Abstract
In recent years attention has been focused on the utilization of microorganisms as alternatives for industrial and nutritional applications. Considerable research has been devoted to techniques for growth, extraction, and purification of high-value lipids for their use as biofuels and biosurfactants as well as high-value metabolites for nutrition and health. These successes argue that the elucidation of the mechanisms underlying the microbial biosynthesis of such molecules, which are far from being completely understood, now will yield spectacular opportunities for industrial scale biomolecular production. There are important additional questions to be solved to optimize the processing strategies to take advantage of the assets of microbial lipids. The present review describes the current state of knowledge regarding lipid biosynthesis, accumulation, and transport mechanisms present in single-cell organisms, specifically yeasts, microalgae, bacteria, and archaea. Similarities and differences in biochemical pathways and strategies of different microorganisms provide a diverse toolset to the expansion of biotechnologies for lipid production. This paper is intended to inspire a generation of lipid scientists to insights that will drive the biotechnologies of microbial production as uniquely enabling players of lipid biotherapeutics, biofuels, biomaterials, and other opportunity areas into the 21st century.
Keywords: lipid droplets, microbial lipids, triacylglycerols, sterol esters, polyhydroxyalkanoate
Introduction
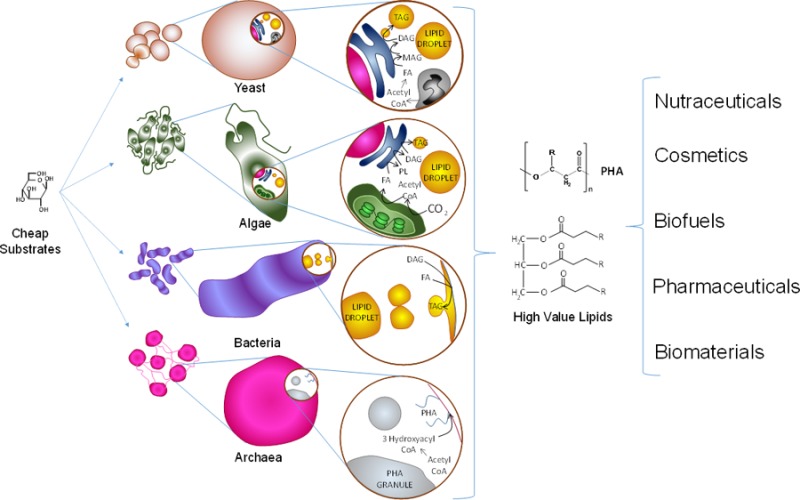
Single-cell microorganisms (SCM) constitute an emerging alternative to source high-value lipids for a series of growing markets demanding low-cost, high-quality alternatives. SCM as a broad class display a series of advantages when compared to plants and animals as lipid sources. In addition to being more genetically accessible, SCM are capable of producing greater diversity and storing higher percentages of lipids. Therefore, their productivity per volume and energy input can be up to 5 or 6 times that of plants and even more when compared to animal sources.1,2 Additionally, single-cell microorganism architecture and circuitry tend to be more straightforward than those in plant or animal cells. For example, there are fewer organelle compartments in fungal cells than in plant cells,3 and some microalgal metabolic pathways (e.g., fatty acid synthesis) are encoded by single-copy genes, whereas plants have multiple genes encoding proteins with redundant enzymatic activities.4 SCM can be readily grown under controlled conditions, and the key metabolic and biochemical questions can be answered more simply and quickly than in complex multicellular systems. In principle, SCM can achieve greater sustainability to alleviate the increasing problem of sourcing oils for both the fuel and human consumption markets, thus mitigating the continuous increase in commodity oil prices. Microbial lipids can also become sources of safe and clean biomaterials (e.g., biosurfactants) at reduced costs and continuous availability.
A complete mechanistic understanding of cellular machinery is the key to moving from principle to practice in lipid production. The purpose of this paper is to provide an overview of the current knowledge of the different cellular and biochemical mechanisms involved in neutral and polymeric lipid accumulation within four main single-cell microbial groups: yeasts, microalgae, bacteria, and archaea. Special emphasis is given to the production of triacylglycerols (TAG) and polyhydroxyalkanoates (PHA) as the hallmark lipids accumulated in eukaryotes and prokaryotes, respectively. Insights are provided on the mechanistic differences existing in each group during the transformation of substrates (e.g., acetyl coenzyme A), formation of intermediate pools (e.g., fatty acyl chains), allocation to intracellular lipid pools (e.g., polar lipid pools or neutral lipid pools), and the architecture of lipid accumulation storage (e.g., lipid droplets).
In microorganisms, different types of neutral lipids are accumulated by different types of SCM. For example, in oleaginous yeast, TAG and/or sterol esters (SE) are the primary neutral lipids accumulated inside the cell,5 whereas in specific types of bacteria and archaea polyhydroxyalkanoates (PHA), and to a lesser extent TAG and wax esters,6,7 are preferentially stored. These lipids are currently viewed as valuable potential biofuel sources, high-value compounds in the food and pharmaceutic industries, building blocks for biomaterials, potential tools to treat chronic diseases, and natural alternatives for the production of oleochemicals, among many other uses. Neutral lipids follow distinct cellular accumulation strategies compared to polar lipids, such as phospholipids (PL), galactolipids, and sulfolipids among others. However, in most cases, both classes share key biosynthetic steps.
Depending on cell needs, the cell is capable of transforming polar lipids into neutral lipids and vice versa due to the different pools present in strategic cellular locations. For instance, polar lipids are pooled and may be mobilized from the different cell membranes. Key membranes involved in the accumulation of lipids include the plasma membrane in both eukaryotes and prokaryotes, and in the case of eukaryotes the endoplasmic reticulum (ER), peroxisome, mitochondrial, plastidial (and their variants, such as apicoplastidial), or even thylakoid membranes, depending on the organism. Key intermediates, such as acetyl coenzyme A (Ac-CoA), malonyl coenzyme A (mal-CoA), acyl coenzyme A (acyl-CoA), and glycerol-3-phosphate (G3P) are also pooled in strategic cellular locations such as the cytosol, plastidial stroma, and mitochondrial intermembrane space. The distribution of the different biosynthetic enzymes plays a key role in achieving the correct lipid traffic toward accumulation. The accumulation of intracellular neutral lipid is a highly reductive process, requiring more NADPH and ATP than the storage of glucose and glucose derivatives.8
Accumulation of Lipids in Single-Cell Microorganisms: An Overview
Accumulation of lipids is a biological process that fulfills several roles in microorganisms. Storage of lipids contributes to cell growth, cell division, stress response, and as energy storage for survival.9 In some cases, lipid accumulation plays a role in pathogenity.7,10−13 Some SCM have the capacity to accumulate lipids >20% of their weight when exposed to an environmental stress, such as the lack of a key nutrient.14−16 These are referred to as oleaginous SCM (OSCM).17
When growing on limiting concentrations of a key nutrient (typically nitrogen) and with sufficient or excess carbon sources, OSCM will stop their replication processes and utilize the available carbon to synthesize and store it in the way of reduced lipids. In the case of TAG and SE, the accumulation process has four main stages. In the first stage, OSCM shift their metabolic machinery to generate a pool of Ac-CoA, as well as NADPH, which is the reducing power driving fatty acid (FA) synthesis forward18,19 The strategies vary among different classes of OSCM, and details will be discussed in the following sections. In the second stage, Ac-CoA is carboxylated to produce mal-CoA, transferred to acyl carrier protein (ACP), and further transformed into an acyl ACP by sequential turns within the fatty acid synthase (FAS). There are two types of FAS: Type I is an integrated enzyme complex (common in eukaryotic cytoplasm), and type II is a dissociated version in which all subunits are independent (common in prokaryotes and in organelles of prokaryotic origin in primary and secondary endosymbionts, such as plastids and mitochondria). Type I can be subdivided in type IA, present in fungi (α6β6 complex in the cytoplasm), and type IB, present in animals (as cytoplasmic α2 dimers). For reviews describing type I FAS and type II FAS, the reader is referred to the works of Schweizer and Lu et al., respectively.20,21 In the third step, depending on the cell’s needs and the type of SCM, the acyl chain might end in a specific type of lipid pool, such as membrane lipid (coupling the acyl chain to G3P, Ac-CoA, or other glycerol-based backbone to form PL or other types of polar lipid) or can eventually be incorporated into a neutral lipid droplet core (via the construction of TAG or SE molecules). The nature, location, and dynamics of this stage vary among species and are explained more in detail in the following sections. In this stage, further elongation and desaturation of the acyl chains may also occur, and these phenomena vary throughout the different domains of life. Finally, the fourth stage involves the formation of intracellular lipid droplets (LD). Variations occur depending on the organism and the environmental conditions.
In the case of prokaryotes, accumulation of TAG or SE is less common. In some members of the Gram-positive actinomycete group,6,22 such as Mycobacterium, Rhodococcus, Nocardia, Dietzia, and Streptomyces, TAG are readily accumulated. Interestingly, bacterial TAG biosynthesis occurs frequently in the environment because actinomycetes are the most abundant microorganisms in soil.22 Accumulation of TAG in Gram-negative bacteria has been reported only in species from the genus Acinetobacter, but only as a minor component of the neutral accumulated lipid. Wax esters are the main compound stored in these species.6 For the rest of the Gram-negative bacteria, the main purpose of fatty acid synthesis is to become membrane lipid precursors.6,18,23−25 However, several studies have successfully transformed model bacteria such as Escherichia coli by inserting the required genes to become oleaginous, with remarkable success.26−28 In addition, TAG have been reported as major components of the neutral lipid of the cyanobacterium Nostoc commune, although no indications were obtained for the storage of lipid inclusions in the cells.6,29 In the case of archaea, TAG accumulation of archaea is yet to be reported.11
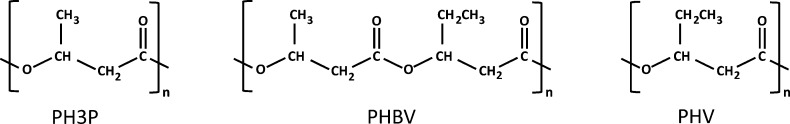
It is more common in prokaryotes to accumulate PHA, comprising specialized lipids such as poly(3-hydroxybutyrate) (P3HB), polyhydroxyvalerate (PHV), or copolymers such as poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV). Figure 1 shows the structures of such lipids. PHA possess thermoplastic and elastomeric properties and are recyclable materials that can be easily degraded into carbon dioxide and water.30 The uses and applications of these polyesters have been thoroughly reviewed elsewhere.30 The most promising are as film formation, paper coating,31 foils and diaphragms,32 and multiple-use packages, because the melted polymers have low viscosity, permitting the injection molding of objects with thin walls. The end product is very hard and can be used at temperatures from −30 to 120 °C.30
Figure 1.
Chemical structures of three different types of PHA, synthesized and accumulated in certain prokaryotes.
Polyhydroxyalkanoate accumulation in prokaryotes can also be divided into four stages. The first stage involves the generation of a suitable cytoplasmic Ac-CoA pool. The second one involves the synthesis of the hydroxyalkanoate (HA) monomer or monomers, which can change depending on the species and substrates. The third stage consists of polymerizing or copolymerizing the monomers into PHA chains, and the fourth one involves the formation of the intracellular PHA LD.
In both prokaryotes and eukaryotes, accumulation of neutral lipids results in the formation of intracellular fat bodies typically called lipid droplets (LD). LD are also called lipid bodies, fat bodies, or adiposomes, and in the case of plants they have also been termed oleosomes, spherosomes,22,33 or plastoglobules, the latter if their location is inside the plastid.11 For the present discussion, the term lipid droplet will be used. In most of the eukaryotes LD are intracellular sphere-like particles composed of a core of neutral lipids, such as TAG or SE or both, coated by a monolayer of polar lipids, mainly PL, and decorated with a series of proteins.11,33−35 The polar heads face the cytoplasm, and the nonpolar tails face the inside, where the neutral lipids reside. The inserted proteins within the polar lipid monolayer play functional, structural, and regulatory roles in the life cycle of LD.35−37 In the case of polyhydroxyalkanoate-accumulating prokaryotes, they form LD, which are also called lipid granules or carbonosomes.38,39 In some higher organisms (e.g., humans), lipid droplet interaction with other cellular organelles, and their dynamics is closely related to progression of metabolic diseases, such as obesity, fatty liver, type 2 diabetes mellitus, and atherosclerosis.36
Therefore, it has become apparent that LD are not just a static storage compartment in the cell, but rather a dynamic organelle that interacts with other organelles within the cell and actively participates in the intricate cellular processing symphony,40,41 to the point that some authors consider LD as specialized organelles in cells.42
As described in later sections, strategies for lipid accumulation in SCM vary among the different types of SCM, and it is important to understand these differences to improve and/or create productive strategies that can become economically as well as technologically feasible for the generation of biofuels as well as nutritional lipid metabolites such as long-chain polyunsaturated fatty acids (LC-PUFA).
The Case of Yeast
Yeasts are single-cell eukaryotic fungi that display heterotrophic behavior. Thus, they are capable of metabolizing carbon from simple sugars or from other simple compounds such as glycerol. When oleaginous yeasts encounter an environmental stress, such as a limiting nutrient (e.g., nitrogen), they shift their metabolic machinery to stop synthesizing proteins and nucleic acids and, thus, begin to allocate the available carbon in the form of reduced lipids. Other examples of limiting nutrients can be phosphorus or magnesium.6 In practice to date, nitrogen has been extensively used because it seems to yield higher lipid accumulation when compared to other nutrients.43 The carbon to nitrogen ratio is a useful tool to construct nitrogen-rich or nitrogen-depleted media. For example, it has been shown that for yeasts, poor lipid accumulation occurs in media in which the carbon to nitrogen (C:N) ratio is <20, whereas ideal lipid production occurs in a C:N ratio range of 30–80. The optimal C:N ratio for lipid accumulation varies greatly with the microbial species, strain, and carbon sources present in the growth medium.44 In yeasts, oleaginicity depends on the ability to produce Ac-CoA, the necessary precursor of fatty acids, in an effective manner.45,46
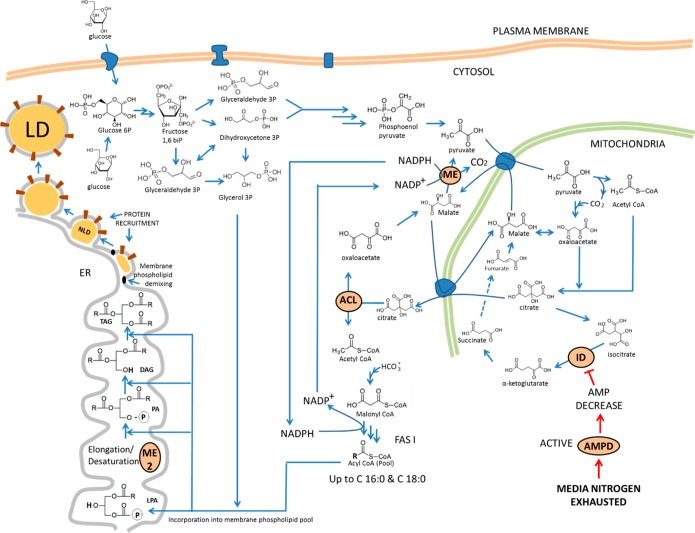
The conversion of 1 mol of Ac-CoA to fatty acids requires the formation of 2 mol of NADPH, which constitutes the reducing power necessary to drive the reaction forward.45−47 The metabolic steps for lipid accumulation in yeast can be divided into four main stages, as mentioned before (see Figure 2).18,42,45−48
Figure 2.
