray data were obtained on whole seedlings and all five peroxidases are expressed mainly in roots, we tested whether the up-regulation measured in roots alone would be higher than in the whole seedlings (Fig. 2). When the up-regulation observed in whole seedlings was compared with that in roots alone, a significantly higher up-regulation in the roots was observed for AT5G05340 Peroxidase (PER) 52 and AT5G06720 (PER 53; Fig. 2). In most cases, the fold change observed by the qRT-PCR assay for whole seedlings was somewhat higher than that found by microarray. The genes encoding these five peroxidases all share common WRKY and other promoter motifs (Supplemental Table S2). WRKY46 (AT2G46400) is significantly up-regulated in induced R2-4A mutants (Supplemental Table S1A).
Figure 2.
Transcript abundance of five genes encoding type III wall peroxidases in whole seedlings and roots increases in R2-4A mutants suppressed in their expression of APY1 and APY2. A, qRT-PCR results of the transcript abundance changes of five peroxidase genes in light-grown 6-d-old seedlings. B, qRT-PCR results of the transcript abundance changes of five peroxidase genes in dark-grown 3.5-d-old seedlings. Fold changes for each gene in microarray analysis are also presented in each graph. Error bars represent sd. At least three biological repeats were performed for this experiment. An asterisk indicates value is significantly different (P < 0.05) from value of adjoining bar.
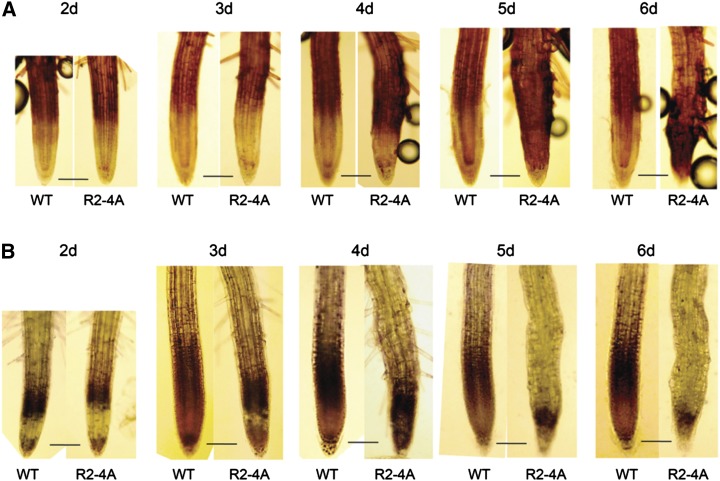
Apyrase Suppression Induces the Accumulation of Hydrogen Peroxide and Lignin in Roots
Because microarray data revealed that cell wall peroxidases are up-regulated in R2-4A, we tested how they function in R2-4A. Type III wall peroxidases use H2O2 substrates to induce protein cross links and lignin formation in cell walls. As assayed by 3,3′-diaminobenzidine tetrahydrochloride (DAB) staining, the H2O2 level is increased in R2-4A roots (Fig. 3A). After germination, wild-type roots maintain the same level of H2O2 from 2 to 6 d; however, R2-4A roots show increased H2O2 levels during their 3- to 6-d period of growth. They also accumulate lignin at this time (Fig. 4). Lignin is accumulated in secondary cell walls, such as in mature vascular tissue (Supplemental Fig. S2). Normally, lignin formation does not occur in the walls of cells in the meristematic, elongation, or early differentiated cell zones of wild-type roots. However, R2-4A seedlings develop mature vascular tissue near their root tip (Supplemental Fig. S2D) and accumulate lignin in not only vascular tissue but also the endodermis, cortex, and epidermis (Fig. 4, B, D, and F).
Figure 3.
Higher reactive oxygen species (ROS) levels are induced in R2-4A roots when APY1 and APY2 are suppressed. A, H2O2 levels in wild-type (WT) and R2-4A roots from 2 to 6 d after estradiol treatment. DAB was used to stain H2O2. B, Superoxide levels in WT and R2-4A roots from 2 to 6 d. Nitroblue tetrazolium was used to stain superoxide. Images are representative of at least three biological repeats. Bar = 100 μm. [See online article for color version of this figure.]
Figure 4.
Higher lignin levels are induced in R2-4A roots when APY1 and APY2 are suppressed. Lignin staining of 6-d-old roots with phloroglucinol in wild-type (A) and R2-4A (B) roots. Magenta color indicates lignin. Cross sections of wild-type (C) and R2-4A (D) roots stained with phloroglucinol. Cross sections are from regions ∼200 to 600 μm from root tip, where vascular tissue is not fully matured in wild type. Lignin autofluorescence of wild type (E) and R2-4A (F). Images are representative of at least three biological repeats. Bars in A and B = 100 μm. Bars in C and D = 10 μm. Bars in E and F = 50 μm.
Apyrase Suppression Increases Cross Linking in Cell Walls
The most dramatic growth change that occurs when apyrases are suppressed in light-grown R2-4A seedlings is inhibition of root growth. One way to account for the growth differences between wild-type and mutant roots would be by the degree of cross linking of homogalacturonan (HG) present in the cell walls of the plant. A commonly reported HG cross-linking mechanism involves the formation of a borate ester that covalently links two rhamnagalacturonan-II (RG-II) monomers found on HG (Supplemental Fig. S3; Ishii et al., 1999; Vincken et al., 2003; O’Neill et al., 2004). The ester forms at O-2 and O-3 within apiosyl residues found on side chain A of RG-II. If this cross linking occurred when the expression of APY1 and APY2 was suppressed, the mutant root samples would contain fewer methyl ester groups and more boron. To investigate this question, we examined the chemical differences between wild-type and mutant root samples with Fourier transform infrared (FTIR) and x-ray photoelectron spectroscopy (XPS).
Although the infrared (IR) spectrum of cell walls is a complicated composition of bands from many functional groups, it was possible to identify differences between the wild-type and mutant samples through FTIR spectroscopy. Figure 5A shows the average of 13 FTIR spectra for both wild-type and R2-4A mutant root samples, combining data from both biological and instrumental repeats. We identified 13 peaks that appeared in both wild-type and mutant spectra (Fig. 5A). The most noticeable differences in the spectra were the decreases in intensities of peaks at 1,741 and 1,251 cm−1 for the mutant samples (Fig. 6A, red spectrum) compared with wild type (Fig. 5A, black spectrum). These peaks correspond to carbonyl (C = O) and ether (C-O) intensities, respectively, and are associated with methyl ester groups on the polysaccharide chains (Nivens et al., 2001; Pau-Roblot et al., 2002). This decrease of methyl esters indicated that cross linking was occurring at those sites in the cell wall, which is consistent with the mechanism for boron cross linking proposed by Ridley et al. (2001). The peak at 1,648 cm−1 is from the amide 1 (C = O stretching) mode, and although it can be characteristic of peptide secondary structure, it does not provide any relevant information about wall cross linking.
Figure 5.