Schematic representation of metabolic steps involved in lipid accumulation in yeasts. The diagram also includes the connection between fatty acid synthesis, triacylglycerol synthesis, and lipid droplet formation. Abbreviations: ME, malic enzyme (cytosolic); ME 2, malic enzyme (ER membrane, responsible for fatty acid desaturation); ACL, ATP:citrate lyase; ID, isocitrate dehydrogenase; NLD, nascent lipid droplet; LD, lipid droplet; ER, endoplasmic reticulum; LPA, lysophosphatidic acid; PA, phosphatidic acid, FAS I, fatty acid synthase I; DAG, diacylglycerol; TAG, triacylglycerol; AMPD, AMP deaminase.
Stage 1. Production of Acetyl CoA and NADPH
This stage begins with depletion of nitrogen in the medium. The cell responds by activating AMP deaminase and inducing an acute decrease of cellular AMP content. In mitochondria of oleaginous species, the decrease in cellular AMP levels causes isocitrate dehydrogenase (IDH) activity to decrease or even stop.19 As a consequence, the production of α-ketoglurarate drops and the tricarboxylic cycle (TCA) is dramatically reduced or even stopped. Therefore, isocitrate will begin to build up inside the mitochondria. To reverse this situation, aconitase transforms isocitrate back to citrate (CIT), leading to an accumulation of CIT in mitochondria. CIT is transported by an antiport protein, known as citrate/malate translocase (CMT), from the mitochondria to the cytoplasm. In the cytoplasm, CIT is cleaved to form oxaloacetate and Ac-CoA, by ATP:citrate lyase (ACL). ACL is an enzyme that is exclusively present in oleaginous species.18,42,49,50 Cytosolic oxaloacetate is reduced by malate dehydrogenase (MD) to form malate. Cytosolic malate is converted to pyruvate by malic enzyme (ME), encoded by ME1. This reaction generates NADPH51,52 and is coupled to a series of parallel reactions as well. The first is the carboxylation of pyruvate to form oxaloacetate by pyruvate carboxylase inside mitochondria. Mitochondrial oxaloacetate is further reduced to form malate by MD. This malate is then transported outside the mitochondria and decarboxylated by ME. This reaction takes NADP+ and converts it to NADPH. This NADPH is the reducing power required to convert Ac-CoA into fatty acids. Thus, pyruvate carboxylase, MD, and ME are known as the transhydrogenation machinery.19,42 The resulting pyruvate completes the cycle and goes inside the mitochondria (Figure 2). ME is present in all fungi, but its role in supplying NADPH for de novo lipogenesis is preponderant in oleaginous species.18 It has been found that, upon nitrogen limitation, ME changes from isoform D to isoform E, which supplies NADPH for de novo lipogenesis.18
Stage 2. Biosynthesis of Fatty Acyl Chains
Ac-CoA is converted by acetyl-CoA carboxylase (Acc) to mal-CoA, which is then used to synthesize fatty acids. In yeast, cytosolic Acc is encoded by ACC1 and mitochondrial Acc is encoded by HFA1. The mitochondrial version closely resembles in both its molecular mass and amino acid sequence the cytoplasmic one.20 Acc uses biotin as a cofactor to transfer CO2 to Ac-CoA in a two-step process. It is a trifunctional enzyme, harboring a biotin carboxyl carrier protein domain, a biotin–carboxylase domain (where CO2 binds to biotin), and a carboxyl-transferase domain (where CO2 is transferred to Ac-CoA to yield mal-CoA).53 The fatty acyl chain is built in a type I cytosolic fatty acid synthase (FAS I). It is an α6β6 complex encoded by two genes, FAS1 (β subunit) and FAS2 (α subunit).53 It is suggested to contain 6 equivalent sites of FA synthesis with a total of 42 catalytic domains, organized in a ring-like structure.54 However, variations to this architecture exist and probably may affect the performance in de novo lipogenesis.42 Loading of substrates to FAS varies among life domains. For example, Ac-CoA is always the basis of FA biosynthesis and is the substrate of β-ketoacyl ACP synthase (KSa) in bacteria.20,21 Yeasts load Ac-CoA first to KSb through ACP before condensation.20 The sequence of reactions in yeast FAS II is condensation of Ac-CoA with mal-CoA (via KS), reduction (via ketoacyl reductase, KR), dehydration (via a dehydratase, DH) and further reduction (via enoyl reductase EAR) in a repetitive manner until palmitoyl-ACP is formed. Release of the acyl moiety is a key step that differs from species and is currently a subject of intense research. Yeasts employ malonyl-palmitoyl transacylase (MPT) to transfer the acyl chain from acyl ACP to acyl-CoA.18,53 In algae, the acyl chain can be released in three ways, depending on the cell’s needs. First, it can be hydrolyzed from ACP as a free fatty acid by means of a thioesterase (TE). Second, it can be transferred either to G3P or monoacylglycerol-3-phosphate (MAG3P) through an acyltransferase (AT) in the chloroplast.8,55,56 Lastly, if the acyl chain is destined to leave the plastid, it can also be released in the acyl-CoA form by acyl-coenzyme A synthetases (ACS).8,57 It has been found that ACS play a key role in regulating in each compartment the internal acyl-CoA pools by esterification of FA to CoA. The localization of the pools is maintained due to Acyl-CoA’s not being able to cross the intracellular membranes. The final fatty acid composition of different types of microalgae is in great part dependent on the activity of these enzymes. For example, microalgal chain length specific TE have been reported, which can release acyl moieties of specific length (e.g., C 12:0 or even C 8:0). This type of fatty acid is more suitable for the production of gasoline and jet fuel.56
In contrast, Gram-positive bacteria first form acyl-phosphate from acyl-CoA (via PlsX), which is then transferred onto G3P by PlsY. Gram-negative species use PlsB acyltransferase only to load an acyl group directly onto G3P from acyl ACP.18,51,58
Stage 3. Allocation of Acyl Moieties to either Polar or Neutral Lipid Pools
In yeast, acyl-CoAs released from cytosolic FAS I system are channeled to the ER membrane. There, a series of esterifications to a G3P backbone occur, also known as the Kennedy pathway. The first step consists of esterifying the acyl moiety from acyl-CoA to G3P or dihydroxyacetone 3-phosphate via glycerol-3-phosphate acyl transferase (G3PAT), on the sn-1 position. G3PAT is encoded in yeast by GAT1 and AYR1, when the acceptor is G3P or dihydroxyacetone 3-phosphate, respectively.18 Gat1 does not exhibit a particular preference for acyl-CoA, whereas AYR1 prefers palmitoyl-CoA, thus defining FA composition. A second acyl moiety can be attached on the sn-2 position to generate phosphatidate (PA), catalyzed by acyl-CoA:lysophosphatidic acyltransferase (LPAAT). PA plays a key role in the regulation of ACC1, FAS1, and FAS2 genes, as it is involved in an autoregulatory loop directed by the concentrations of inositol and choline in the cytosol and PA in the ER membrane.18,20,53,59−62 Expression of upstream activating sequence (UASINO)-operated genes changes in parallel to PA, inositol, and choline concentrations. In the case when PA is going to be transformed into TAG, it has to be transformed into diacylglycerol (DAG) first. This can happen in two ways. First, the phosphate group can be directly hydrolyzed by a phosphatidate phosphatase (PAP) to create DAG. PAP is encoded by PAH1 and requires a magnesium ion to perform hydrolysis.60,62 It is regulated by lipids, nucleotides, and phosphorylation. The second one involves synthesizing cytidine diphosphate DAG (CDP-DAG) from PA, catalyzed by CDP-DAG kinase, encoded by CDS1.18,42 CDP-DAG is the precursor of the different PL in the ER membrane. It thus stays as either phosphatidylcholine (PC), phosphatidylethanolamine (PE), or another type of PL. Eventually, DAG is generated by cleavage of the PL via phospholipase C. Both alternatives are in metabolic interlock with each other. This creates a positive feedback system that channels PA into the TAG pathway if CDP-DAG concentration is higher than DAG concentration or directs PA to the membrane pool in the opposite situation.18,60−62 Lastly, DAG is transformed into TAG by esterifying an acyl moiety from Acyl CoA catalyzed by an Acyl CoA:DAG acyltransferase (ADAT), encoded by either DGA1 or LRO1,42 depending on whether DAG is generated directly from PA via PAP or it comes from the ER membrane PL pool, respectively.
Further elongation and desaturation of fatty acids also occur in the ER membrane. Desaturation requires fatty acids to be attached to the ER membrane PL. Here another ME, different from the cytosolic malic enzyme, catalyzes the conversion of malate to pyruvate and the consequent reduction of NADP to NADPH. NADPH then couples to a series of electron transfer reactions involving cytochrome b5 reductase and further activating the desaturase, which adds a double bond on the fatty acid chain at the expense of converting 1 mol of oxygen into a mole of water.3,
Ergosterol is the major sterol present in yeast. For a review in the biosynthesis of ergosterol backbone, the reader is referred to the work of Kristan.64 Yeast SE are synthesized via transesterification of ergosterol with acyl-CoA.5 In yeast, two acyl-CoA cholesterol acyltransferase (ACAT) related enzymes, ARE1P and ARE2P (encoded by ARE1 and ARE2), catalyze the reaction. Microscopic localization of green fluorescent protein hybrids and enzyme measurements showed that both ARE1P and ARE2P are localized to the ER.65 ARE1P esterifies ergosterol and its precursors with nearly equal efficiency with a slight preference for lanosterol, whereas ARE2P uses ergosterol as a preferred substrate.65 An acyl-CoA-independent pathway for the formation of SEs has not been identified in yeast.
Stage 4. Lipid Droplet Biogenesis
Lipid droplet biogenesis is an area of intensive current research, and there is emerging scientific evidence showing that in yeast it takes place between the two membrane leaflets of the ER.34,66,67 By mechanisms still not clearly understood there are spots in the ER membrane where there is a concentration of ADATs DGA1 and LRO1. The first one is responsible for TAG synthesis in the outer leaflet of the endoplasmic reticulum, whereas the second is responsible for TAG synthesis in the inner leaflet of the ER.18,42 The synthesized TAG begin to accumulate, generating a lens-like protrusion and promoting the recruitment of structural proteins. In yeast, PAT proteins (from the initials of perilipin, adipocyte differentiation-related protein, and TIP47)68 accumulate in the outer leaflet (see Figure 3), whereas in plants this role is played by oleosins. When there is enough accumulation of TAG between the leaflets, the outer buds off and the lipid droplet is formed (Figure 3).34,69−71
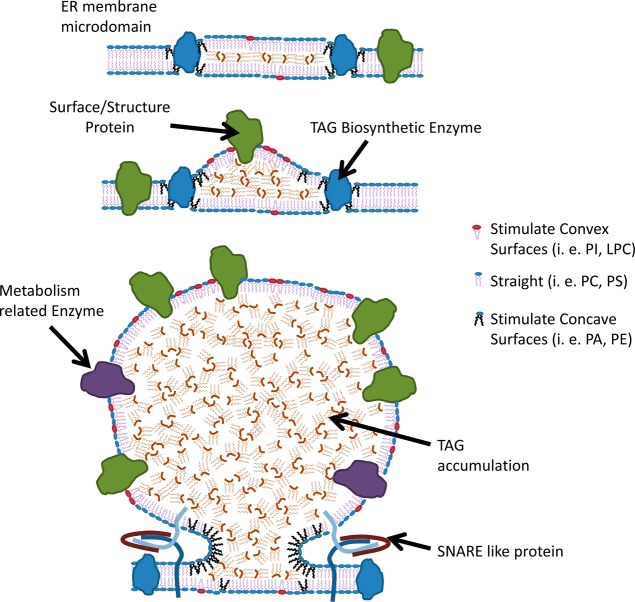
Figure 3.
Model of lipid droplet formation in the ER membrane. Adapted from ref (11).
There seems to be a functional relationship between the lipid droplet membrane and the ER membrane, because several studies have shown that certain functional proteins can migrate between the lipid droplet and ER membranes by mechanisms that do not require energy expenditure.72 In yeast, most ER proteins are also detected on the lipid droplet membrane. For example, experiments using yeast mutants unable to synthesize TAG revealed that LD were not formed. However, the proteins that were present in the wild forms in LD were also present in the ER of the yeast mutants, suggesting a relationship between ER proteins and lipid droplet proteins.69 In some cases, lipid droplet-localized proteins can relocate back to the ER, indicating that some continuity between the two organelles is maintained, even if only transiently, in a way that allows the two-way partitioning of proteins between the two compartments.33
In describing the functions of proteins embedded in the droplet monolayers, two mechanisms have been proposed. The first is a hydrophobic helix that interacts with the monolayer. Examples of this strategy are the multifunctional caveolin protein, which has been localized to both the plasma membrane and LD, and DGA1, the major enzyme catalyzing triacylglycerol synthesis.33 Both have similar topology. Their long internal hydrophobic stretch may enable them to be embedded in either bilayers or monolayers. The second mechanism is best represented by the previously mentioned PAT protein family. They display a four-helix bundle with great similarity to the N-terminal domain of apolipoprotein E.33,73 Upon binding to lipids, the apoE four-helix bundle opens to expose amphipathic helices that can bind the monolayer surfaces of lipoproteins. In an analogous manner, PAT proteins may bind to lipid droplets by embedding hydrophobic helices into the droplet surface. PAT proteins also share a common structural element: N-terminal 11-mer repeats that have an amphipathic helical structure.74 Different PAT proteins attach to different-sized LD. It is still unclear why or how they do it.