Cell walls of Arabidopsis isolated from R2-4A mutants suppressed in their expression of APY1 and APY2 have altered FTIR spectra, indicating fewer methyl ester linkages. A, Averaged FTIR spectra for wild-type (black) and R2-4A mutant (red) samples both computed using 13 spectra. Each of the spectra was obtained from 100 scans at a resolution of 4 cm−1. The averaged spectra were normalized to the maximum intensity peak at 1,056 cm−1 and then baseline corrected. Dashed lines are added to highlight differences in the two spectra at 1,251 and 1,741 cm−1. B, PCA of wild-type (black squares) and mutant (red circles) samples. PC1 is plotted against PC3, accounting for 99.3% of the variance. Each point on the figure represents 1 of 13 peaks selected for analysis (1,035, 1,057, 1,111, 1,161, 1,207, 1,251, 1,319, 1,336, 1,374, 1,431, 1,516, 1,648, and 1,741 cm−1). C, An average spectrum of the 4A mutant samples (black) is shown here with 17 fitted Gaussian curves along with the total Gaussian fit (red). The Gaussian fits associated with stretches arising from methyl ester groups are highlighted in green (1,742 cm−1) and blue (1,234 cm−1), which correspond to C = O and O-C-O vibrations, respectively.
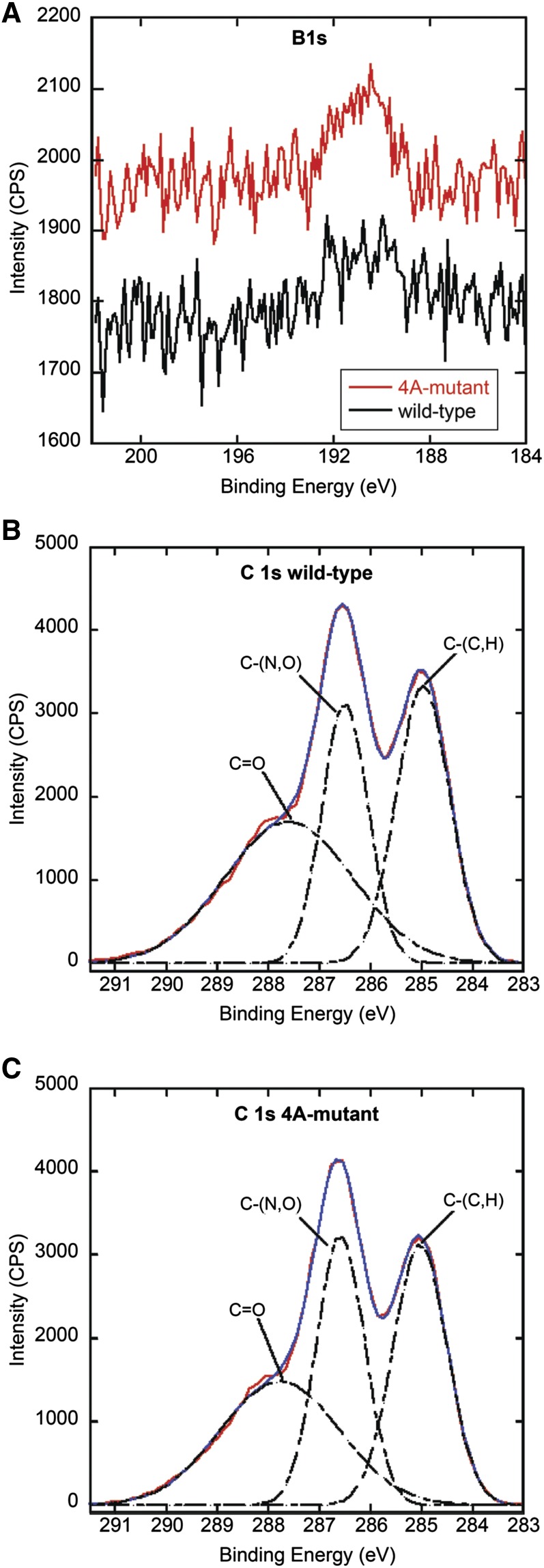
Figure 6.
Cell walls of Arabidopsis isolated from R2-4A mutants suppressed in their expression of APY1 and APY2 have altered x-ray photoelectron spectra, indicating more boron linkages. A, X-ray photoelectron spectra of the B 1s region showing a peak near 190 eV for wild-type (black) and mutant (red) samples. Scans were taken at a resolution of 0.1 eV, a 300-ms dwell time, and a detector aperture of 160 mm. The C 1s region of x-ray photoelectron spectra of wild-type (B) and R2-4A-mutant (C) cell walls taken at a resolution of 0.1 eV, a dwell time of 3,000 ms, and a detector aperture of 20 mm. Spectral decomposition was done by fitting the peaks to Gaussian curves. The baseline corrected result is shown in red, Gaussian fit is in blue, and peak composites are dashed lines. The aliphatic carbon [C-(C,H)] peak was adjusted to 285 eV to account for binding energy shifts caused by the use of a charge neutralizer. Peaks at 286.6 and 287.8 eV correspond to C-(N,O) and C = O functional groups, respectively. When measured with respect to the 285-eV signal, the R2-4A mutant sample shows a 20% increase in the 286.6-eV peak over the wild type. CPS, Copalyl diphosphate synthase.
To determine if the decrease in intensity of the peaks at 1,741 and 1,251 cm−1 could be used to quantitatively differentiate the wild-type and mutant samples, we used principal component analysis (PCA), a rigorous method for distinguishing wild-type and mutant samples from small differences in the complex spectra (Mouille et al., 2003). All 13 spectral peaks listed above were used in the analysis. A standard PCA analysis to calculate a covariant matrix from which the principal components (PCs) were found was done with in-house scripts written in Mathematica. The clearest distinction between wild-type and R2-4A mutant samples was found when comparing the first and third PCs (Fig. 5B), which represented 99.3% of the observed variance between the wild-type and mutant samples. If both samples were identical in composition, the clustering of points in the PC plots would overlap; however, the data shown in Figure 5B clearly display two distinct clusters for the wild-type (black) and mutant (red) samples, showing that the small differences in IR spectra between the wild-type and mutant samples were characteristic of the chemical composition of each sample.
Although PCA can illustrate differences between two spectra, it cannot provide a measure of their significance (Lloyd et al., 2011). To quantify these results, we integrated peaks associated with methyl ester groups involved in HG cross linking. The averaged IR spectrum for the mutant samples was fit using 17 Gaussian curves, where peak position, intensity, and width were optimized using a Levenberg Maquardt algorithm packaged with the OPUS software. For the wild-type samples, the averaged spectrum was fit using 17 peak positions found for the mutant samples, but peak intensity and width were optimized by the fitting routine. The Gaussian fit for the 1,741-cm−1 stretch shown in Figure 5A was found to have an actual peak position of 1,742 cm−1 after deconvolution, and the 1,251-cm−1 peak from the average spectra arose from two overlapping peaks at 1,234 and 1,259 cm−1. The 1,259-cm−1 peak showed no significant change between wild-type and mutant samples, whereas the 1,234-cm−1 peak accounted for the decrease in the peak intensity at 1,251 cm−1 described in Figure 5A.
The peaks at 1,742 and 1,234 cm−1 correspond to the C = O and O-C-O stretches of methyl ester, respectively. An example is shown in Figure 5C, which depicts the averaged spectra of the R2-4A mutant samples along with the Gaussian fits used to isolate the peaks associated with the methyl ester groups. This analysis indicated that the intensity of the peak at 1,742 cm−1 decreased by 21.4% and that the peak at 1,234 cm−1 was reduced by 44.9% compared with the wild-type samples. This decrease in intensity is proportional to the degree of methyl ester reduction, which is in agreement with the hypothesis of an increase in RG-II found on HG.
The reaction scheme in Supplemental Fig. S3 proposes that bioavailable boron mediates the cross linking of HG associated with the root cell wall. We used XPS to measure the content of boron in both wild-type and mutant samples to determine if the mutants contained a greater amount of boron. The boron 1s photoelectron near 190 eV has a low cross section of photoelectron emission (the Kratos XPS software uses a sensitivity factor of 0.159 for B 1s compared with 0.278 for C 1s), and therefore, to increase the sensitivity of our instrument, we opened up the detector aperture to its maximum value of 160 mm. Although it resulted in peaks with significantly greater background noise than the C 1s and O 1s regions, we were still able to detect the B 1s peak in our samples. Representative examples of x-ray photoelectron spectra of the B 1s peak are shown in Figure 6A, and atomic compositions of both wild-type and mutant samples are given in Supplemental Table S3 (Moulder et al., 1992). Because boron is required for the normal growth of a plant, the wild-type sample contained a small amount of boron (0.065 ± 0.092% [w/v]; Fleischer et al., 1998; O’Neill et al., 2004). However, the apyrase-suppressed mutant contained nearly 4-fold more boron (0.245 ± 0.091% [w/v]) than the wild-type sample. We attribute this increase in boron in the chemical composition of the cell wall to boron-mediated cross linking of HG.
An increase in the amount of boron-mediated cross linking in the mutant samples should also yield changes in the relative intensity of various types of carbon bonds in the cell walls. By using the 20-mm aperture, the C 1s region was also studied to understand the changes in carbon bonding. For the C 1s region, the R2-4A mutant and wild-type samples showed similar spectra with three peaks (Fig. 6B). The aliphatic carbon 1s peak is shown at 285 eV and peaks at 286.6 and 287.8 eV correspond to C-(N,O) and C = O functional groups, respectively, which are in agreement with previously published spectra taken from cell walls (Dufrêne et al., 1997; Ramstedt et al., 2011). A quantitative analysis of the C-(C,H) and C-(N,O) signal amplitudes shows that the average intensity ratio C-(N,O)/C-(C,H) is 0.744 and 0.925 for the wild-type (Fig. 6B) and R2-4A mutant (Fig. 6C) samples, respectively. These ratios would support an increase of 1.2-fold more C-O bonds for the R2-4A mutant. Because an increase in boron cross linking requires an increase of C-O bonds (Supplemental Fig. S3), both the B 1s and C 1s spectral measurements support an increase in boron-mediated cross linking in the R2-4A mutant.
Overall, the above FTIR and XPS results indicate that there is a decrease in the presence of methyl ester groups and an increase in the concentration of boron in the R2-4A mutant. To the extent that these results reflect an increase in the amount of boron-mediated cross linking between cell walls, they would indirectly reflect higher methylesterase activity in the R2-4A mutant. Consistent with this conclusion, one of the pectin methylesterase (PME) inhibitor proteins that can bind PMEs and inhibit their activity, PMEI4 (AT4G25250), is down-regulated >3.9-fold in estradiol-induced R2-4A seedlings (Supplemental Table S1B).
Apyrase Suppression Results in Increased [eATP] in Growth Media
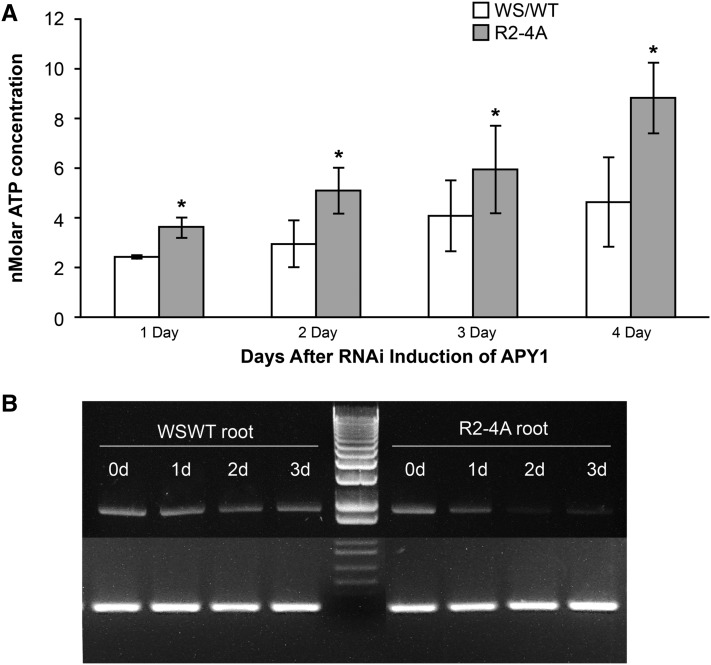
In ecotype Wassilewskija (Ws) of Arabidopsis wild-type and induced R2-4A samples, the [ATP] began to differ 1 d after transfer to liquid growth conditions with estradiol (Fig. 7A). In growth media of R2-4A plants, the [ATP] was higher than that of wild-type plants at all points tested, and the difference became greater coincident with decreased APY1 expression (Fig. 7B). This difference steadily increased up to 2-fold through 4 d, in parallel with decreased APY1 expression. In contrast, the [eATP] in the growth media of Ws wild-type and uninduced R2-4A plants did not differ through 4 d of growth (data not shown). The [ATP] measured in the media is a very large dilution of the [ATP] likely to be present immediately to the outside of the plasma membrane (probably more than 1,000-fold; Song et al., 2006). The R2-4A plants grown in liquid culture had the expected root length and root tip phenotypes previously described (Liu et al., 2012). During their growth in liquid culture, the primary roots of Ws wild-type and uninduced R2-4A seedlings both grew an average of 18 mm, whereas the primary roots of induced R2-4A seedlings grew an average of 1.5 mm (Supplemental Table S4). The experiment was performed three times, with six biological replicas of each sample included each time. Results were similar between all experiments; data from one experiment is shown in Figure 7A.
Figure 7.
Media [eATP] rises with increased suppression of apyrase expression in R2-4A seedlings. A, Time course of increase of [ATP] in seedling growth media based on luciferase assay luminescence compared with the ATP standard curve. Error bars represent sd. An asterisk indicates statistical significance based on P value > 0.05 in a Student’s t test. B, Time course of loss of APY1 transcripts during continuous treatment of R2-4A mutants with estradiol inducer of RNAi construct. WS/WT, Ws cultivar, wild-type.
DISCUSSION
The significant root growth inhibition and root morphology changes observed in the primary roots of seedlings suppressed in AtAPY1 and AtAPY2 expression (Wu et al., 2007) are reminiscent of the growth inhibition effects observed in seedlings responding to biotic and abiotic stresses, some of which seem to be mediated by ethylene and 1-aminocyclopropane-1-carboxylic acid (Tsang et al., 2011). Many different stresses can induce similar responses in plants, the so-called stress-induced morphogenic responses (Potters et al., 2009). The stress-induced morphogenic responses in roots involve common physiological processes, such as altered ROS production, hormone transport changes, inhibition of elongation and consequent formation of root hairs near the apex, and blocked cell division in the primary meristem (Potters et al., 2007). All of these changes have been documented in the roots of estradiol-induced R2-4A seedlings (Wu et al., 2007; Liu et al., 2012), and therefore, it is not surprising that the gene expression changes in these mutants would be characteristic of plants subjected to stress conditions.
The striking finding in our GO analyses is that, although the R2-4A seedlings were grown under sterile conditions, their gene expression changes in response to apyrase suppression remarkably resembled the transcriptomic changes that plants undergo when under biotic attack.