In the specific case of perilipins (members of the PAT family), there are five groups, and their abundance has been correlated with the abundance of TAG in the lipid droplet as well.35 Perilipins play crucial roles in regulating triacylglycerol hydrolysis by protein–protein interactions, suggesting physiological and regulatory functions.35 It is important to mention that perilipin-like proteins are not found in plants. As was already mentioned, functionally similar oleosins and caleosins are present in plants instead.11
There is increasing evidence that phospholipid demixing occurs during the birth of LD.69 The accumulation of lipids that promote concavity decreases the energy burden required for budding. It has been shown that lipid droplet PL contain more lysophospholipids and less sphingomyelin and PA compared to the total membrane.33,69 In silico studies proposed a packing parameter S to quantify the degree of convexity/concavity in PL.69 Lysophosphatidylcholine and phosphatidylinositol (PI) promote a convex shape where their lipid footprint areas are much smaller than their headgroup areas. Their S values are <1. Conversely, PE and PA induce a concave shape. These PL have S values >1.69 Lipids with S values <1 adopt a convex surface, favoring lipid droplet formation. This is diagrammed in Figure 4.
Figure 4.
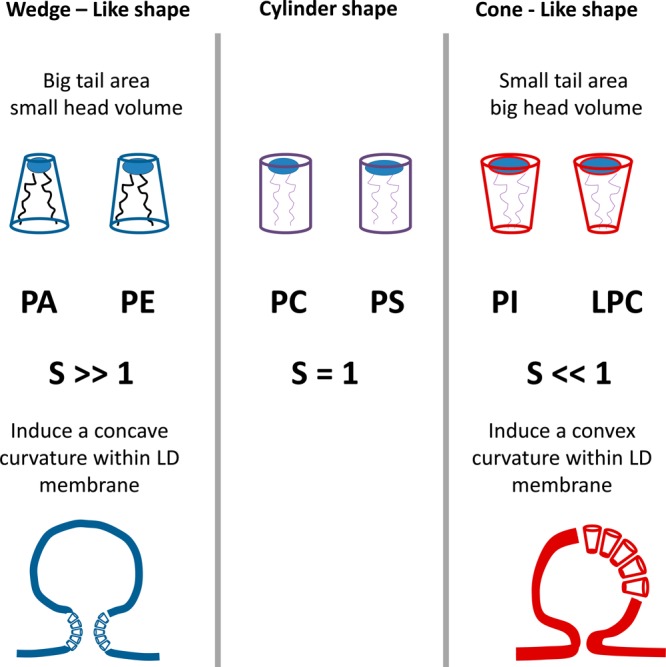
Configurations adopted by different phospholipids, which can affect the curvature during lipid droplet formation.69
The Case of Microalgae
Eukaryotic microalgae are classified into nine divisions: Glaucophyta, Rhodophyta (red algae), Heterokontophyta, Haptophyta, Cryptophyta, Dinophyta (dinoflagellates), Euglenophyta, Chlorarachniophyta, and Chlorophyta (green algae)55,75,76 All of these divisions include single-cell strains, which can be either motile, nonmotile, or both. In terms of their nutritional strategies, microalgae can be divided into obligate heterotrophs, obligate photoautotrophs, facultative mixotrophs, and obligate mixotrophs. Most algal divisions contain colorless heterotrophic species that can obtain organic carbon from the external environment either by taking up dissolved substances (osmotrophy) or by engulfing bacteria and other cells as particulate prey (phagotrophy).76 In most cases, lipid accumulation strategies in microalgae and some single-cell protists (both primary and secondary endosymbionts) follow the four-stage lipid accumulation strategy described for yeast. However, several differences are present in each stage. In addition, further studies are demonstrating that lipid biosynthesis pathways in microalgae are not a simple mirror image of what happens in higher plants, which have been more thoroughly studied.77,78 It seems likely that regulation of triacylglycerol synthesis and breakdown in microalgae tends to obey a stress response phenomenon, whereas in plants it follows a developmental phenomenon. For a review comparing lipid metabolism between microalgae and plants, see Liu.4
Stage 1. Production of Acetyl-CoA and NADPH
Microalgae differ from yeast in the location of the acetyl-CoA pools within the cell. Microalgae display plastidial and cytosolic acetyl-CoA pools, which are key for lipid accumulation. In contrast, yeast’s main acetyl-CoA pool used for lipid accumulation is located in the cytosol. There is an additional acetyl-CoA pool present in mitochondria as well as a mitochondrial lipid biosynthesis pathway employing a type II FAS (different from plastidial FAS II) both in algae and in yeasts.18,77,79−83 They play important roles in different cell processes, such as RNA processing, mitochondrial lipoic acid synthesis, and protein lipoylation. However, mitochondrial lipid synthesis is not the main avenue for lipid accumulation and will not be covered in the present work. For a review of mitochondrial lipid biosynthesis and its relevance in cell function, the reader is referred to the work of Hiltunen and co-workers.79−81
Plastids present in algae play a key role in de novo lipid biosynthesis (for a review of plastid evolution and diversification, see the work of Keeling84). In photoautotrophic microalgae, photosynthesis provides an endogenous source of plastidial acetyl-CoA, although more than one pathway may contribute to maintaining the acetyl-CoA pool.55 For instance, in photoautotrophic microalgae plastidial pyruvate can be sourced via transformation of photosynthesis-derived glyceraldehyde-3-phosphate to phosphoenolpyruvate (PEP). PEP is irreversibly converted to pyruvate (see Figure 5) by pyruvate kinase (PK).55,85 Finally, the plastidial pyruvate dehydrogenase complex (PDH) catalyzes the oxidative decarboxylation of pyruvate to produce plastidial Ac-CoA, CO2, and NADH. PDH contains three components: E1 (pyruvate dehydrogenase, composed of E1α and E1β subunits), E2 (dihydrolipoyl acyltransferase), and E3 (dihydrolipoamide dehydrogenase). It is possible that photosynthesis-derived pyruvate is the major contributor to plastidial Ac-CoA for de novo fatty acid synthesis. However, in mixotrophic grown cultures of heterokonts such as Nannochloropsis sp., incorporation of acetate directly into lipids occurs.8,86−89 The acetyl-CoA synthetase (ACSIN) converts acetate to plastidial Ac-CoA. Additionally, a study showed that under nitrogen deprivation Chlamydomonas is capable of changing its metabolism from converting acetate to glucose to a more direct incorporation of acetate into fatty acids by down-regulating glyoxylate cycle activity and gluconeogenesis.4,90
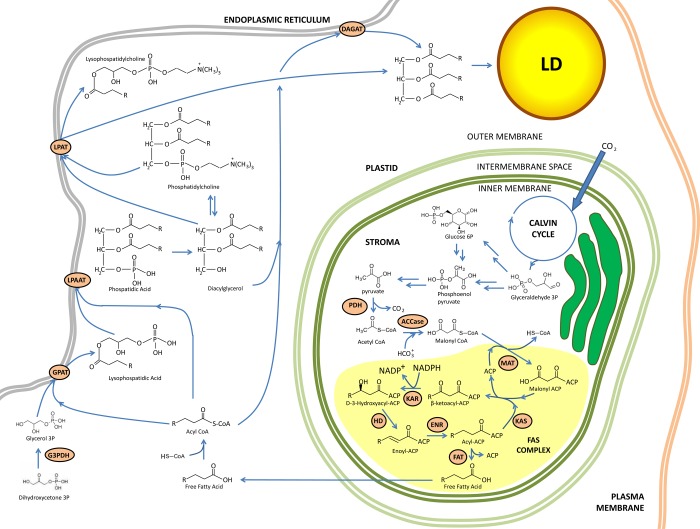
Figure 5.
Simplified overview of lipid biosynthesis and lipid droplet formation in photosynthetic algae. Adapted from ref (56). Abbreviations: ER, endoplasmic reticulum; ACCase, acetyl-CoA carboxylase; ACP, acyl carrier protein; DAGAT, diacylglycerol acyltransferase; DHAP, dihydroxyacetone phosphate; ENR, enoyl-ACP reductase; FAT, fatty acyl-ACP thioesterase; G3PDH, glycerol-3-phosphate dehydrogenase; GPAT, glycerol-3-phosphate acyltransferase; HD, 3-hydroxyacyl-ACP dehydratase; KAR, 3-ketoacyl-ACP reductase; KAS, 3-ketoacyl-ACP synthase; LPAAT, lysophosphatidic acid acyltransferase; LPAT, lysophosphatidylcholine acyltransferase; MAT, malonyl-CoA:ACP transacylase; PDH, pyruvate dehydrogenase complex.
In microalgae, the cytosolic Ac-CoA pool is mainly fueled by the release of mitochondrial CIT to the cytosol and further cleaved into oxaloacetate and Ac-CoA by ACL. Cytosolic Ac-CoA is the building block used for LC-PUFA elongation in the ER. Concomitant production of NADPH by ME generates the reductive power necessary to drive plastidial de novo fatty acid synthesis forward. Availability of NADPH can increase the reaction velocity of Acc 2 (see stage 2) and ACL.8 Fatty acid synthesis is an energy-demanding process due to the activity of elongases and desaturases. For instance, the formation of a C18 FA requires 54 NADPH from oxygenic photosynthesis.8 Other functions of malic enzyme in algae may include delivery of CO2 from the TCA for the plastidial ribulose-1,5-bisphosphate carboxylase (Rubisco). Studies also suggest the existence of a plastidial ME (absent in yeast), which can provide electrons for plastidial FA synthesis.8,91−93 For a review about plastidial ME see the work of Maier et al.94
Some microalgae are capable of accumulating intracellular starch, such as the thoroughly studied microalgal model Chlamydomonas. Starch synthesis is an example of how carbon partitioning might play a key role in lipid accumulation. One study showed that in wild-type Chlamydomonas, TAG accumulated only after the maximum amount of starch was reached, whereas starchless Chlamydomonas mutants initiated TAG accumulation earlier and reached a higher level than wild-type strain.4,95 Therefore, it is possible to engineer high TAG algal strains by eliminating competing carbon utilization pathways such as starch synthesis to maximize lipid biosynthesis.
Stage 2. Biosynthesis of Fatty Acyl Chains
In algae, de novo fatty acid biosynthesis occurs primarily in the plastid. The committed step in plastidial fatty acid synthesis is the conversion of plastidial Ac-CoA to mal-CoA by Acc1. The three domains of the homomeric Acc1 are located on a multifunctional polypeptide encoded by a nuclear gene.8,96 It works via biotin carboxylation and subsequent carboxyl transfer to Ac-CoA.55,97 Some microalgae contain only Acc1, such as I. galbana. Others, such as T. pseudonana and P. tricornutum, contain two homomeric Accases, Acc1 (described above) and a cytosolic Acc (Acc2), which uses cytosolic Ac-CoA to generate mal-CoA. The latter plays a role in LC-PUFA elongation in the ER membrane.8,98
FAs are synthesized in microalgae’s plastid via a dissociated type II FAS, containing discrete, monofunctional enzymes encoded by distinct genes.99 In plastidial FAS II mal-CoA is loaded to ACP via malonyl-CoA:ACP transacylase encoded by FABD. Malonyl-ACP is used in the cyclic condensation reactions to extend the acyl group to palmitoyl ACP or stearoyl ACP. The acyl ACP can be released from FAS II in several ways: It can be hydrolyzed by a fatty acyl-ACP thioesterase located in the chloroplast envelope, forming a free fatty acid, or it can be transesterified from ACP to CoA via (ACS), or it can even be coupled to either G3P or MAG3P through an AT in the chloroplast.8,55
There are some heterotrophic microalgal species that contain a cytosolic type I FAS, synthesized from one or two polypeptides,8,99 different from plastidial type II FAS. For example, Aurantiochytrium(100) contains a type I FAS, which synthesizes saturated C14:0 and C16:0. The synthesized free FAs and the absence of genes homologous to a type II TE may indicate integration of TE activity into the synthase.101
Several attempts have been made to overexpress specific enzymes in the lipid biosynthetic pathways. In the cases of Acc and KS (KS III), overexpression failed to increase lipid accumulation.102
Stage 3. Allocation of Acyl Moieties to either Polar or Neutral Lipid Pools
The released acyl moieties as free fatty acids destined to stay within the plastid may be further desaturated (typically to hexadecatrienoic acid HDT, C16:3n-4) and coupled to a monogalactosyldiacylglycerol (MGDG) backbone. MGDG, along with other types of galactosylglycerides (GG), is a major component of photosynthetic membranes in microalgae and in some cases may be even more abundant than PL.8,75 In plants, the FA combination of GG can be traced back to their biosynthetic pathways; the so-called eukaryotic molecular species (C18/C18) of GG are synthesized outside the chloroplast in the eukaryotic pathway, and the prokaryotic molecular species (C18/C16) are synthesized in the plastid via the prokaryotic pathway.8,103 Microalgae differ from plants in that most C20 FAs are synthesized outside the chloroplast and are present in both the eukaryotic-like (C20/C20, C18/C18) and prokaryotic-like (C18/C16, C20/C16) molecular species.8,75,104,105 Therefore, microalgae’s GG are referred to as “C20/C20” or “C20/MLC” (where MLC means medium to long chain) rather than eukaryotic- and prokaryotic-like GG.