One possible explanation for this finding is that both biotic stress and suppression of APY1/APY2 expression release ATP into the ECM. There are reports documenting ATP release from wound sites (Song et al., 2006; Torres et al., 2008) and cells stimulated by elicitors (Kim et al., 2006; Wu et al., 2008). This paper and others (Wu et al., 2007; Clark et al., 2010a, 2010b) document that blocking apyrase expression by RNAi or antibodies results in an increase in the [eATP]. To the extent that the release of ATP into the ECM is a signal indicating to plants that they are undergoing a biotic attack, it would not be surprising that the increased [eATP] resulting from apyrase suppression in R2-4A mutants would induce gene expression changes characteristic of those changes that happen when plants experience pathogen or other biotic stresses.
The increased [eATP] resulting from apyrase activity inhibition can occur within minutes (Wu et al., 2007), and the release of ATP from wounded cells or cells rendered leaky by pathogen attack would also be expected to occur within minutes. The signaling changes induced by eATP, which include increased [Ca2+]cyt, increased NO and ROS production, and increased mitogen-activated protein (MAP) kinase expression (Tanaka et al., 2010; Clark and Roux, 2011; Choi et al., 2014), are also induced by pathogen stress, further supporting a functional connection between increased eATP and the induction of biotic stress responses. In this regard, the data in the work by Chivasa et al., (2009), which show a significant [eATP] drop 5 or more h after salicylic acid or elicitor treatment, may indicate that a decrease in [eATP] can be a later consequence of biotic stress in plants.
Although one might expect that the signaling pathways linking different hormone treatments to growth changes would converge on similar gene expression changes, Nemhauser et al., (2006) found that the transcript abundance changes induced by abscisic acid, auxin, brassinosteroid, cytokinin, ethylene, GAs, and jasmonates were remarkably dissimilar from each other. Nemhauser et al., (2006) concluded that different hormones regulate growth and other responses in plants by distinctly different networks of genes. The transcriptional changes that link apyrase suppression to growth inhibition are also distinct from the transcriptional changes that occur in response to growth-inhibitory levels of any one of the hormones. Statistical techniques similar to those techniques used by Nemhauser et al., (2006) revealed that the overall pattern of gene expression changes induced by apyrase suppression is different from patterns observed when auxin, ethylene, abscisic acid, or brassinosteroids inhibit growth (data not shown).
Superoxide (O2−) is rapidly converted to H2O2 in cells, and peroxidases use H2O2 as a substrate to catalyze diverse products that affect root growth (Gapper and Dolan, 2006; Nakagami et al., 2006; Dunand et al., 2007; Tsukagoshi et al., 2010; Tsukagoshi, 2012). Because the inhibition of primary root growth and up-regulation of several type III wall peroxidases occur together in roots of estradiol-induced R2-4A mutants, we assayed ROS levels in these roots. With estradiol treatment, the balance of O2− and H2O2 levels in the R2-4A primary root starts to change near when apyrase suppression peaks (∼3 d), and by 6 d, the level of H2O2 is much higher than that of O2− (Fig. 3). Maintenance of O2− levels is important for normal cell growth (Gapper and Dolan, 2006; Dunand et al., 2007), and when root H2O2 levels are increased, the progression of meristematic cells through S phase and M phase is decreased (Tsukagoshi, 2012).
Along with increased H2O2 in roots, there is an increased abundance of transcripts encoding type III peroxidases (Fig. 2; Table I) that can catalyze a broad range of physiological processes, like cross linking of phenolic compounds to proteins and polysaccharides and lignin deposition (Passardi et al., 2004; Passardi et al., 2005; Almagro et al., 2009; Djebali et al., 2011; Zhang et al., 2012). Peroxidase genes are up-regulated by plants in response to biotic stresses, and their expression is associated with increased resistance to insect herbivores and pathogens (Hu et al., 2012; Suzuki et al., 2012). Five of the genes most up-regulated when APY1 and APY2 are suppressed encode type III wall peroxidases that are coexpressed in roots and up-regulated together by abiotic stresses known to inhibit root growth. The genes encoding these five peroxidases all share common WRKY motifs (Supplemental Table S2), and therefore, it is noteworthy that WRKY46 (AT2G46400), with expression that enhances basal resistance against Pseudomonas spp. pathogens (Hu et al., 2012), is also significantly up-regulated in induced R2-4A mutants. Clearly, peroxidase up-regulation occurs in plant responses to both biotic and abiotic stresses, and therefore, increased wall peroxidase expression could serve as a key convergence point in the signaling pathways that cross over in stress signaling networks (Fujita et al., 2006). The up-regulation of peroxidases is often correlated with growth inhibition through peroxidase-catalyzed wall cross-linking reactions, lignin formation, and/or the ROS changes induced by these enzymes (Pedreira et al., 2011). We confirmed that the apyrase-suppressed mutants have much more extensive lignin formation than wild-type plants, and this difference continues to increase with time (Fig. 4, B, D, and F).
Among genes encoding extracellular proteins, those genes encoding peroxidases undergo the largest changes in transcript abundance when APY1 and APY2 are suppressed. However, there are scores of other genes for extracellular proteins that undergo significant changes—so many, in fact, that they are significantly overrepresented in the GO terms for cellular components. Supplemental Table S6 5 lists 21 genes encoding proteins that function in the ECM and are up-regulated in apyrase-suppressed plants and 48 genes that are down-regulated in these plants. Unexpectedly, an expansin-like B-1 protein is up-regulated in apyrase-suppressed mutants. Expansins typically promote growth, but the mutants have greatly reduced growth. Expansin-like B proteins have sequence similarity to α-expansins and β-expansins, but they have not yet been experimentally shown to induce cell wall loosening. Additionally, the HisPheAsp motif in the catalytic site of domain 1 of expansins is not conserved in the expansin-like B-1 protein, which might indicate that it functions differently than α-expansins and β-expansins (Sampedro and Cosgrove, 2005).
Paralleling these wall changes are two other wall changes documented by FTIR and XPS data. These assays reveal decreased pectin methyl ester bonds, which can result in increased Ca2+ cross linking of wall pectins (Palin and Geitmann, 2012), and increased boron accumulation, which can inhibit root growth (Reid et al., 2004; Aquea et al., 2012). These results support the conclusion that there is higher PME activity in these roots, which could result from the decreased expression of PMEI4 (also observed in induced R2-4A mutants). PMEI4 is one of the 10 most down-regulated genes in roots when their growth is inhibited by 1-aminocyclopropane-1-carboxylic acid (Markakis et al., 2012).
The decrease by 21.4% and 44.9% of the 1,740-cm−1 and 1250-cm−1 FTIR peaks, respectively, indicates that methyl esters have decreased proportionally; nevertheless, these magnitudes are larger than the relative increase (3.8-fold) in boron signal seen by the XPS and larger than the 1.2-fold increase of C-O signal. This finding means that the boron-mediated cross linking does not account for all of the methyl ester loss expected to occur in the cell wall and indicates that boron-mediated cross linking is just one of multiple pathways in which the cell wall cross links (Jolie et al., 2010). Other cross-linking mechanisms that do not use boron were not investigated and should be taken into consideration for a more quantitative analysis that would more completely account for the decrease in methyl ester groups.
Taken together, increased H2O2 level, increased expression of wall peroxidases, increased lignin and boron accumulation in cell walls, and decreased pectin methyl ester bonds would all contribute to root growth inhibition by gradually decreasing the number and size of cells in meristematic and elongation zones. These changes, then, are likely transduction changes that help link apyrase suppression to the inhibition of growth seen in the R2-4A mutants.