When acyl ACP’s are esterified to either G3P or MAG3P, they can join the plastidial PL pool. In plants two ATs catalyze these reactions. The first is a soluble enzyme that prefers oleoyl-ACP as substrate. The second one resides on the inner chloroplast envelope membrane and preferentially selects palmitoyl-ACP.78 In some algal species, such as C. reinhardtii and P. lutheri, PC is absent in both plastidial and extraplastidial phospholipid pools. Instead, they contain the non-phosphorus betaine lipid diacylglyceryl-N,N,N-trimethylhomoserine (DGTS), which has similar physicochemical properties, as a major membrane component.106−110 This is a significant difference against higher plants. It has been suggested that DGTS may have a role in lipid droplet formation similar to that of PC in higher plants.106 However, they contain other types of phospholipids, such as PA, PE, and PI.8,75
Acyl ACPs synthesized in plastidial type II FAS can be transesterified with CoA via ACS. These enzymes regulate the acyl pools in different cellular compartments. Thus, stearoyl ACP can leave the FAS II complex via transesterification by a series of ACS responsible for maintaining both intraplastidial and cytosolic acyl-CoA pools.8,111 In both cases, acyl-CoA is destined to leave the plastid and enter the ER membrane for further oxygen-dependent elongation and desaturation, to become long-chain polyunsaturated fatty acids (LC-PUFA), such as eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA). Two types of desaturases can be distinguished in microalgae. The first are front-end desaturases containing an N-terminal cytochrome b5 domain and insert the new double bond between the FA carboxyl group and a possible existing double bond. One example is the high substrate specific plastidial-located Δ12 desaturase, identified in P. tricornutum, which desaturates palmitoleic acid C16:1n-7 to hexadecadienoic acid 16:2n-4.8 The second group comprises the less common ω6/ω3 desaturases capable of inserting a new double bond between the FA methyl end and a pre-existing double bond.8,112
Stearoyl CoA can be desaturated by an ER membrane-bound Δ9 desaturase to oleyl-CoA and then linked to a glycerol backbone for further processing. Some microalgae contain a plastidial Δ9 desaturase, capable of direct desaturation of stearoyl ACP to be then transesterified to CoA and sent to the ER for elongation and desaturation. In most microalgae, n-3 LC-PUFA are more abundant than n-6 LC-PUFA,113 whereas in filamentous fungi, such as mortierella and mucor, n-6 LC-PUFA are more common.3,19 In microalgae the most common pathways for EPA and DHA synthesis are the n-3 and n-6 pathways, but variations of the theme occur. In the n-3 pathway, oleic acid already linked to a glycerol backbone in the ER membrane is desaturated to linoleic acid (LA), via a Δ12 desaturase (encoded by FAD2). A third double bond inserted by an n-3 Δ15 desaturase gives α-linolenic acid (LNA), which is further desaturated to produce stearidonic acid (SA) by a Δ6 desaturase. SA is then elongated to C20:4 n-3 (eicosatetraenoic acid, EA) and finally desaturated by a Δ5 desaturase to produce EPA. In most DHA-producing microalgae, EPA undergoes an additional elongation and a Δ4 desaturation to create DHA.113 This is a different, less complicated strategy for DHA synthesis from EPA, compared to the Sprecher pathway, present in mammals, which involves an additional elongation and a β-oxidation. In contrast to the n-3 pathway, some microalgae synthesize EPA via the n-6 pathway, following Δ6 desaturation of LA to γ-linolenic acid (GLA), elongation to dihomo-γ-linolenic acid (DGLA), creation of a fourth double bond by Δ5 desaturase to produce arachidonic acid (ARA), and a final desaturation to create EPA via a n-3 Δ17 desaturase.113 Some microalgae express Δ9 elongase and Δ8 desaturase, allowing them to synthesize EPA in a different way compared to the n-3 and n-6 pathways.113 In a variation of the n-3 pathway, LNA is elongated by a Δ9 elongase to eicosatrienoic acid (ETA) and desaturated by the Δ8 desaturase, creating EA. On the other hand, a variation of the n-6 pathway using this pair of enzymes involves Δ9 elongation of LA to eicosadienoic acid (EDA) and Δ8 desaturation to DGLA.75 Thus, microalgae can switch from n-6 fatty acids to n-3 fatty acids via the n-3 Δ15 desaturase and the n-3 Δ17 desaturase. In some species of the Thraustochytrids, it is also possible to switch from n-6 docosapentaenoic acid (DPA) to DHA, via an n-3 Δ4 desaturase.
In addition to these pathways, there is a different n-3 LC PUFA biosynthetic pathway present in one of the three genera of heterotrophic Thraustochytrids, namely Auranthiochytrium, based on an anaerobic polyketide synthase pathway (PKS).8,114 A large multifunctional enzyme complex carries out the multitude of individual reactions, utilizing mal-CoA and producing free n-3 LC-PUFAs. The major free FAs DPA (22:5n-6) and DHA (22:6n-3) are then activated to acyl-CoA and incorporated into TAGs.8,101 Because PKS does not require aerobic desaturation, the pathway is energetically favorable compared to the membrane-bound desaturases and elongases.115
TAG synthesis in microalgae follows the Kennedy pathway in the ER in a similar fashion as yeast (see Figure 5). However, an acyl-CoA-independent mechanism for triacylglycerol synthesis in some plants and yeast has been reported.55,116 This pathway uses PL as acyl donors and DAG as the acceptor, and the reaction is catalyzed by the enzyme phospholipid:DAG acyltransferase.
There are two diacylglycerol acyltransferase families identified in Chlamydomonas, involved in the final step of triacylglycerol synthesis: type one (DGAT), encoded by DGAT1, and type two (DGTT), encoded by five DGTT genes, which do not share sequence similarity. Two independent studies showed the expression levels of DGAT1 and DGTT1 increased considerably following nitrogen deprivation.4,90,117 However, it remains unclear which of these DGATs are primarily responsible for the accumulation of TAGs under this condition or whether individual isoforms have specific roles.
Triacylglycerol synthesis can also occur within the plastid, through a series of acyl-ACP esterifications to plastidial G3P, catalyzed by plastidial ATs similar to the Kennedy pathway in the ER.118 The involvement of GG (the major polar lipid family in plastidial membranes in several microalgae) in this process remains to be elucidated.
During light–dark cycles, many microalgae initiate triacylglycerol storage during the day and deplete those stores at night to support cellular ATP demands and/or cell division.56 This cycling has to be taken into account when scaling up processes for production of lipids from algae. This variable may be key to the overall success of an open pond process or a closed photoreactor process.
Stage 4. Lipid Droplet Biogenesis
In contrast to yeast, LD can emerge from plastidial membranes. They can grow facing the cytosol or toward the inside of the plastid, facing the stroma, which is the major aqueous fluid surrounding the thylakoids inside the chloroplast.4,118 The mechanisms underlying the orientation of lipid droplet growth in plastid membranes are not well understood. When LD grow toward the inside of the plastid (facing the stroma), they can also be called plastoglobules (PTG). PTG can be considered to be functionally equivalent to cytosolic LD, but differ in three major respects. First, they are confined to the stroma. Second, they can assume several different forms including rods, fibers, and globules; and third, they are bound by a specific family of proteins, variously termed plastoglobulins, plastid lipid-associated proteins, and fibrillins.11 Oleosins (the major lipid droplet proteins present in plants) are not present in green algae, but a major lipid droplet protein was identified by applying proteomics in Chlamydomonas and shown to modulate lipid droplet size.1 Orthologues of the major lipid droplet protein are present in other green algae. A wide variety of parameters affect the abundance of LD in Chlamydomonas, and there is ample evidence that turnover of LD plays crucial roles in cellular lipid or carbon homeostasis.119
The Case of Bacteria
In bacteria, the most frequent types of neutral and polymeric lipids synthesized and accumulated are PHA, TAG, wax esters (WE), and, to a lesser extent, SE. This review focuses on the first two. Most bacteria are capable of synthesizing either PHA or TAG. However, a special situation occurs in bacteria such as Rhodococcus ruber, and other related bacteria, capable of accumulating both types of lipids from unrelated carbon sources such as glucose.7,11
The mechanisms of wax ester accumulation have been reviewed elsewhere.120,121 In a nutshell, an acyl-CoA is transformed to an aldehyde via an NADPH-dependent acyl-CoA reductase (encoded by Acr1), which is then reduced to an alcohol via an also NADPH-dependent fatty aldehyde reductase. The alcohol is then transesterified to an acyl-CoA via an enzyme displaying both wax ester synthase (WS) and acyl-CoA:diacylglycerol acyltransferase (DGAT) activities (abbreviated WS/DGAT).7
The only report of the presence of an sterol ester-synthesizing enzyme in prokaryotes was provided by Thornton et al.122
Accumulation of TAG in Bacteria
To date, triacylglycerol biosynthesis has been detected only in aerobic heterotrophic bacteria and in cyanobacteria.6 In most bacteria, accumulation of TAG and other neutral lipids, such as WE, is stimulated by a carbon source present in excess, together with limited nitrogen in the medium.
Biosynthesis of Fatty Acyl Chains
Structurally, fluorescence staining experiments showed that lipid biosynthesis starts at peripheral lipid domains close to the cytoplasm membrane.22,123 Biochemically, fatty acid biosynthesis begins in bacteria with Acc, a heterotetrameric enzyme encoded by four genes, accA, accB, accC, and accD.21 Fatty acids in bacteria are synthesized via a dissociated type II FAS. Each monofunctional protein is encoded by a specific gene.
Ac-CoA is converted to mal-CoA and transferred to ACP by malonyl-CoA:ACP transacylase (FabD in bacteria) to form malonyl-ACP. A first condensation of malonyl-ACP with acetyl-CoA by β-ketoacyl-ACP synthase III (FabH) to form β-ketobutyryl-ACP and CO2 initiates a cycle that can elongate the fatty acyl-ACP by two carbon units for each cycle until a saturated fatty acid of 16 or 18 carbons is made. KSI (FabB) and KSII (FabF) are responsible for the subsequent elongation cycles of the growing acyl-ACP chain.
To balance chain initiation with growth and utilization in bacterial FASII, KSIII, enoyl-ACP reductase (FabI in bacteria), and β-ketoacyl-ACP reductase (Fab G in bacteria) are under negative feedback control by long-chain acyl-ACPs.18 Long-chain acyl-ACPs directly control reductase activities; consequently, FASII is biased to catalyze forward if these products are withdrawn from the system by any conversion, including phospholipid or triacylglycerol synthesis.
In some bacteria, FAS II yields unsaturated fatty acids, which play a key role in bacterial membrane fluidity and function.21,124 Unlike ER desaturases present in microalgae or yeast, which introduce double bonds into the completed fatty acid chains at the expense of oxygen, the bacterial FAS II system can also desaturate fatty acids anaerobically, because it does not require molecular oxygen.21 FabA introduces the double bond at the 10-carbon intermediate, forming cis-2-decenoyl-ACP. It additionally isomerizes it into trans-3-decenoyl-ACP, which is further elongated by FabB.18,21 Two genes, fabA and fabB, are the key players in this pathway and occur together in bacteria that produce unsaturated fatty acids.
Some Rhodococcus and Nocardia bacteria are capable of incorporating branched or phenylic groups into their intracellular TAG if the corresponding substrate is fed to the medium. For example, Nocardia globerula strain 432 accumulated TAG containing the branched fatty acid 4,8,12-trimethyltridecanoic acid after cells were fed pristane (a branched alkane),6 and TAG with a subfraction containing phenyldecanoic acid residues were detected in cells of R. opacus PD630 after phenyldecane was fed as sole carbon source.6,125
Allocation of Acyl Moieties to either Polar or Neutral Lipid Pools
The enzymes involved in the esterification of the glycerol moiety probably act via sequential acylation of the sn-1, -2, and -3 positions of G3P, with the removal of the phosphate group occurring before the final acylation step.6 The first esterification is catalyzed by a G3PAT using either acyl-CoA or acyl ACP to form lysophosphatidic acid. A second acylation to lysophosphatidic acid gives PA, which is the first branchpoint for the synthesis of TAG and PL, because it can be converted to CDP-DAG, the precursor of the different PL species in bacterial membranes. DAG itself is also at a metabolic branchpoint that divides phospholipid and triacylglycerol formations, because it acts as a precursor for TAG, PC, and PE biosynthesis. In addition, DAG can also be derived from PL by the action of phospholipase C7,126 The distribution of acyl groups on the hydroxyl groups of the glycerol backbone is nonrandom, as has been demonstrated for R. opacus PD630.6,127 The shorter and saturated fatty acids were esterified to the hydroxyl group at position 2, whereas unsaturated fatty acids were preferentially found at position 3. This distribution in bacterial TAG is different from the TAG of mammals and plants, where the longer unsaturated fatty acids are found at position sn-2.128
The final step in wax ester and triacylglycerol biosyntheses in bacteria is catalyzed by WS/DGAT.7,128 WS/DGAT is encoded by atfA and is not related to any known AT involved in the formation of TAG and WE in eukaryotes. It has also been shown that WS/DGAT is localized at the bacterial cytoplasmic membrane, presumably attached to the inner leaflet of the membrane, probably via ionic interactions.22 For a detailed review about WS/DGAT, the reader is referred to the work of Waltermann et al.7 Alternative pathways for triacylglycerol synthesis that do not involve DAG in bacteria have also been reported and may involve an enzyme similar to that reported by Dalquist in yeast,116 which catalyzes the formation of TAG from the transesterification of an acyl donor (e.g., acyl-CoA) to PL.6 This follows observations that following a double knockout of WS/DGAT genes in A. borkumensis, cells were still capable of substantial triacylglycerol accumulation in LD.11,130
Triacylglycerol/Wax Ester Lipid Droplet Biogenesis
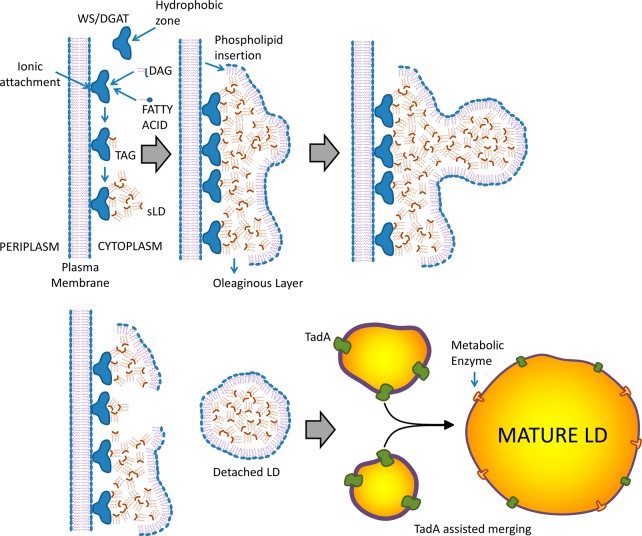
Triacylglycerol/wax ester lipid droplet biogenesis presumably begins by allocation of newly formed TAG in a hydrophobic zone within WS/DGAT. As time goes by, more WS/DGAT attaches to the membrane, and cumulative synthesis advances, presumably leading to the formation of very small triacylglycerol agglomerates, depicted by Wältermann et al. as small lipid droplets (sLD).22 sLD apparently recruit PL from the membrane by a mechanism that is still not well understood, forming a layer toward the cytoplasmic side (see Figure 6). The agglomeration of phospholipid-coated sLD appears in microscopy as an oleaginous layer just parallel to the membrane.22 Then, accumulation of sLD in a given point gives birth to LD, which are coated by PL and detach from the oleaginous layer. These droplets will acquire more TAG and grow. It has been suggested that acquisition of additional TAG is by merging with other LD freshly synthesized from the oleaginous layer via coalescence22 and/or through a protein identified as TadA.11 An interesting study by Ding and co-workers36 examined the proteome of Rhodococcus sp. RHA 1 (a Gram-positive bacteria capable of accumulating TAG)36 grown both in nitrogen-abundant (not favoring lipid accumulation) and nitrogen-depleted conditions (favoring lipid accumulation). Using a combination of techniques, including LC-MS, SDS-PAGE, and immunoblot assays, they reported 228 lipid droplet-associated proteins, which clustered primarily into metabolism-related enzymes, transcriptional regulators, ribosome proteins, and cell division-related proteins. Interestingly, they identified two major proteins, ro02104 and PspA, which constituted about 15% of the total lipid droplet protein. According to their findings, the structure predicted for ro02104 resembles that of apolipoproteins, the structural proteins of plasma lipoproteins in mammals.
Figure 6.
Suggested mechanisms of neutral LD formation in bacteria. Adapted from ref (22).