Among the wall changes induced by apyrase suppression, increased lignification and boron cross linking (associated with decreased pectin methyl ester groups) are especially relevant. These changes stiffen the wall, which in addition to mediating the dramatically decreased growth of primary roots in the induced R2-4A mutants, could also help account for the reported association of increased eATP with pathogen resistance. Fungal elicitors, like apyrase suppression, induce an increased [eATP] in plants (Kim et al., 2006; Wu et al., 2008) and also, both induce increased cross linking in cell walls (Almagro et al., 2009), a change that impedes pathogen penetration into cells. Insect herbivores secrete apyrases as part of their attack arsenal (Wu et al., 2012), and to the extent that increased [eATP] induces plant defense genes, as indicated by earlier reports (Jeter et al., 2004; Song et al., 2006; Choi et al., 2014), the apyrases secreted by plant attackers would block this induction.
The suppression of APY1 and APY2 results in a strong inhibition of polar auxin transport in shoots and roots (Liu et al., 2012); however, surprisingly, none of the genes significantly up- or down-regulated were members of the PIN-formed, ATP-binding cassetteB, or AUXIN1/LIKE-AUX1 family of auxin transport facilitators (Peer et al., 2011; Supplemental Table S6). These results point to the likelihood that the rapid growth changes induced by apyrase suppression, which are observed in less than 15 min in pollen tubes, are mediated initially by posttranscriptional changes in these transporters. Although our assays of gene expression changes in R2-4A mutants were carried out within hours after the level of APY1 transcripts dropped to their low point after their RNAi -mediated suppression, these transcriptomic changes are surely downstream of earlier signaling changes induced by apyrase suppression.
A major breakthrough in the field of eATP signaling has been the recent identification of a receptor for eATP in Arabidopsis (Choi et al., 2014). This receptor is a lectin receptor kinase and was given the name Does Not Respond to Nucleotides 1 (DORN1). It is needed for ATP-induced rapid changes in [Ca2+]cyt, downstream changes in MAP kinase, and gene expression changes related to wound responses. Relevant to data reported here, this lectin receptor kinase plays an important role in fungal pathogen resistance (Bouwmeester et al., 2014). Increased ligand concentration often induces changes in the level or activity of its receptor (Chen et al., 2007). Although suppression of apyrase, which increases the [eATP], does not significantly alter the transcript level of DORN1, it will be valuable to learn whether apyrase suppression alters the activity, turnover, or rate of endocytosis of DORN1.
Wall peroxidases can catalyze the oxidative degradation of auxin (Krylov and Dunford, 1996; Gazarian et al., 1998; Potters et al., 2009), and by promoting increased lignification of cell walls in the Casparian strip, they could alter auxin transport, which passes through this region as it cycles between shootward and rootward transport in primary roots (Naseer et al., 2012). It seems likely, however, that these wall changes are kinetically downstream of the NO and ROS changes that take place in less than 30 min when they are induced by increased [eATP] (Salmi et al., 2013). As noted by Liu et al. (2012), all the growth and morphology changes induced by the suppression of APY1 and APY2 cannot be mimicked by increasing the [eATP]. Additional research would be needed to determine how many of the gene expression changes induced by the suppression of APY1 and APY2 in R2-4A seedlings are attributable to changes induced by the functions of these apyrases (e.g. Golgi functions) that are not related to signaling changes induced by increased [eATP].
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Plant materials used in microarrays were Arabidopsis (Arabidopsis thaliana) ecotype Ws and RNAi mutant line R2-4A. The RNAi construct, generation of the R2-4A line, and growth conditions of the seedlings were as previously described (Wu et al., 2007). For estradiol treatment, estradiol was added into Murashige and Skoog (MS) medium to reach a final concentration of 4 µm. Plates were placed upright in a culture chamber and grown at 23°C under 24-h fluorescent light after 3 d vernalization at 4°C, and seedlings were harvested at 6 d.
Total RNA Isolation and NimbleGen Microarray Experiments
Each microarray had four different treatments: induced RNAi mutant (R2-4A), apy2 single knockout with a noninduced RNAi construct, wild type treated with estradiol (wild type + estradiol), and wild type only. Three repeats were carried out in each microarray. The effects of estradiol treatment were also analyzed in both microarrays, and the comparison between wild type + estradiol and wild type showed low gene expression changes in the whole gene expression profile (data not shown).
Seedlings were collected and frozen in liquid nitrogen. Plant tissues were ground to fine powder with a chilled mortar and pestle. Total RNA was isolated using the Spectrum Plant Total RNA Kit (Sigma) according to the manufacturer’s instruction. Four different samples were used in each microarray experiment (one for each of four different treatments noted above), and three replicates were prepared from each of the samples. Total RNA was sent to NimbleGen and hybridized to Arabidopsis 4-plex arrays (A4511001-00-01). Complementary DNA (cDNA) synthesis, sample labeling, array hybridization, scanning, data extraction, and preliminary data analysis were performed at NimbleGen.
qRT-PCR
Total RNA was extracted using the Spectrum Plant Total RNA Kit (Sigma) according to the manufacturer’s instruction. After treated with DNase I (Invitrogen), total RNA (1 µg) was reversely transcribed to cDNA using the SuperScript II Reverse Transcriptase Kit (Invitrogen). cDNA was amplified in real-time PCR reactions using SYBR Green PCR Master Mix (Applied Biosystems) with gene-specific primers (see “Supplemental Data”). Real-time PCR was carried out in triplicate on each sample using an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). The relative expressions of genes were normalized to the level of a reference gene protein phosphatase 2A. The changes of transcripts were calculated using the method of Shi and Chiang (2005).
Significance Analysis of Microarrays
To determine the transcriptomic changes caused by estradiol treatment of R2-4A seedlings in each set of microarray data, the comparison between the mutant and wild-type plants treated with estradiol was conducted. Those minor changes only caused by estradiol treatment of wild-type plants and the single knockout (apy2) background were excluded. Significance analysis of microarrays (SAM; Tusher et al., 2001) was used to find statistically significant genes in our microarray experiment. SAM considers the relative change of each gene expression level with respect to the sd of repeated measurements and then assigns a score to each gene. This analysis uses a nonparametric test and a permutation idea to estimate the percentage of genes identified by chance (the false discovery rate). It does not require the assumptions of equal variances or independence of genes. SAM is run as an Excel add-in and available for download online at http://www-stat.stanford.edu/~tibs/SAM/.
Histochemistry
Wild type and R2-4A were grown vertically for 6 d in continuous light. For comparison of H2O2 and superoxide levels in roots, DAB (D3939; SIGMA) and nitroblue tetrazolium (N5514; SIGMA) were used, respectively. Lignin staining was performed using phloroglucinol (P3502; SIGMA) in ethanol and HCl. For cross section, 6-d-old seedlings were fixed with 4% paraformaldehyde in PBS overnight and embedded in LR White (14381; EMS). After sectioning, the specimens were stained with phloroglucinol.
The method used to observe autofluorescence of lignin, clearing root, and visualization was the method by Hosmani et al., 2013, and 6-d-old seedlings were assayed. After the clearing steps, seedlings were immersed in 25% glycerol and mounted on glass slides with 50% glycerol. Autofluorescence of lignin was observed with excitation at 488 mm (Leica DM IRE2 confocal microscope).