Accumulation of PHA in Bacteria
In bacteria PHA comprise a complex class of storage polyesters, with almost 150 different hydroxyalkanoic acids known as constituents.6 A wide variety of Gram-positive as well as Gram-negative bacteria synthesize PHA. Examples include Pseudomonas, Bacillus, Ralstonia, Aeromonas, and Rhodobacter, among others. PHA are divided into two groups on the basis of the number of constituent carbon atoms in their monomer units: short chain length PHAs (SCL PHA) and medium chain length PHAs (MCL PHA). Monomers of SCL PHA are 3–5 carbon atoms long, compared to 6–14 carbon atoms in MCL PHA. In addition, SCL PHA are stiff and brittle with a high degree of crystallinity, whereas MCL PHA are flexible and have low crystallinity, tensile strength, and melting point.30
Production of Acetyl-CoA and Synthesis of Hydroxyalkanoate Monomers
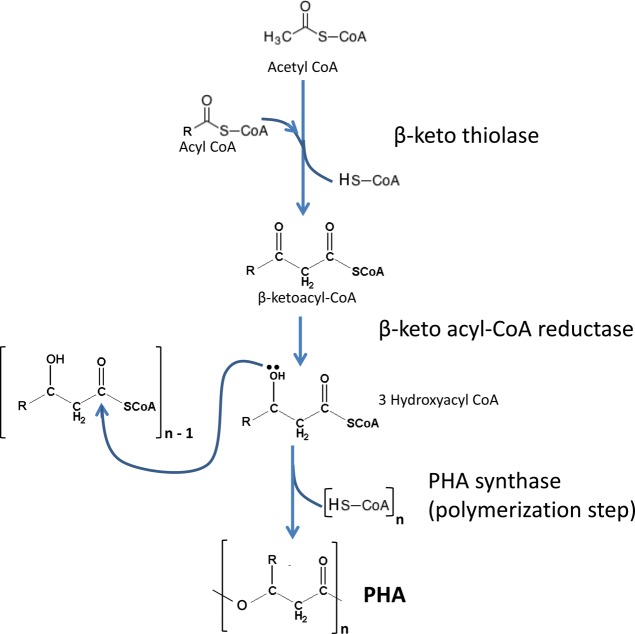
Accumulation of PHA starts with the creation of hydroxyalkanoate monomers from three different biosynthetic pathways. The first one involves incorporation of two acetyl-CoAs to form acetoacetyl-CoA, by the enzyme β-ketothiolase.11,30,131,132 Then acetoacetyl-CoA reductase converts acetoacetyl-CoA into 3-hydroxybutyryl-CoA. This pathway creates hydroxybutyrate (HB) monomers exclusively and is used by bacteria such as Cupriavidus necator and Azotobacter beijerinckii (see Figure 7).30,131,132 On a second pathway involving de novo fatty acid biosynthesis, monomers of different lengths can be formed by transesterifying 3-hydroxyacyl-ACP, an intermediate of FAS II to 3-hydroxyacyl-CoA, presumably by the enzyme acyl-ACP-CoA transacylase, encoded by phaG. This enzyme is the key link between de novo fatty acid synthesis and polyhydroxyalkanoate biosynthesis.30,133 This pathway is of biotechnological interest because it helps generate monomers for polyhydroxyalkanoate synthesis from structurally unrelated and simple, inexpensive carbon sources such as glucose or related simple sugars. On a third embodiment, monomers of different lengths can also be sourced from fatty acid β-oxidation pathway either by conversion of 2-enoyl-CoA by an R-specific enoyl-CoA hydratase, encoded by PhaJ, or by reduction of 3-ketoacyl-CoA, presumably by FabG or PhaB.30 In this case, the monomer composition is related to the carbon source used.
Figure 7.
Biosynthesis of PHA.
Polymerization and Copolymerization into Polyhydroxyalkanoate Chains
The resulting R-3-hydroxyacyl-CoA monomers are polymerized by polyhydroxyalkanoate synthase. The ability of microorganisms to synthesize a particular form of polyhydroxyalkanoate is mainly due to the substrate specificity of polyhydroxyalkanoate synthases. These enzymes are divided into four classes, depending on their structure and specificity. Class I enyzmes utilize CoA thioesters of 3-hydroxyalkanoates (3-HAs), 4-HAs, and 5-HAs comprising three to five carbon atoms.134 Species such as Pseudomonas putida and R. eutropha possess this class of enzymes. Members of class II display major specificity for monomers ranging from 6 to 14 carbon atoms. Enzymes from both classes consist of a single subunit of an average size of 60–70 kDa and are encoded by PhaC. In contrast, class III and class IV synthases are encoded by two genes, PhaC/PhaE, and PhaC/PhR respectively, and consist of two subunits. Class III members are capable of polymerizing preferably monomers ranging from three to five carbons, yet can utilize monomers from six to eight carbons as well.134 Species such as Allochromatium vinosum contain class III synthases, whereas class IV has been reported only in Bacillus sp.38 All polyhydroxyalkanoate synthases share a conserved cysteine as a catalytic site to which the growing polyhydroxyalkanoate chain is covalently attached. The active-site cysteine, histidine, and aspartate constitute a catalytic triad similar to esterases.38 Literature is available for an in-depth analysis of polyhydroxyalkanoate synthases.135−141
Polyhydroxyalkanoate Lipid Droplet Biogenesis
Two models currently exist that may explain the formation of in vivo polyhydroxyalkanoate LD (which are also called granules or carbonosomes38,39): the micellar and the budding models. The first one is based on the assumption that the polyhydroxyalkanoate synthase is present in the cell as a soluble enzyme, distributed throughout the cytoplasm. Once polymerization of substrate molecules (CoA-thioesters of suitable hydroxyalkanoic acids) starts, the nascent polyester chain converts the initially soluble enzyme into an amphipathic molecule and the increasingly hydrophobic polyhydroxyalkanoate chains aggregate into a micelle-like structure. Polyhydroxyalkanoate synthase remains attached to the surface of the granule and therefore becomes insoluble (see Figure 8). In this model, PL and proteins of the surrounding layer would gradually become incorporated as the self-assembled polyhydroxyalkanoate inclusion increases in size.38,39 This model requires the polyhydroxyalkanoate granule to be localized in the cytoplasm at all stages of formation. In contrast, the budding model assumes that polyhydroxyalkanoate synthase is associated with the inner face of the cytoplasmic membrane, either inherently or as soon as a polyhydroxyalkanoate chain emerges from the enzyme. In this case, biosynthesis of the polyester would be directed into the intermembrane space where the extending chains would accumulate until eventually the granules detach from the membrane and polyhydroxyalkanoate-specific surface proteins can be attached to the growing granules.38,39 Although the micelle model is supported by the fact that polyhydroxyalkanoate granules can be produced in vivo in the absence of membranes, most of the recently emerging evidence is in favor of the budding model.39 The major structural proteins present in polyhydroxyalkanoate granules are phasins (Phas). Phas may constitute approximately 5% (w/w) of total cellular proteins,142,143 and they play a main structural role in preventing polyhydroxyalkanoate granules from aggregating and in preventing the nonspecific attachment of other proteins to polyhydroxyalkanoate granules.142−144 In addition, phasins are presumably involved in the regulation of polyhydroxyalkanoate synthesis, polyhydroxyalkanoate degradation, polyhydroxyalkanoate granule size control,142,143,145,146 the formation of networks on the polyhydroxyalkanoate granule surface,147 and the distribution of polyhydroxyalkanoate granules during cell division. Another important group of proteins present in the external granule layer is the depolymerases, which are responsible of catalyzing polyhydroxyalkanoate breakdown. This is a relevant step in the role of polyhydroxyalkanoate accumulation as a survival mechanism in the absence of suitable energy/carbon sources, as was demonstrated many years ago in R. eutropha.148 Two groups have been identified: intracellular (PhaZs) and extracellular depolymerases. PhaZs have been investigated much less than extracellular depolymerases, and the mechanism by which intracellular native polyhydroxyalkanoate granules can be reutilized is still not well understood.39 Interestingly, the gene coding for PhaZs is located between two copies of phaC1 and phaC2 (both polyhydroxyalkanoate synthase genes) in all investigated bacteria that accumulate MCL PHA.38 In contrast, secreted depolymerases are used by most bacteria to assimilate PHA present in the environment from, for example, other nonliving cells.149
Figure 8.
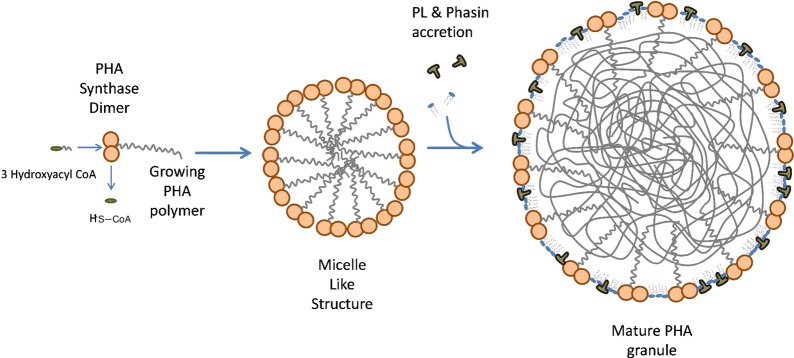
Proposed mechanism of PHA containing lipid droplet formation in bacteria (micelle model). Adapted from ref (22).
Polyhydroxyalkanoate granule synthesis and phasin production are tightly regulated by the effectiveness of the transcriptional regulator PhaR. Genes encoding proteins homologous to PhaR are widely distributed among SCL PHA producing bacteria, indicating an important role in the regulation of SCL PHA biosynthesis.39,150,151
The Case of Archaea
The discovery of the Archaea domain in 1977 revealed a novel class of microorganisms encountered in exceptional ecological niches such as high (thermophiles and hyperthermophiles) or low (psychrophiles) temperatures, acidic media (acidophiles an thermoacidophiles), anaerobic atmosphere (methanogens), and high salinity (halophiles).152,153 The unique chemical structure of their core membrane lipids is in part responsible for their adaptation to such hostile environments. Archaeal membrane lipids, in contrast to those of bacteria and eukaryotes, are made up of saturated chains containing methyl branches, attached to glycerol by ether linkages with a stereochemistry in the 2-position of the glycerol opposite that of conventional mesophilic lipids. (For a review on archaeal ether lipid structures, the reader is referred to the work of Jacquemet et al.152) Moreover, Archaea do not synthesize fatty acyl esters, which are the most common constituents of LD; instead, their lipids are based on isoprenoid chains.11 Therefore, no accumulation of TAG has been reported yet in Archaea. Despite these differences, however, evidence of polyhydroxyalkanoate accumulation was first reported in Haloarchaea back in 1972.154,155 The strains were called at that time “Halobacterium sp. from the Dead Sea”, but later identified as Haloarcula marismortui.156 Since then, strains of several other haloarchaeal genera, including Haloferax, Halobiforma, and Haloquadratum, have been found to accumulate PHA.142,156 As in bacteria, Archaea produce PHA under conditions of nutrient limitation but where carbon is available in excess.157−163 The mechanisms of polyhydroxyalkanoate accumulation within Archaea are beginning to be understood, and work is underway to elucidate the type of proteins involved in archaeal polyhydroxyalkanoate accumulation. Genes involved in polyhydroxyalkanoate biosynthesis in Haloarchaea were not recognized until recently, when the polyhydroxyalkanoate synthase genes were identified and characterized for Haloarcula marismortui and Haloferax mediterranei.158,159,161−165 These archaeal polyhydroxyalkanoate synthases are all composed of two subunits, PhaE and PhaC, and they are homologous to class III bacterial polyhydroxyalkanoate synthases but have a longer C-terminal extension in the PhaC subunit.164 The close similarity of archaeal and bacterial type III polyhydroxyalkanoate synthase genes and the lack of other polyhydroxyalkanoate gene types in archaea suggest that archaeal PHA originated from the horizontal transfer of an ancestral type III gene from a bacterium.11 Some authors suggest this transfer to have occurred already before Permian times.154
Genome-wide analysis of H. marismortui ATCC 43049164,165 revealed eight paralogues of a short-chain dehydrogenase/reductase, responsible for reduction of acetoacetyl-CoA to (R)-3-hydroxybutyryl-CoA (3-HB CoA), a monomer used by polyhydroxyalkanoate synthase to produce polyhydroxybutyrate (PHB). Another study164 demonstrated that a similar paralogue in H. hispanica, namely fabG1, encodes a PHA-specific acetoacetyl-CoA reductase responsible for providing 3-HB-CoA for polyhydroxyalkanoate biosynthesis in Haloarcula species. The authors concluded that the polyhydroxyalkanoate biosynthesis pathway from Ac-CoA, catalyzed by β-ketoacyl thiolase, acetoacetyl-CoA reductase, and polyhydroxyalkanoate synthase, as distributed in bacteria, likely also exists in the domain of Archaea. Nonetheless, they pointed out that for PHB-accumulating haloarchaeal Natrialba strain 56,159,160 no enzyme activity of acetoacetyl-CoA reductase or β-ketoacyl thiolase was detected in the crude extract, indicating that a different metabolic route toward production of PHB might be employed.
Polyhydroxyalkanoate synthase, putative enoyl-CoA hydratase, and two structural phasin-like proteins have been identified in haloarchaeal polyhydroxyalkanoate granule surfaces.142 The phasin-like proteins in Haloarchaea share some structural features with bacterial Phas, such as the presence of hydrophobic domains and a high α-helix content.142,166 After a genome-wide investigation into the 12 Haloarchaea that harbored the PhaP gene, most of these Archaea were found to possess a similar pha cluster, with five genes, namely, maoC-gap12-phaP-phaE-phaC, having the same organization as that in H. mediterranei.142 The extensive existence of this pha gene cluster was suggested as an indication of an evolutionarily conserved pha gene cluster unique to Haloarchaea.142
A promising approach is to develop archaeal species as industrial scale polyhydroxyalkanoate producers. In particular, several halophilic Archaea have the advantages of utilizing much cheaper carbon sources (including waste materials), as well as having less strict sterilization requirements, plus easier and more efficient methods for polyhydroxyalkanoate extraction.134
Future Perspectives
Microorganisms provide an exciting platform for the development of lipid technologies. In the course of evolution, they have developed elegant pathways to synthesize a wide array of lipids, providing a versatile and cost-effective approach for sourcing lipids to virtually all sectors of industry. The understanding of the biochemical and cellular mechanisms of lipid production, accumulation, and secretion will provide valuable insights on innovations to overcome the hurdles in microbial lipid utilization.
Ongoing studies using different “omics” approaches will provide a holistic view of flows and interactions between the different metabolic pathways involved in lipid accumulation. Tools such as next-generation sequencing, transcriptome analysis, proteomics, and mass spectrometry are already clearing out the missing links in single-cell lipid biosynthesis. This information will permeate in the creation and scaleup of more efficient processes.