Cell Wall Isolation
Ws wild-type and R2-4A seedlings were grown on vertical plates in continuous light for 6 d. Cell wall alcohol insoluble residues were prepared with a modified online protocol (http://cellwall.genomics.purdue.edu/techniques/7.html). Briefly, roots of 6-d-old light-grown seedlings were harvested and immediately ground into fine powder in liquid N with a mortar and pestle. The powder was treated for 5 min at 70°C with 50 mm Tris⋅HCl (pH 7.0) containing 1% SDS. After centrifugation, the pellet was washed with water, collected on a nylon mesh filter, ball-milled (Retsch) for 10 min, and then boiled in 95% ethanol for 5 min. The samples were filtered through the nylon mesh and successively washed two times with water (70°C) and 50% ethanol (70°C); then, they were washed with 70% ethanol (room temperature) and dried using a SpeedVac concentrator.
FTIR Spectroscopy
Cell wall powder was investigated by FTIR in potassium bromide pellets. A mixture containing the 1% (w/w) isolated cell wall powder in dry KBr was pressed into pellets ∼1 cm in diameter and 0.5 mm thick for measurement. Vibrational spectra were obtained on a Bruker Vertex 70 FTIR spectrometer in a typical absorption/transmission configuration, with pellet samples mounted perpendicular to the IR beam. Before each measurement, the sample compartment was purged with dry N2 (grams) for 10 min. A mercury cadmium telluride detector was used to collect 100 scans at a resolution of 4 cm−1 from 500 to 4,000 cm−1. Repeat spectra were collected on single pellets by translating the sample vertically in the IR beam. Three different preparations of cell walls (biological repeats) were measured, and four measurements per pellet were taken (instrumental repeats) for a total of 13 individual spectra for both wild-type and mutant samples. These spectra were normalized with respect to the highest intensity peak at ∼1,056 cm−1, and an average spectrum was determined for both wild-type and mutant samples. These spectra were then baseline corrected with a rubber band algorithm. For quantification of IR peaks, these averaged and baseline-corrected spectra were each fit to 17 Gaussian curves, which were then integrated. All spectrum manipulations were performed with the instrument software package OPUS (Bruker Optics, Inc.).
XPS
XPS was used to measure chemical differences between the composition of the powdered wild-type and mutant samples. A densely packed layer of powders of each sample was placed on the XPS sample holder with copper tape and introduced to the XPS sample chamber. A Kratos Azis Ultra XPS with a monochromatic Al Kα source maintained at a pressure below 2 × 10−9 Torr was used to collect binding energy measurements from 0 to 1,200 eV at a resolution of 1 eV with an illumination spot approximately 300 × 700 μm. Ejected photoelectrons were collected in a hemispherical electron energy analyzer that was positioned normal to the sample surface. Spectra of the C 1s and O 1s were collected for 3,000 ms with the detector aperture set to 20 mm. Because of the low abundance and low cross section of photoelectron emission, photoelectrons from the B 1s orbital were collected for 300 ms with the detector aperture set to 160 mm. Although this decreased the detector resolution, it provided a high enough count rate for detection of B 1s photoelectrons. Spectra of the C 1s and O 1s regions are shown under small aperture conditions; however, quantification of the chemical composition of the sample for all elements studied is reported under large aperture conditions for consistency. A charge neutralizer was used for all measurements, and all peak positions are reported with respect to the C 1s peak at 285 eV.
All peaks were fit to a Shirley baseline, and the integrated area (Ix) of the photoelectron peaks of interest was calculated using the Kratos XPS software. The areas of these peaks were then scaled by a sensitivity factor, Sx, of 0.278, 0.780, and 0.159 for C 1s, O 1s, and B 1s peaks, respectively. The atomic fraction of each species, Cx, was then calculated using Equation 1:
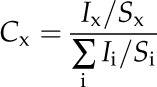 |
(1) |
The atomic fraction was then multiplied by 100 to obtain the percent composition of each element.
To determine whether there were any changes in the type of bonds that carbon was making in the cell walls, high-resolution spectra of the C 1s region were collected with the aperture set to 2 mm. These spectra were baseline corrected and fit to Gaussian functions using the OPUS software package. Individual contributions to the deconvoluted spectra were integrated to calculate the relative abundance of C-(C,H), C-(N,O), and C = O bonds.
Assay of [eATP] in Growth Media of Apyrase-Suppressed Seedlings
Seeds were planted on standard MS media with 1% Suc and 1% agar. Twenty 3-d-old seedlings per well were transferred to liquid culture in six-well plates, which had 3 mL MS media with 1% Suc per well. Each plant line was grown in 4 µm estradiol prepared in dimethyl sulfoxide (DMSO) to induce suppression of APY1 and control media with an equal volume of DMSO only. Each six-well plate contained six biological replicas of the same plant line and treatment, and the treatments were wild type and R2-4A in DMSO only and wild type and R2-4A in 4 µm E. One six-well plate was prepared for each of the days after planting that media samples were collected.
After transfer to liquid culture, seedlings were grown for the indicated number of days with constant shaking and constant light at 22–25°C. Two hours before media collection, the plates were removed from the shaker and kept still. Three individual samples of 50 µL media were removed from each well and flash frozen in liquid nitrogen. Samples were stored in −40°C until ATP measurements were made. After media collection, pictures were taken of several seedlings per treatment to measure root length and verify expected phenotypes (Supplemental Table S4).
The [ATP] in the growth media was determined using the ENLITEN ATP Assay System from Promega. Reactions were carried out on 96-well plates, and luminescence was detected using the Mithras LB940 microplate reader (Berthold Technologies). Media samples were thawed, placed on the plate quickly, and kept on ice for ≤30 min before ENLITEN reagent was added. In each reaction, 40 µL ENLITEN reagent was added to 10 µL media sample. An ATP standard curve from 10−7 to 10−12 m was included on each plate. Lumen measurements were evaluated by the Dixon’s Q test for outliers. In all cases, at least four replica samples were included in the average.
Sequence data from this article can be found in The Arabidopsis Information Resource data libraries under the accession numbers provided in the figures and tables. APY1 and APY2 accession numbers are AT3G04080 and AT5G18280, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. GO Biological Processes category analysis of genes significantly up- or down-regulated after APY suppression.
Supplemental Figure S2. Lignin-stained vascular tissue in wild-type and R2-4A primary roots.
Supplemental Figure S3. Diagram of homogalacturonan segment before and after boron-mediated cross-linking.
Supplemental Table S1. Genes up- or down-regulated when APY is suppressed.
Supplemental Table S2. Common promoter motifs in five wall peroxidases.
Supplemental Table S3. Comparison of carbon, oxygen, and boron content of the primary roots of wild-type and estradiol-induced R2-4A mutants.
Supplemental Table S4. Average primary root length of seedlings grown in liquid culture for media ATP concentration tests.
Supplemental Table S5. Among genes that change expression when APY is suppressed, those in the GO term “Extracellular Region” are overrepresented.
Supplemental Table S6. No significant differences in gene expression changes of auxin transporters/carriers in estradiol-induced R2-4A mutants.
Supplementary Material
Acknowledgments
We thank the Texas Advanced Computing Center for the use of resources, Guy Thompson for critical reading of the manuscript, and Marianna Grenadier for preparation of the figures.
Glossary
- DMSO
dimethyl sulfoxide
- FTIR
Fourier transform infrared
- IR
infrared
- MAP
mitogen-activated protein
- MS
Murashige and Skoog
- PME
pectin methylesterase
- Ws
ecotype Wassilewskija of Arabidopsis
- RNAi
RNA interference
- qRT-PCR
quantitative real-time-PCR
- ECM
extracellular matrix
- eATP
extracellular ATP
- [eATP]
concentration of extracellular ATP
- [ATP]
concentration of ATP
- cDNA
complementary DNA
Footnotes
This work was supported by the National Science Foundation (grant nos. IOS 0718890 and IOS 1027514 to G.C. and S.J.R.) and by the Army Research Office (grant no. W911NF–10–1–0280 to I.F.G. and J.W.D.). L.J.W. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and is an Alfred P. Sloan Foundation Research Fellow.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Almagro L, Gómez Ros LV, Belchi-Navarro S, Bru R, Ros Barceló A, Pedreño MA. (2009) Class III peroxidases in plant defence reactions. J Exp Bot 60: 377–390 [DOI] [PubMed] [Google Scholar]
- Aquea F, Federici F, Moscoso C, Vega A, Jullian P, Haseloff J, Arce-Johnson P. (2012) A molecular framework for the inhibition of Arabidopsis root growth in response to boron toxicity. Plant Cell Environ 35: 719–734 [DOI] [PubMed] [Google Scholar]
- Bouwmeester K, Han M, Blanco-Portales R, Song W, Weide R, Guo LY, van der Vossen EAG, Govers F. (2014) The Arabidopsis lectin receptor kinase LecRK-I.9 enhances resistance to Phytophthora infestans in Solanaceous plants. Plant Biotechnol J 12: 10–16 [DOI] [PubMed] [Google Scholar]
- Chen YF, Shakeel SN, Bowers J, Zhao XC, Etheridge N, Schaller GE. (2007) Ligand-induced degradation of the ethylene receptor ETR2 through a proteasome-dependent pathway in Arabidopsis. J Biol Chem 282: 24752–24758 [DOI] [PubMed] [Google Scholar]
- Chiu TY, Christiansen K, Moreno I, Lao JM, Loqué D, Orellana A, Heazlewood JL, Clark G, Roux SJ. (2012) AtAPY1 and AtAPY2 function as Golgi-localized nucleoside diphosphatases in Arabidopsis thaliana. Plant Cell Physiol 53: 1913–1925 [DOI] [PubMed] [Google Scholar]
- Chivasa S, Murphy AM, Hamilton JM, Lindsey K, Carr JP, Slabas AR. (2009) Extracellular ATP is a regulator of pathogen defence in plants. Plant J 60: 436–448 [DOI] [PubMed] [Google Scholar]
- Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, Stacey G. (2014) Identification of a plant receptor for extracellular ATP. Science 343: 290–294 [DOI] [PubMed] [Google Scholar]
- Clark G, Roux SJ. (2009) Extracellular nucleotides: ancient signaling molecules. Plant Sci 177: 239–244 [Google Scholar]
- Clark G, Roux SJ. (2011) Apyrases, extracellular ATP and the regulation of growth. Curr Opin Plant Biol 14: 700–706 [DOI] [PubMed] [Google Scholar]
- Clark G, Torres J, Finlayson S, Guan XY, Handley C, Lee J, Kays JE, Chen ZJ, Roux SJ. (2010a) Apyrase (nucleoside triphosphate-diphosphohydrolase) and extracellular nucleotides regulate cotton fiber elongation in cultured ovules. Plant Physiol 152: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G, Wu M, Wat N, Onyirimba J, Pham T, Herz N, Ogoti J, Gomez D, Canales AA, Aranda G, et al. (2010b) Both the stimulation and inhibition of root hair growth induced by extracellular nucleotides in Arabidopsis are mediated by nitric oxide and reactive oxygen species. Plant Mol Biol 74: 423–435 [DOI] [PubMed] [Google Scholar]
- Djebali N, Mhadhbi H, Lafitte C, Dumas B, Esquerre-Tugaye MT, Aouani ME, Jacquet C. (2011) Hydrogen peroxide scavenging mechanisms are components of Medicago truncatula partial resistance to Aphanomyces euteiches. Eur J Plant Pathol 131: 559–571 [Google Scholar]
- Dufrêne YF, van der Wal A, Norde W, Rouxhet PG. (1997) X-ray photoelectron spectroscopy analysis of whole cells and isolated cell walls of gram-positive bacteria: comparison with biochemical analysis. J Bacteriol 179: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand C, Crèvecoeur M, Penel C. (2007) Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytol 174: 332–341 [DOI] [PubMed] [Google Scholar]
- Fleischer A, Titel C, Ehwald R. (1998) The boron requirement and cell wall properties of growing and stationary suspension-cultured chenopodium album L. cells. Plant Physiol 117: 1401–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franza T, Expert D. (2013) Role of iron homeostasis in the virulence of phytopathogenic bacteria: an ‘à la carte’ menu. Mol Plant Pathol 14: 429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442 [DOI] [PubMed] [Google Scholar]
- Gapper C, Dolan L. (2006) Control of plant development by reactive oxygen species. Plant Physiol 141: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazarian IG, Lagrimini LM, Mellon FA, Naldrett MJ, Ashby GA, Thorneley RNF. (1998) Identification of skatolyl hydroperoxide and its role in the peroxidase-catalysed oxidation of indol-3-yl acetic acid. Biochem J 333: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Riewe D, Fernie AR. (2010) The central regulation of plant physiology by adenylates. Trends Plant Sci 15: 98–105 [DOI] [PubMed] [Google Scholar]
- Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, Salt DE. (2013) Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc Natl Acad Sci USA 110: 14498–14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YR, Dong QY, Yu DQ. (2012) Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci 185-186: 288–297 [DOI] [PubMed] [Google Scholar]
- Ishii T, Matsunaga T, Pellerin P, O’Neill MA, Darvill A, Albersheim P. (1999) The plant cell wall polysaccharide rhamnogalacturonan II self-assembles into a covalently cross-linked dimer. J Biol Chem 274: 13098–13104 [DOI] [PubMed] [Google Scholar]
- Jeter CR, Tang WQ, Henaff E, Butterfield T, Roux SJ. (2004) Evidence of a novel cell signaling role for extracellular adenosine triphosphates and diphosphates in Arabidopsis. Plant Cell 16: 2652–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolie RP, Duvetter T, Van Loey AM, Hendrickx ME. (2010) Pectin methylesterase and its proteinaceous inhibitor: a review. Carbohydr Res 345: 2583–2595 [DOI] [PubMed] [Google Scholar]
- Kim SY, Sivaguru M, Stacey G. (2006) Extracellular ATP in plants: visualization, localization, and analysis of physiological significance in growth and signaling. Plant Physiol 142: 984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles AF. (2011) The GDA1_CD39 superfamily: NTPDases with diverse functions. Purinergic Signal 7: 21–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov SN, Dunford HB. (1996) Detailed model of the peroxidase-catalyzed oxidation of indole-3-acetic acid at neutral pH. J Phys Chem 100: 913–920 [Google Scholar]
- Liu X, Wu J, Clark G, Lundy S, Lim M, Arnold D, Chan J, Tang W, Muday GK, Gardner G, et al. (2012) Role for apyrases in polar auxin transport in Arabidopsis. Plant Physiol 160: 1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AJ, William Allwood J, Winder CL, Dunn WB, Heald JK, Cristescu SM, Sivakumaran A, Harren FJ, Mulema J, Denby K, et al. (2011) Metabolomic approaches reveal that cell wall modifications play a major role in ethylene-mediated resistance against Botrytis cinerea. Plant J 67: 852–868 [DOI] [PubMed] [Google Scholar]