It is thus important to project the potential applications and envision the frontiers to which this valuable toolset can lead the diverse fields of lipid technology. For example, in the field of biotherapeutics, microbial-based approaches have the flexibility to construct platforms to manufacture personalized lipid therapies starting as simply as LC PUFA combinations, appropriate to the metabolic phenotype of each individual167 and eventually becoming as complex and selective as personalized cancer interventions. This approach of personal medicine will carry improved benefits in the treatment of a range of conditions: in immunological diseases from infections to autoimmunities; in metabolic conditions from diabetes to cardiometabolic diseases; and in microbiota dysbiosis from IBS to IBD. Microbial technologies can also provide an alternative pathway for sourcing promising lipids and fatty acids that are unavailable in the market at present. For example, oil-producing microbes can be considered as appropriate vehicles in which foreign (plant) genes could be cloned for the production of commercially attractive fatty acids such as nervonic acid (C24:1 Δ15) obtained from Honesty (Lunaria),3 which is used in small amounts in the treatment of particular neuropathies. Another example is sterculic acid {ω-(2n-octylcycloprop-1-enyl)-octanoic acid},168 which has been considered as a treatment for certain cancers of the bowel. One more promising example is represented by the non-methylene-interrupted fatty acids (e.g., C20:3 Δ5, Δ11, Δ14), which can be obtained from Juniperus chinensis seed oil and is known to reduce the amount of arachidonic acid in certain phospholipid pools and, thus, act to alter eicosanoid signaling.169
In the field of energy and biofuels, understanding the mechanisms by which each group of microorganisms generates highly combustible lipids will foster the production of cost-effective fuels through sustainable processing and with improved performance. It is yet to be tested whether the biochemical mechanisms for amphipathic lipid secretion present in some microorganisms would work for the secretion of neutral lipids. This concept, if brought to practice, will significantly reduce the processing costs and render high-quality combustible lipids for the production of biofuels.
In the case of the food industry, a microbial-based approach will allow food companies to expand their core businesses by creating new product portfolios out of their current byproducts. Two promising examples are the production of cocoa butter analogues3 and the production of high-value oils out of spent agricultural materials.
In the field of biomaterials, microbial lipids are already being used for producing biosurfactants and bioplastics, replacing synthetic analogues due to their improved biodegradability and reduced cost. Companies are now emerging worldwide taking advantage of the microbial lipid toolset. Products such as Metabolix PHA and Biomer are examples on how microbial-derived plastics are gaining momentum in the market.
Microbial biosurfactants are beginning to generate improved results against their synthetic homologues. Their specific modes of action, low toxicity, relative ease of preparation, and widespread applicability are increasing their use in applications such as emulsifiers, wetting and foaming agents, functional food ingredients, detergents in petroleum, petrochemicals, environmental management, agrochemicals, cosmetics and pharmaceuticals, commercial laundry detergents, mining and metallurgical industries (for an overview of microbial surfactant applications, see Mukherjee171). The scope of applications goes to such an extent that the petroleum extractive industry is already researching the use of microbial lipid-based biosurfactants for increasing oil recovery yields from subterranean depots.171,172 The great era of chemistry culminating in the restructuring of the human condition in the 20th century has been defined by the principles of reductionism and simplicity. In contrast, the 21st century heralds the era of complexity for which the inherent biological diversity and information content of microorganisms will be key to adding value to all industrial chains.
A search performed on July 6, 2013, with the terms “microbial lipids” in the U.S. Patent Office “quick search” option retrieved 11905 results. Figure 9 shows a steady increase in U.S. patent publication since 2008, showing the increasing interest in the generation of intellectual property around this area of research.
Figure 9.
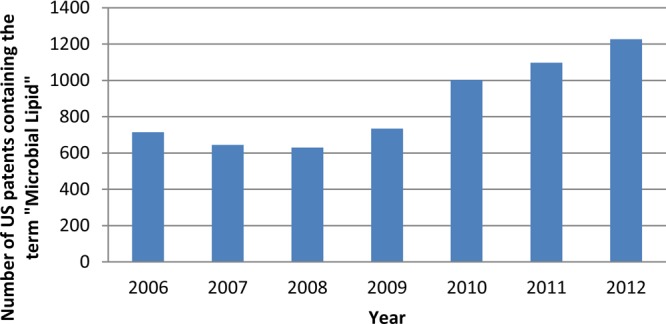
Number of U.S. patents containing the term “microbial lipids” per year.
Notice the increasing trend starting in 2008. The estimated patents by the end of 2013 according to the current trend will be around 1420, almost doubling the number registered in 2009.
The next step in microbial lipid technology is the integration of processes and applications to generate comprehensive, sustainable solutions. The immediate challenge is to combine desired metabolic pathways present in different species or strains to generate superior microbial species. Benefits will include improved yields, targeted lipidomic profiles, and structural features to synthesize in a sustainable and cost-effective way the lipid structures necessary to satisfy the breadth of needs of 21st century industry. For example, an appropriate integration of microbial-based lipid technologies will allow delivery of smart biofuels and even personalized edible oils derived from microorganisms and reporter-equipped biodegradable containers also derived from microorganisms. Currently the most used packaging material for commercial edible oils is polyethylene terephthalate, which is virtually a nondegradable plastic. With the appropriate use of microbial PHA, it could be possible to see in the same facility the production of high-value microbial oils (rich in a combination of LC PUFA), along with the production of polyhydroxyalkanoate-based bottles for its commercialization. Furthermore, additional microbial biotechnology platforms will be integrated to transform the byproducts either into biomass nutrients to make the process sustainable or even into another portfolio of high-value products, as a way to increase the business profitability (Figure 10). This would be a proposed example of a sustainable microbial-based industry.
Figure 10.
Conceptual flowchart for an integral, sustainable microbial-based edible oil process.
Microbial lipid technologies are a valuable toolset available for the future generation of scientists, which will help them push the boundaries of production solutions ultimately toward a more sustainable society.
We gratefully acknowledge funding from the National Mexican Council of Science and Technology (CONACYT) Fellowship 291795, Institute of Health U24 DK097154, National Institutes of Health awards R01HD059127, R01HD065122, R01HD061923, R21AT006180, and R01AT007079.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Merchant S.; Kropat J.; Liu B.; Shaw J. TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 2012, 23, 352–363. [DOI] [PubMed] [Google Scholar]
- Singh A.; Singh P.; Murphy J. Mechanism and challenges in commercialization of algal biofuels. Bioresour. Technol. 2011, 102, 26–34. [DOI] [PubMed] [Google Scholar]
- Ratledge C.Microbial lipids: commercial realities or academic curiosities. In Industrial Applications of Single Cell Oils, 1st ed.; Kyle D., Ratledge C., Eds.; AOCS Press: Champaign, IL, USA, 1992; pp 1–15. [Google Scholar]
- Liu B.; Benning C. Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotechnol. 2013, 24, 300–309. [DOI] [PubMed] [Google Scholar]
- Athenstaedt K.; Daum G. The life cycle of neutral lipids: synthesis, storage and degradation. Cell. Mol. Life Sci. 2006, 63, 1355–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez H. M.; Steinbüchel A. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 2002, 60, 367–376. [DOI] [PubMed] [Google Scholar]
- Wältermann M.; Stöveken T.; Steinbüchel A. Key enzymes for biosynthesis of neutral lipid storage compounds in prokaryotes: properties, function and occurrence of wax ester synthases/acyl-CoA:diacylglycerol acyltransferases. Biochimie 2007, 89, 230–242. [DOI] [PubMed] [Google Scholar]
- Mülroth A.; Li K.; Rokke G.; Winge P.; Olsen Y.; Hohmann-Marriott M. F.; Vadstein O.; Bones A. M. Pathways of lipid metabolism in marine algae, co-expression network, bottlenecks and candidate genes for enhanced production of EPA and DHA in species of Chromista. Mar. Drugs 2013, 11, 4662–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K. D.; Dyer J. M.; Mullen R. T. Biogenesis and functions of lipid droplets in plants. Thematic Review Series: Lipid droplet synthesis and metabolism: from yeast to man. J. Lipid Res. 2012, 53, 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar J.; Wilson E. H.; Masek K.; Hunter C. A.; Striepen B. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl. Acad. Sci. U.S.A. 2006, 1033513192–13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 2012, 249, 541–585. [DOI] [PubMed] [Google Scholar]
- Daniel J.; Deb C.; Dubey V. S.; Sirakova T. D.; Abomoelak B.; Morbidoni H. R.; Kolattukudy P. E. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 2004, 186, 5017–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton N. J.; Christensen H.; Minnikin D. E.; Adegbola R. A.; Barer M. R. Intracellular lipophilic inclusions of Mycobacteria in vitro and in sputum. Microbiology 2002, 148, 2951–2958. [DOI] [PubMed] [Google Scholar]
- Heredia-Arroyo T.; Wei W.; Hu B. Oil accumulation via heterotrophic/mixotrophic Chlorella protothecoides. Appl. Biochem. Biotechnol. 2010, 162, 1978–1995. [DOI] [PubMed] [Google Scholar]
- Skrupski B.; Wilson K. E.; Goff K. L.; Zou J. Effect of pH on neutral lipid and biomass accumulation in microalgal strains native to the Canadian prairies and the Athabasca oil sands. J. Appl. Phycol. 2013, 25, 937–949. [Google Scholar]
- Ratha S. K.; Prasanna R.; Prasad R. B.N.; Sarika C.; Dhar D. W.; Saxena A. K. Modulating lipid accumulation and composition in microalgae by biphasic nitrogen supplementation. Aquaculture 2013, 392 – 395, 69–76. [Google Scholar]
- Ratledge C. Resources conservation by novel biological processes. Part 1: Grow fats from wastes. Chem. Soc. Rev. 1979, 8, 283–296. [Google Scholar]
- Kosa M.; Ragauskas A. Lipids from heterotrophic microbes: advances in metabolism research. Trends Biotechnol. 2011, 29253–61. [DOI] [PubMed] [Google Scholar]
- Wynn J.; Hamid A.; Li Y.; Ratledge C. Biochemical events leading to the diversion of carbon into storage lipids in the oleaginous fungi Mucor circinelloides and Mortierella alpine. Microbiology 2001, 147, 2857–2864. [DOI] [PubMed] [Google Scholar]
- Schweizer E.; Hofmann J. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol. Mol. Biol. Rev. 2004, 683501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.-J.; Zhang Y.-M.; Rock C. O. Product diversity and regulation of type II fatty acid synthases. Biochem. Cell Biol. 2004, 82, 145–155. [DOI] [PubMed] [Google Scholar]
- Wältermann M.; Hinz A.; Robenek H.; Troyer D.; Reichelt R.; Malkus U.; Galla H.; Kalscheuer R.; Stöveken T.; Landenberg P.; Steinbüchel A. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol. Microbiol. 2005, 553750–763. [DOI] [PubMed] [Google Scholar]
- White S. W.; Zheng J.; Zhang Y. M.; Rock C. O. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 2005, 74, 791–831. [DOI] [PubMed] [Google Scholar]
- Schujman G. E.; Mendoza D. Regulation of type II fatty acid synthase in Gram-positive bacteria. Curr. Opin. Microbiol. 2008, 11, 148–152. [DOI] [PubMed] [Google Scholar]
- Zhang Y. M.; Rock C. O. Transcriptional regulation in bacterial membrane lipid synthesis. J. Lipid Res. 2009, 50, S115–S119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen E. J.; Kang Y.; Bokinsky G.; Hu Z.; Schirmer A.; McClure A.; Del Cardayre S. B.; Keasling J. D. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 2010, 4637280559–562. [DOI] [PubMed] [Google Scholar]
- Clomburg J. M.; Gonzalez R. Biofuel production in E. coli: the role of metabolic engineering and synthetic biology. Appl. Microbiol. Biotechnol. 2010, 862419–434. [DOI] [PubMed] [Google Scholar]
- Handke P.; Lynch S. A.; Gill R. T. Application and engineering of fatty acid biosynthesis in Escherichia coli for advanced fuel. Metab. Eng. 2011, 13128–37. [DOI] [PubMed] [Google Scholar]
- Taranto P. A.; Keenan T. W.; Potts M. Rehydration induces rapid onset of lipid biosynthesis in desiccated Nostoc commune (cyanobacteria). Biochim. Biophys. Acta 1993, 1168, 228–237. [DOI] [PubMed] [Google Scholar]
- Philip S.; Keshavarz T.; Roy I. Polyhydroxyalkanoates: biodegradable polymers with a range of applications. J. Chem. Technol. Biotechnol. 2007, 82, 233–247. [Google Scholar]
- Lauzier C.; Monasterios C.; Saracovan I.; Marchessault R.; Ramsay B. Film formation and paper coating with poly(β-hydroxyalkanoate), a biodegradable latex. Tappi J. 1993, 76571–77. [Google Scholar]
- Babel W.; Riis V.; Hainich E. Mikrobielle thermoplaste: biosynthese, eigenschaften und anwendung. Plaste Kautschuk 1990, 37, 109–115. [Google Scholar]
- Walther T.; Farese R. The life of lipid droplets. Biochim. Biophys. Acta 2009, 1791, 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanova M.; et al. Influence of squalene on lipid particle/droplet and membrane organization in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 2012, 1821, 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieler A.; Brubaker S.; Vick B.; Benning C. A lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol. 2012, 158, 1562–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y.; Yang L.; Zhang S.; Wang Y.; Du Y.; Pu J.; Peng G.; Chen Y.; Zhang H.; Yu J.; Hang H.; Wu P.; Yang F.; Yang H.; Steinbüchel A.; Liu P. Identification of the major functional proteins of prokaryotic lipid droplets. J. Lipid Res. 2011, 53, 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz R.; Zehmer J.; Zhu M.; Chen Y.; Serrero G.; Zhao Y.; Liu P. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 2007, 6, 3256–3265. [DOI] [PubMed] [Google Scholar]