- Markakis MN, De Cnodder T, Lewandowski M, Simon D, Boron A, Balcerowicz D, Doubbo T, Taconnat L, Renou JP, Höfte H, et al. (2012) Identification of genes involved in the ACC-mediated control of root cell elongation in Arabidopsis thaliana. BMC Plant Biol 12: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Höfte H. (2003) Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform InfraRed (FT-IR) microspectroscopy. Plant J 35: 393–404 [DOI] [PubMed] [Google Scholar]
- Moulder JF, Stickle WF, Sobol PE, Bomben KD. (1992) Handbook of X-Ray Photoelectron Spectroscopy. Physical Electronics, Inc., Chanhassen, MN [Google Scholar]
- Nakagami H, Soukupová H, Schikora A, Zárský V, Hirt H. (2006) A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol Chem 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N. (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA 109: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong FX, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nivens DE, Ohman DE, Williams J, Franklin MJ. (2001) Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol 183: 1047–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill MA, Ishii T, Albersheim P, Darvill AG. (2004) Rhamnogalacturonan II structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu Rev Plant Biol 55: 109–139 [DOI] [PubMed] [Google Scholar]
- Palin R, Geitmann A. (2012) The role of pectin in plant morphogenesis. Biosystems 109: 397–402 [DOI] [PubMed] [Google Scholar]
- Passardi F, Cosio C, Penel C, Dunand C. (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24: 255–265 [DOI] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C. (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9: 534–540 [DOI] [PubMed] [Google Scholar]
- Pau-Roblot C, Petit E, Sarazin C, Courtois J, Courtois B, Barbotin JN, Séguin JP. (2002) Studies of low molecular weight samples of glucuronans with various acetylation degree. Biopolymers 64: 34–43 [DOI] [PubMed] [Google Scholar]
- Pedreira J, Herrera MT, Zarra I, Revilla G. (2011) The overexpression of AtPrx37, an apoplastic peroxidase, reduces growth in Arabidopsis. Physiol Plant 141: 177–187 [DOI] [PubMed] [Google Scholar]
- Peer WA, Blakeslee JJ, Yang HB, Murphy AS. (2011) Seven things we think we know about auxin transport. Mol Plant 4: 487–504 [DOI] [PubMed] [Google Scholar]
- Potters G, Pasternak TP, Guisez Y, Jansen MAK. (2009) Different stresses, similar morphogenic responses: integrating a plethora of pathways. Plant Cell Environ 32: 158–169 [DOI] [PubMed] [Google Scholar]
- Potters G, Pasternak TP, Guisez Y, Palme KJ, Jansen MAK. (2007) Stress-induced morphogenic responses: growing out of trouble? Trends Plant Sci 12: 98–105 [DOI] [PubMed] [Google Scholar]
- Ramstedt M, Nakao R, Wai SN, Uhlin BE, Boily JF. (2011) Monitoring surface chemical changes in the bacterial cell wall: multivariate analysis of cryo-x-ray photoelectron spectroscopy data. J Biol Chem 286: 12389–12396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RJ, Hayes JE, Post A, Stangoulis JCR, Graham RD. (2004) A critical analysis of the causes of boron toxicity in plants. Plant Cell Environ 27: 1405–1414 [Google Scholar]
- Ridley BL, O’Neill MA, Mohnen D. (2001) Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57: 929–967 [DOI] [PubMed] [Google Scholar]
- Roux SJ, Steinebrunner I. (2007) Extracellular ATP: an unexpected role as a signaler in plants. Trends Plant Sci 12: 522–527 [DOI] [PubMed] [Google Scholar]
- Rudnick G. (2008) Vesicular ATP transport is a hard (V)NUT to crack. Proc Natl Acad Sci USA 105: 5949–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmi ML, Clark G, Roux SJ. (2013) Current status and proposed roles for nitric oxide as a key mediator of the effects of extracellular nucleotides on plant growth. Front Plant Sci 4: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. (2005) The expansin superfamily. Genome Biol 6: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller M, Massalski C, Kurth T, Steinebrunner I. (2012) The Arabidopsis apyrase AtAPY1 is localized in the Golgi instead of the extracellular space. BMC Plant Biol 12: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Chiang VL. (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39: 519–525 [DOI] [PubMed] [Google Scholar]
- Song CJ, Steinebrunner I, Wang XZ, Stout SC, Roux SJ. (2006) Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis. Plant Physiol 140: 1222–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinebrunner I, Wu J, Sun Y, Corbett A, Roux SJ. (2003) Disruption of apyrases inhibits pollen germination in Arabidopsis. Plant Physiol 131: 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Dowd PF, Johnson ET, Hum-Musser SM, Musser RO. (2012) Effects of elevated peroxidase levels and corn earworm feeding on gene expression in tomato. J Chem Ecol 38: 1247–1263 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Gilroy S, Jones AM, Stacey G. (2010) Extracellular ATP signaling in plants. Trends Cell Biol 20: 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WQ, Brady SR, Sun Y, Muday GK, Roux SJ. (2003) Extracellular ATP inhibits root gravitropism at concentrations that inhibit polar auxin transport. Plant Physiol 131: 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J, Rivera A, Clark G, Roux SJ. (2008) Participation of extracellular nucleotides in the wound response of Dasycladus Vermicularis and Acetabularia Acetabulum (Dasycladales, Chlorophyta). J Phycol 44: 1504–1511 [DOI] [PubMed] [Google Scholar]
- Tsang DL, Edmond C, Harrington JL, Nühse TS. (2011) Cell wall integrity controls root elongation via a general 1-aminocyclopropane-1-carboxylic acid-dependent, ethylene-independent pathway. Plant Physiol 156: 596–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H. (2012) Defective root growth triggered by oxidative stress is controlled through the expression of cell cycle-related genes. Plant Sci 197: 30–39 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincken JP, Schols HA, Oomen RJ, McCann MC, Ulvskov P, Voragen AG, Visser RG. (2003) If homogalacturonan were a side chain of rhamnogalacturonan I. Implications for cell wall architecture. Plant Physiol 132: 1781–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasinghe RR, Swanson SJ, Okada SF, Garrett MB, Kim SY, Stacey G, Boucher RC, Gilroy S, Jones AM. (2009) Touch induces ATP release in Arabidopsis roots that is modulated by the heterotrimeric G-protein complex. FEBS Lett 583: 2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Steinebrunner I, Sun Y, Butterfield T, Torres J, Arnold D, Gonzalez A, Jacob F, Reichler S, Roux SJ. (2007) Apyrases (nucleoside triphosphate-diphosphohydrolases) play a key role in growth control in Arabidopsis. Plant Physiol 144: 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Peiffer M, Luthe DS, Felton GW. (2012) ATP hydrolyzing salivary enzymes of caterpillars suppress plant defenses. PLoS ONE 7: e41947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Liu YS, Wu JY. (2008) The signaling role of extracellular ATP and its dependence on Ca2+ flux in elicitation of Salvia miltiorrhiza hairy root cultures. Plant Cell Physiol 49: 617–624 [DOI] [PubMed] [Google Scholar]
- Zhang KW, Bhuiya MW, Pazo JR, Miao YC, Kim H, Ralph J, Liu CJ. (2012) An engineered monolignol 4-O-methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis. Plant Cell 24: 3135–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.