- Jendrossek D. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J. Bacteriol. 2009, 191103195–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grage K.; Jahns A. C.; Parlane N.; Palanisamy R.; Rasiah I. A.; Atwood J. A.; Rehm B. H. A. Bacterial polyhydroxyalkanoate granules: biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules 2009, 10, 660–669. [DOI] [PubMed] [Google Scholar]
- Goodman J. The gregarious lipid droplet. J. Biol. Chem. 2008, 283, 28005–28009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S.; Martin S.; Parton R. Lipid droplet-organelle interactions; sharing the fats. Biochim. Biophys. Acta 2009, 1791, 441–447. [DOI] [PubMed] [Google Scholar]
- Zhu Z.; Zhang S.; Liu H.; Shen H.; Lin X.; Yang F.; Zhou Y.; Jin G.; Ye M.; Zou H.; Zhao Z. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat. Commun. 2012, 3, 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreton R.Physiology of lipid accumulation. In Single Cell Oil, 1st ed.; Moreton R., Ed.; Longman Scientific: New York, USA, 1988; pp 1–32. [Google Scholar]
- Beopoulos A.; Cescut J.; Haddouche R.; Uribelarrea J.-L.; Molina-Jouve C.; Nicaud J.-M. Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 2009, 48, 375–387. [DOI] [PubMed] [Google Scholar]
- Ratledge C. Regulation of lipid accumulation in oleaginous micro-organisms. Biochem. Soc. Trans. 2002, 3061047–1050. [DOI] [PubMed] [Google Scholar]
- Ratledge C. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie 2004, 86, 807–815. [DOI] [PubMed] [Google Scholar]
- Ratledge C.Biochemistry, stoichiometry, substrates and economics. In Single Cell Oil, 1st ed.; Moreton R., Ed.; Longman Scientific: New York, USA, 1988; pp 33–64. [Google Scholar]
- Vorapreeda T.; Thammarongtham C.; CHeevadhanarak S.; Laoteng K. Alternative routes of acetyl-CoA synthesis identified by comparative genomic analysis: involvement in the lipid production of oleaginous yeast and fungi. Microbiology 2012, 158, 217–228. [DOI] [PubMed] [Google Scholar]
- Morin N.; Cescut J.; Beopoulos A.; Lelandais G.; Le Berre V.; Uribelarrea J.; Molina-Jouve C.; Nicaud J. Transcriptomic analyses during the transition from biomass production to lipid accumulation in the oleaginous yeast Yarrowia lipolytica. PLoS One 2011, 6111–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beopoulos A.; Mrozova Z.; Thevenieau F.; Le Dall M.; Hapala I.; Papanikolaou S.; Chardot T.; Nicaud J. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2008, 74247779–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Adams I.; Ratledge C. Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 2007, 153, 2013–2025. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Frómeta R.; Gutierrez A.; Torres-Martinez S.; Garre V. Malic enzyme activity is not the only bottleneck for lipid accumulation in the oleaginous fungus Mucor circinelloides. Appl. Microbiol. Biotechnol. 2013, 97, 3063–3072. [DOI] [PubMed] [Google Scholar]
- Tehlivets O.; Scheuringer K.; Kohlwein S. D. Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta 2007, 1771, 255–270. [DOI] [PubMed] [Google Scholar]
- Kolodziej S. J.; Penczek P. A.; Schroeter J. P.; Stoops J. K. Structure-function relationships of the Saccharomyces cerevisiae fatty acid synthase, three-dimensional structure. J. Biol. Chem. 1996, 271, 28422–28429. [DOI] [PubMed] [Google Scholar]
- Hu Q.; Sommerfeld M.; Jarvis E.; Ghirardi M.; Posewitz M.; Seibert M.; Darzins A. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 2008, 54, 621–639. [DOI] [PubMed] [Google Scholar]
- Radakovits R.; Jinkerson R.; Darzins A.; Posewitz M. Genetic engineering of algae for enhanced biofuel production. Eukaryotic Cell 2010, 94486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot P. H.; Scholte H. R.; Hulsmann W. C. Fatty acid activation: specificity, localization and function. Adv. Lipid Res. 1976, 14, 75–126. [DOI] [PubMed] [Google Scholar]
- Schujman G. E.; Mendoza D. Transcriptional control of membrane lipid synthesis in bacteria. Curr. Opin. Microbiol. 2005, 8, 149–153. [DOI] [PubMed] [Google Scholar]
- Chen M.; Hancock L. C.; Lopes J. M. Transcriptional regulation of yeast phospholipid biosynthetic genes. Biochim. Biophys. Acta 2007, 1771, 310–321. [DOI] [PubMed] [Google Scholar]
- Rajakumari S.; Grillitsch K.; Daum G. Synthesis and turnover of non-polar lipids in yeast. Prog. Lipid Res. 2008, 47, 157–171. [DOI] [PubMed] [Google Scholar]
- Carman G. M.; Han G.-S. Regulation of phospholipid synthesis in yeast. J. Lipid Res. 2009, 50, S69–S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G. M.; Han G.-S. Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 2009, 284, 2593–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristan K.; Lanisnik Rizner T. Steroid-transforming enzymes in fungi. J. Steroid Biochem. Mol. Biol. 2012, 129, 79–91. [DOI] [PubMed] [Google Scholar]
- Zweytick D.; Leitner E.; Kohlwein S. D.; Yu C.; Rothblatt J.; Daum G. Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 2000, 267, 1075–1082. [DOI] [PubMed] [Google Scholar]
- Long A.; Manneschmidt A.; VerBrugge B.; Dortch M.; Minkin S.; Prater K.; Biggerstaff J.; Dunlap J.; Dalhaimer P. Lipid droplet de novo formation and fission are linked to the cell cycle in fission yeast. Traffic 2012, 13, 705–714. [DOI] [PubMed] [Google Scholar]
- Choudhary V.; Jacquier N.; Schneiter R. The topology of the triacylglycerol synthesizing enzyme Lro1 indicates that neutral lipids can be produced within the luminal compartment of the endoplasmic reticulum. Commun. Integr. Biol. 2011, 46781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londos C.; Sztalryd C.; Tansey J. T.; Kimmel A. R. Role of PAT proteins in lipid metabolism. Biochimie 2005, 87, 45–49. [DOI] [PubMed] [Google Scholar]
- Zanghellini J.; Wodlei F.; Grünberg H. Phospholipid demixing and the birth of a lipid droplet. J. Theor. Biol. 2010, 264, 952–961. [DOI] [PubMed] [Google Scholar]
- Wolinski H.; Kolb D.; Hermann S.; Koning R.; Kohlwein S. A role for seipin in lipid droplet dynamics and inheritance in yeast. J. Cell Sci. 2011, 124, 3894–3904. [DOI] [PubMed] [Google Scholar]; Adeyo O.; Horn P.; Lee S.; Binns D.; Chandrahas A.; Chapman K.; Goodman J. The yeast lipin orthologue Pah 1p is important for biogenesis of lipid droplets. J. Cell Biol. 2011, 19261043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyo O.; Horn P.; Lee S.; Binns D.; Chandrahas A.; Chapman K.; Goodman J. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 2011, 19261043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozaquel-Morais B.; Madeira J.; Maya-Monteiro C.; Masuda C.; Montero-Lomeli M. A new fluorescence-based method identifies protein phosphatases regulating lipid droplet metabolism. PLoS One 2010, 5101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C.; Wardell M.; Weisgraber K.; Mahley R.; Agard D. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science 1991, 252, 1817–1822. [DOI] [PubMed] [Google Scholar]
- Hickenbottom S.; Kimmel A.; Londos C.; Hurley J. Structure of a lipid droplet protein: the PAT family member TIP47. Structure 2004, 12, 1199–1207. [DOI] [PubMed] [Google Scholar]
- Guschina I. A.; Harwood J. L. Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid Res. 2006, 45, 160–186. [DOI] [PubMed] [Google Scholar]
- Barsanti L.; Gualtieri P.. Algae Anatomy, Biochemistry Biotechnology, 1st ed.; Taylor & Francis: Boca Raton, FL, USA, 2006; pp 1–33. [Google Scholar]
- Li-Beisson Y.; Shorrosh B.; Beisson F.; Andersson M. X.; Arondel V.; Bates P. D.; Baud S.; Bird D.; DeBono A.; Durret T. P.; Franke R. B.; Graham I. A.; Katayama K.; Kelly A. A.; Larson T.; Markham J. E.; Miquel M.; Molina I.; Nishida I.; Rowland O.; Samuels L.; Schmid K. M.; Wada H.; Welti R.; Xu C.; Zallot R.; Ohlrogge J.. Acyl-lipid metabolism. In The Arabidopsis Book; e-ISSN 1543-8120, DOI: 10.1199/tab.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J.; Browse J. Lipid biosynthesis. Plant Cell 1995, 7, 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen J. K.; Schonauer M. S.; Autio K. J.; Mittelmeier T. M.; Kastaniotis A. J.; Dieckmann C. L. Mitochondrial fatty acid synthesis type II: more than just fatty acids. J. Biol. Chem. 2009, 284, 9011–9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen J. K.; Autio K. J.; Schonauer M. S.; Samuli Kursu V. A.; Dieckmann C. L.; Kastaniotis A. J. Mitochondrial fatty acid synthesis and respiration. Biochim. Biophys. Acta 2010, 1797, 1195–1202. [DOI] [PubMed] [Google Scholar]
- Hiltunen J. K.; Chen Z.; Haapalainen A. M.; Wierenga R. K.; Kastaniotis A. J. Mitochondrial fatty acid synthesis – an adopted set of enzymes making a pathway of major importance for the cellular metabolism. Prog. Lipid Res. 2010, 49, 27–45. [DOI] [PubMed] [Google Scholar]
- Wada H.; Shintani D.; Ohlrogge J. Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 1591–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueguen V.; Macherel D.; Jaquinod M.; Douce R.; Bourguignon J. Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J. Biol. Chem. 2000, 275, 5016–5025. [DOI] [PubMed] [Google Scholar]
- Keeling P. J. The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. B 2010, 365, 729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre C.; Froehlich J. E.; Moll M. R.; Benning C. A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2006–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F.; Toussaint J. P.; Cooksey K. E.; Fields M. W.; Gerlach R.; Peyton B. M.; Carlson R. P. Physiological and molecular analysis of carbon source supplementation and pH stress-induced lipid accumulation in the marine diatom Phaeodactylum tricornutum. Appl. Microbiol. Biotechnol. 2013, 97, 3625–3642. [DOI] [PubMed] [Google Scholar]
- Kitano M.; Matsukawa R.; Karube I. Enhanced eicosapentaenoic acid production by Navicula saprophila. J. Appl. Phycol. 1998, 10, 101–105. [Google Scholar]
- Schneider J. C.; Roessler P. Radiolabeling studies of lipids and fatty-acids in Nannochloropsis (Eustigmatophyceae), an oleaginous marine alga. J. Phycol. 1994, 30, 594–598. [Google Scholar]
- Hofmann M.; Eichenberger W. Radiolabeling studies on the lipid metabolism in the marine brown alga Dictyopteris membranacea. Plant Cell Physiol. 1998, 39, 508–515. [Google Scholar]
- Miller R.; Wu G.; Deshpande R. R.; Vieler A.; Gartner K.; Li X.; Moellering E. R.; Zauner S.; Cornish A. J.; Liu B. Changes in transcription abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 2010, 154, 1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.-K.; Niu Y.-F.; Ma Y. H.; Xue J.; Zhang M. H.; Yang W. D.; Liu J. S.; Lu S. H.; Guan Y.; Li H. Y. Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol. Biofuels 2013, 6, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellou S.; Aggelis G. Biochemical activites in Chlorella sp. and Nannochloropsis salina during lipid and sugar synthesis in a lab-scale open pond simulating reactor. J. Biotechnol. 2012, 164, 318–329. [DOI] [PubMed] [Google Scholar]
- Kroth P. G.; Chiovitti A.; Gruber A.; Martin-Jezequel V.; Mock T.; Parker M. S.; Stanley M. S.; Kaplan A.; Caron L.; Weber T. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS One 2008, 3, e1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A.; Zell M. B.; Maurino V. G. Malate decarboxylases: evolution and roles of NAD(P)-ME isoforms in species performing C4 and C3 photosynthesis. J. Exp. Bot. 2011, 6293061–3069. [DOI] [PubMed] [Google Scholar]
- Fan J.; Yan C.; Andre C.; Shanklin J.; Schwender J.; Xu C. Oil accumulation is controlled by carbon precursor supply for fatty acid synthesis in Chlamydomonas reinhardtii. Plant Cell Physiol. 2012, 53, 1380–1390. [DOI] [PubMed] [Google Scholar]
- Huerlimann R.; Heimann K. Comprehensive guide to acetyl-carboxylases in algae. Crit. Rev. Biotechnol. 2013, 33, 49–65. [DOI] [PubMed] [Google Scholar]
- Roessler P. G. Purification and characterization of acetyl CoA carboxylase from the diatom Cyclotella cryptica. Plant Physiol. 1990, 92, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolau B. J.; Ohlrogge J. B.; Wurtele E. S. Plant biotin-containing carboxylases. Arch. Biochem. Biophys. 2003, 414, 211–222. [DOI] [PubMed] [Google Scholar]
- Ryall K.; Harper J. T.; Keeling P. J. Plastid-derived type II fatty acid biosynthetic enzymes in chromists. Gene 2003, 313, 139–148. [DOI] [PubMed] [Google Scholar]
- Nagano N.; Sakaguchi K.; Taoka Y.; Okita Y.; Honda D.; Ito M.; Hayashi M. Detection of genes involved in fatty acid elongation and desaturation in Thraustochytrid marine eukaryotes. J. Oleo Sci. 2011, 609475–481. [DOI] [PubMed] [Google Scholar]
- Metz J. G.; Kuner J.; Rosenzweig B.; Lippmeier J. C.; Roessler P.; Zirkle R. Biochemical characterization of polyunsaturated fatty acid synthesis in Schizochytrium: release of the products as free fatty acids. Plant Physiol. Biochem. 2009, 47, 472–478. [DOI] [PubMed] [Google Scholar]
- Dehesh K.; Tai H.; Edwards P.; Byrne J.; Jaworski J. G. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 2001, 125, 1103–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan P. G.; Slack C. R. Cellular organization of glycerolipid metabolism. Plant Physiol. 1982, 33, 97–132. [Google Scholar]
- Khozin-Goldberg I.; Didi-Cohen S.; Shayakhmetova I.; Cohen Z. Biosynthesis of eicosapentaenoic acid (EPA) in the freshwater eustigmatophyte Monodus subterraneus (Eustigmatophyceae). J. Phycol. 2002, 38, 745–756. [Google Scholar]
- Khozin I.; Adlerstein D.; Bigongo C.; Heimer Y. M.; Cohen Z. Elucidation of the biosynthesis of eicosapentaenoic acid in the microalga Porphyridium cruentum. II. Studies with radiolabeled precursors. Plant Physiol. 1997, 114, 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khozin-Goldberg I.; Cohen Z. Unraveling algal lipid metabolism: recent advances in gene modification. Biochimie 2011, 93, 91–100. [DOI] [PubMed] [Google Scholar]
- Thompson G. A. Lipids and membrane function in green algae. Biochim. Biophys. Acta 1996, 1302, 17–45. [DOI] [PubMed] [Google Scholar]
- Riekhof W. R.; Sears B. B.; Benning C. Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1(Cr). Eukaryotic Cell 2005, 4, 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering E. R.; Miller R.; Benning C.. Molecular genetics of lipid metabolism in the model green alga Chlamydomonas reinhardtii. In Lipids in Photosynthesis: Essential and Regulatory Functions; Wada H., Murata N., Eds.; Springer Science: Dordrecht, The Netherlands, 2009; pp 139–155. [Google Scholar]
- Moellering E. R.; Benning C.. Glycerolipid biosynthesis. In The Chlamydomonas Sourcebook: Organellar and Metabolic Processes, 2nd ed.; Stern D., Harris E. H., Eds.; Elsevier: New York, 2008; Vol. 2, pp 41–68. [Google Scholar]
- Fulda M.; Shockey J.; Werber M.; Wolter F. P.; Heinz E. Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid β-oxidation. Plant J. 2002, 32, 93–103. [DOI] [PubMed] [Google Scholar]
- Sperling P.; Heinz E. Desaturases fused to their electron donor. Eur. J. Lipid Sci. Technol. 2001, 103, 158–180. [Google Scholar]
- Alves Martins D.; Custodio L.; Barreira L.; Pereira H.; Ben-Hamadou R.; Varela J.; Abu-Salah K. M. Alternative sources of n-3 long-chain polyunsaturated fatty acids in marine microalgae. Mar. Drugs 2013, 11, 2259–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz J. G.; Roessler P.; Facciotti D.; Levering C.; Dittrich F.; Lassner M.; Valentine R.; Lardizabal K.; Domergue F.; Yamada A. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 2001, 293, 290–293. [DOI] [PubMed] [Google Scholar]
- Harwood J. L.; Guschina I. A. The versatility of algae and their lipid metabolism. Biochimie 2009, 91, 679–684. [DOI] [PubMed] [Google Scholar]
- Dahlqvist A.; Stähl U.; Lenman M.; Banas A.; Lee M.; Sandager L.; Ronne H.; Stymne S. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 6487–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle N. R.; Page M. D.; Liu B.; Blaby I. K.; Casero D.; Kropat J.; Cokus S.; Hong-Hermesdorf A.; Shaw J.; Karpowicz S. J. Three acyltransferases and a nitrogen responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 2012, 287, 15811–15825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W.-L.; Ansari W.; Schoepp N. G.; Hannon M. J.; Mayfield S. P.; Burkart M. D. Modifications of the metabolic pathways of lipid and triacylglycerol production in microalgae. Microb. Cell Factories 2011, 10, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieler A.; Wu G.; Tsai C.; Bullard B.; Cornish A.; Harvey C.; Reca I.; Thornburg C.; Achawanantakun R.; Buehl C.; Campbell M.; Cavalier D.; Childs K.; Clark T.; Deshpande R.; Erickson E.; Ferguson A.; Hanee W.; Kong Q.; Li X.; Liu B.; Lundback S.; Peng C.; Roston R.; Sanjaya S.; Simpson J.; TerBush A.; Warakanont J.; Zäuner S.; Farre E.; Hegg E.; Jiang N.; Kuo M.; Lu Y.; Niyogi K.; Ohlrogge J.; Osteryoung K.; Shrachar-Hill Y.; Sears B.; Sun Y.; Takahashi H.; Yandell M.; Shiu S.; Benning C. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genetics 2012, 8111–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishige T.; Tani A.; Sakai Y.; Kato N. Wax ester production by bacteria. Curr. Opin. Microbiol. 2003, 6, 244–250. [DOI] [PubMed] [Google Scholar]
- Manila-Perez E.; Lange A. B.; Hetzler S.; Steinbüchel A. Occurrence, production and export of lipophilic compounds by hydrocarbonoclastic marine bacteria and their potential use to produce bulk chemicals from hydrocarbons. Appl. Microbiol. Biotechnol. 2010, 86, 1693–1706. [DOI] [PubMed] [Google Scholar]
- Thornton J.; Howard S. P.; Buckley J. T. Molecular cloning of a phospholipid-cholesterol acyltransferase from Aeromonas hydrophila: sequence homologies with lecithin-cholesterol acyltransferase and other lipases. Biochim. Biophys. Acta 1988, 959, 153–159. [DOI] [PubMed] [Google Scholar]
- Christensen H.; Garton N. J.; Horobin R. W.; Minnikin D. E.; Barer M. R. Lipid domains of mycobacteria studied with fluorescent molecular probes. Mol. Microbiol. 1999, 31, 1561–1572. [DOI] [PubMed] [Google Scholar]
- Cronan J. E.; Vagelos P. R. Metabolism and function of the membrane phospholipids of Escherichia coli. Biochim. Biophys. Acta 1972, 2652560. [DOI] [PubMed] [Google Scholar]
- Alvarez H. M.; Luftmann H.; Silva R. A.; Cesari A. C.; Viale A.; Wältermann M.; Steinbüchel A. Identification of phenyldecanoic acid as a constituent of triacylglycerols and wax esters produced by Rhodococcus opacus PD630. Microbiology 2002, 148, 1407–1412. [DOI] [PubMed] [Google Scholar]
- Pelech S. L.; Vance D. E. Signal transduction via phosphatidylcholine cycles. Trends Biochem. Sci. 1989, 14, 28–30. [Google Scholar]
- Wältermann M.; Steinbüchel A. In vitro effects of sterculic acid on lipid biosynthesis in Rhodococcus opacus strain PD630 and isolation of mutants defective in fatty acid desaturation. FEMS Microbiol. Lett. 2000, 190, 45–50. [DOI] [PubMed] [Google Scholar]
- Lehner R.; Kuksis A. Biosynthesis of triacylglycerols. Prog. Lipid Res. 1996, 35, 169–201. [DOI] [PubMed] [Google Scholar]
- Kalscheuer R.; Stöveken T.; Malkus U.; Reichelt R.; Golyshin P.; Sabirova J.; Ferrer M.; Timmis K.; Steinbüchel A. Analysis of storage lipid accumulation in Alcanivorax borkumensis: evidence for alternative triacylglycerol biosynthesis routes in bacteria. J. Bacteriol. 2007, 1893918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior P. J.; Dawes E. A. Poly-β-hydroxybutyrate biosynthesis and the regulation of glucose metabolism in Azotobacter beijerinckii. Biochem. J. 1971, 125, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior P. J.; Dawes E. A. The regulation of poly-β-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem. J. 1973, 134, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm B. A.; Kruger N.; Steinbüchel A. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. The phaG gene from Pseudomonas putida KT2440 encodes a 3-hydroxy-acyl carrier protein-coenzyme A transferase. J. Biol. Chem. 1998, 273, 24044–24051. [DOI] [PubMed] [Google Scholar]
- Quillaguaman J.; Guzmán H.; Van-Thuoc D.; Hatti-Kaul R. Synthesis and production of polyhydroxyalkanoates by halophiles: current potential and future prospects. Appl. Microbiol. Biotechnol. 2010, 85, 1687–1696. [DOI] [PubMed] [Google Scholar]
- Rehm B. Genetic and biochemistry of polyhydroxyalkanoate-granule self-assembly: the key role of polyester synthases. Biotechnol. Lett. 2006, 28, 207–213. [DOI] [PubMed] [Google Scholar]
- Rehm B. H. Biogenesis of microbial polyhydroxyalkanoate granules: a platform technology for the production of tailor-made bioparticles. Curr. Issues Mol. Biol. 2007, 9, 41–62. [PubMed] [Google Scholar]
- Rehm B. H. Polyester synthases: natural catalysts for plastics. Biochem. J. 2003, 376, 15–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbüchel A.; Hein S. Biochemical and molecular basis of microbial synthesis of polyhydroxyalkanoates in microorganisms. Adv. Biochem. Eng./Biotechnol. 2001, 71, 81. [DOI] [PubMed] [Google Scholar]
- Stubbe J.; Tian J. Polyhydroxyalkanoate (PHA) homeostasis: the role of PHA synthase. Nat. Prod. Rep. 2003, 20, 445–457. [DOI] [PubMed] [Google Scholar]
- Stubbe J.; Tian J.; He A.; Sinskey A. J.; Lawrence A. G.; Liu P. Non-template-dependent polymerization processes: polyhydroxyalkanoate synthases as a paradigm. Annu. Rev. Biochem. 2005, 74, 433–480. [DOI] [PubMed] [Google Scholar]
- Valappil S. P.; Boccaccini A. R.; Bucke C.; Roy I. Polyhydroxyalkanoates in Gram-positive bacteria: insights from the genera Bacillus and Streptomyces. Antonie van Leeuwenhoek 2007, 91, 1–17. [DOI] [PubMed] [Google Scholar]
- Cai S.; Cai L.; Liu H.; Liu X.; Han J.; Zhou J.; Xiang H. Identification of the Haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl. Environ. Microbiol. 2012, 7861946–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbüchel A. Considerations on the structure and biochemistry of bacterial polyhydyroxyalkanoic acid inclusions. Can. J. Microbiol. 1995, 41Suppl. 194–105. [DOI] [PubMed] [Google Scholar]
- Wieczorek R.; Pries A.; Steinbüchel A.; Mayer F. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 1995, 177, 2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handrick R. Unraveling the function of the Rhodospirillum rubrum activator of polyhydroxybutyrate (PHB) degradation: the activator is a PHB-granule-bound protein (phasin). J. Bacteriol. 2004, 186, 2466–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York G. M.; Stubbe J.; Sinskey A. J. New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J. Bacteriol. 2001, 183, 2394–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis D.; Sein V.; Martinez E.; Augustine B. PhaP is involved in the formation of a network on the surface of polyhydroxyalkanoate inclusions in Cupriavidus necator H16. J. Bacteriol. 2008, 190, 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippe H.; Schlegel H. G. Hydrolyse von PHBS durch intracelluläre Depolymerase von Hydrogenomanas H16. Arch. Mikrobiol. 1967, 56, 278–299. [PubMed] [Google Scholar]
- Jendrossek D.; Handrick R. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 2002, 56, 403–432. [DOI] [PubMed] [Google Scholar]
- Liebergesell M.; Steinbüchel A. New knowledge about the PHA-locus and P(3HB) granule-associated proteins in Chromatium vinosum. Biotechnol. Lett. 1996, 186719–724. [Google Scholar]
- Pieper-Fürst U.; Madkour M. H.; Mayer F.; Steinbüchel A. Identification of the region of a 14-kilodalton protein of Rhodococcus ruber that is responsible for the binding of this phasin to polyhydroxyalkanoic acid granules. J. Bacteriol. 1995, 177, 2513–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemet A.; Barbeau J.; Lemiegre L.; Benvegnu T. Archaeal tetraether bipolar lipids: structures, functions and applications. Biochimie 2009, 91, 711–717. [DOI] [PubMed] [Google Scholar]
- Woesze C. R.; Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U.S.A. 1977, 74, 5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legat A.; Gruber C.; Zangger K.; Wanner G.; Stan-Lotter H. Identification of polyhydroxyalkanoates in Halococcus and other halaorchaeal species. Appl. Microb. Cell Physiol. 2010, 87, 119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk R. G.; Ginzburg M. Ultrastructure of two species of halobacterium. J. Ultrastruct. Res. 1972, 41, 80–94. [DOI] [PubMed] [Google Scholar]
- Oren A.; Ginzburg M.; Ginzburg B. Z.; Hochstein L. I.; Volcani B. E. Haloarcula marismortui (Volcani) sp. nov., nom. rev., an extremely halophilic bacterium from the Dead Sea. Int. J. Syst. Evol. Microbiol. 1990, 40, 209–210. [DOI] [PubMed] [Google Scholar]
- Brandl H.; Gross R. A.; Lenz R. W.; Fuller R. C. Physiology and biotechnology of Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 1988, 5481977–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.; Lu Q.; Zhou J.; Xiang H. Molecular characterization of the phaECHm genes, required for biosynthesis of poly(3-hydroxybutyrate) in the extremely halophilic archaeon Haloarcula marismortui. Appl. Environ. Microbiol. 2007, 73, 6058–6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezayen F. F.; Rehm B. H.; Eberhardt R.; Steinbüchel A. Polymer production by two newly isolated extremely halophilic archaea: application of a novel corrosion-resistant bioreactor. Appl. Microbiol. Biotechnol. 2000, 54, 319–325. [DOI] [PubMed] [Google Scholar]
- Hezayen F. F.; Steinbüchel A.; Rehm B. H. Biochemical and enzymological properties of the polyhydroxybutyrate synthase from the extremely halophilic archaeon strain 56. Arch. Biochem. Biophys. 2002, 403, 284–291. [DOI] [PubMed] [Google Scholar]
- Hezayen F. F.; Tindall B. J.; Steinbüchel A.; Rehm B. H. Characterization of a novel halophilic archaeon, Halobiforma haloterrestris gen. nov., sp. nov., and transfer of Natronobacterium nitratireducens to Halobiforma nitratireducens comb. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 2271–2280. [DOI] [PubMed] [Google Scholar]
- Lu Q.; Han J.; Zhou J.; Xiang H. Genetic and biochemical characterization of the poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthase in Haloferax mediterranei. J. Bacteriol. 2008, 190, 4173–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisano A.; Overhage J.; Rehm B. H. The polyhydroxyalkanoate biosynthesis genes are differentially regulated in planktonic- and biofilm-grown Pseudomonas aeruginosa. J. Biotechnol. 2008, 133, 442–452. [DOI] [PubMed] [Google Scholar]
- Han J.; Lu Q.; Zhou L.; Liu H.; Xiang H. Identificaction of the polyhydroxyalkanoate (PHA)-specific acetoacetyl coenzyme A reductase among multiple FabG paralogs in Haloarcula hispanica and reconstruction of the PHA biosynthetic pathway in Haloferax volcanii. Appl. Environ. Microbiol. 2009, 75196168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga N. S.; Bonneau R.; Facciotti M. T.; Pan M.; Glusman G.; Deutsch E. W.; Shannon P.; Chiu Y.; Weng R. S.; Gan R. R.; Hung P.; Date S. V.; Marcotte E.; Hood L.; Mg W. V. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 2004, 14, 2221–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann L. Binding of the major phasin, PhaP1, from Ralstonia eutropha H16 to poly(3-hydroxybutyrate) granules. J. Bacteriol. 2008, 190, 2911–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins S.; Hammock B.; Newman J.; German B. Individual metabolism should guide agriculture toward foods for improved health and nutrition. Am. J. Clin. Nutr. 2001, 74, 283–286. [DOI] [PubMed] [Google Scholar]
- Verma J.; Bhola N.; Aggarwal J. Structure of sterculic acid. Nature 1955, 175, 84–85. [Google Scholar]
- Berger A.Diet-Induced Acyl Modifications of Mouse Phospholipid Classes. Doctoral thesis, University of California, 1992. [Google Scholar]
- Mukherjee A.; Das K.. Microbial surfactants and their potential applications: an overview. In Biosurfactants, 1st ed; Sen R., Ed.; Landes Bioscience and Springer Science: New York, USA, 2010; Vol. 672, pp 54–64. [DOI] [PubMed] [Google Scholar]
- Youssef N.; Simpson D.; Duncan K.; McInerney M.; Folmsbee M.; Fincher T.; Knapp R. In situ biosurfactant production by Bacillus strains injected into a limestone petroleum reservoir. Appl. Environ. Microbiol. 2007, 7341239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]