Abstract
The murine dilute coat color locus encodes an unconventional myosin heavy chain that is thought to be required for the elaboration or maintenance of dendrites or organelle transport in melanocytes and neurons. In previous studies we showed that the d mutation carried by many inbred strains of mice (now referred to as dilute viral, dv), is caused by the integration of an ecotropic murine leukemia virus (Emv-3) into the dilute gene and that phenotypic revertants of dv (termed d+) result from viral excision; a solo viral long terminal repeat (LTR) is all that remains in revertant DNA. In the studies described here we show that Emv-3 sequences are located within an intron of the dilute gene in a region of the C-terminal tail that is differentially spliced. We also show that these Emv-3 sequences result in the production of shortened and abnormally spliced dilute transcripts and that the level of this effect varies among tissues. This tissue-specific effect on dilute expression likely accounts for the absence of neurological abnormalities observed in dv mice. Surprisingly, we also found that the solo viral LTR present in revertant d+ DNA produces a tissue-specific effect on dilute expression, although this effect is less dramatic than with the full-length provirus and produces no obvious mutant phenotype. These findings have important implications for understanding the effects of viral sequences on mammalian gene expression.
Full text
PDF

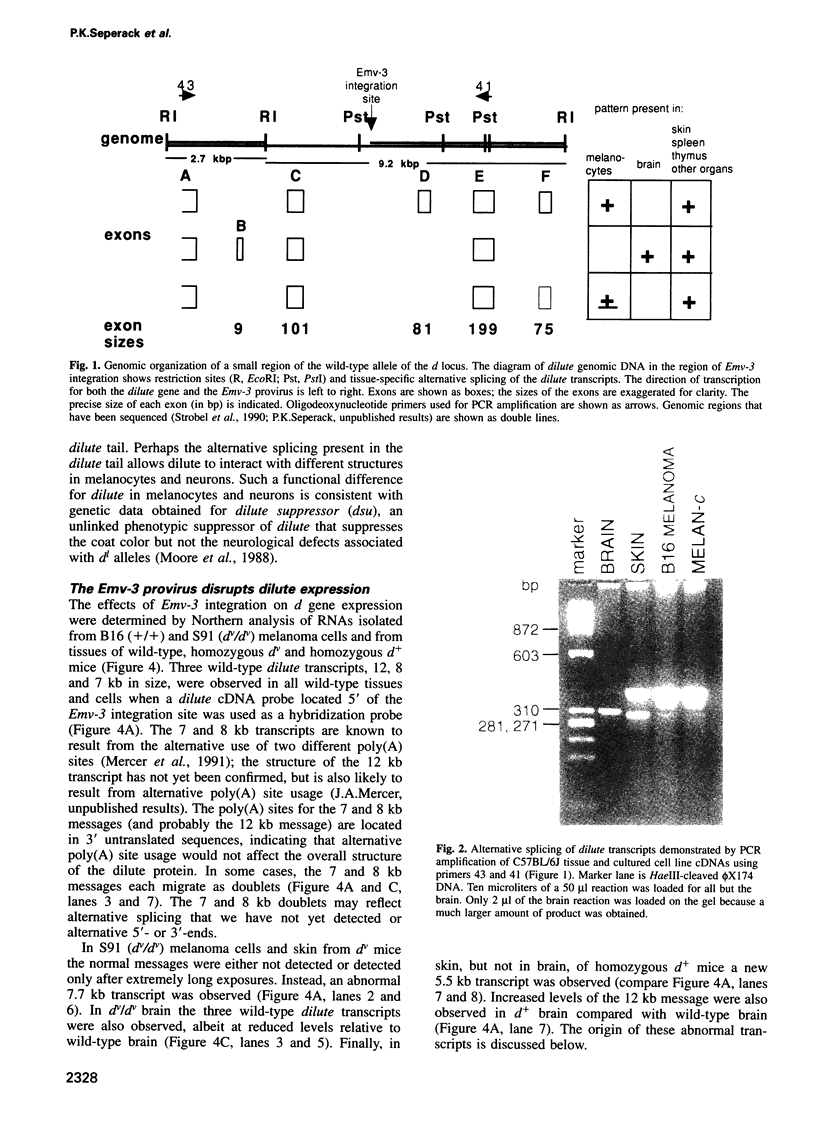

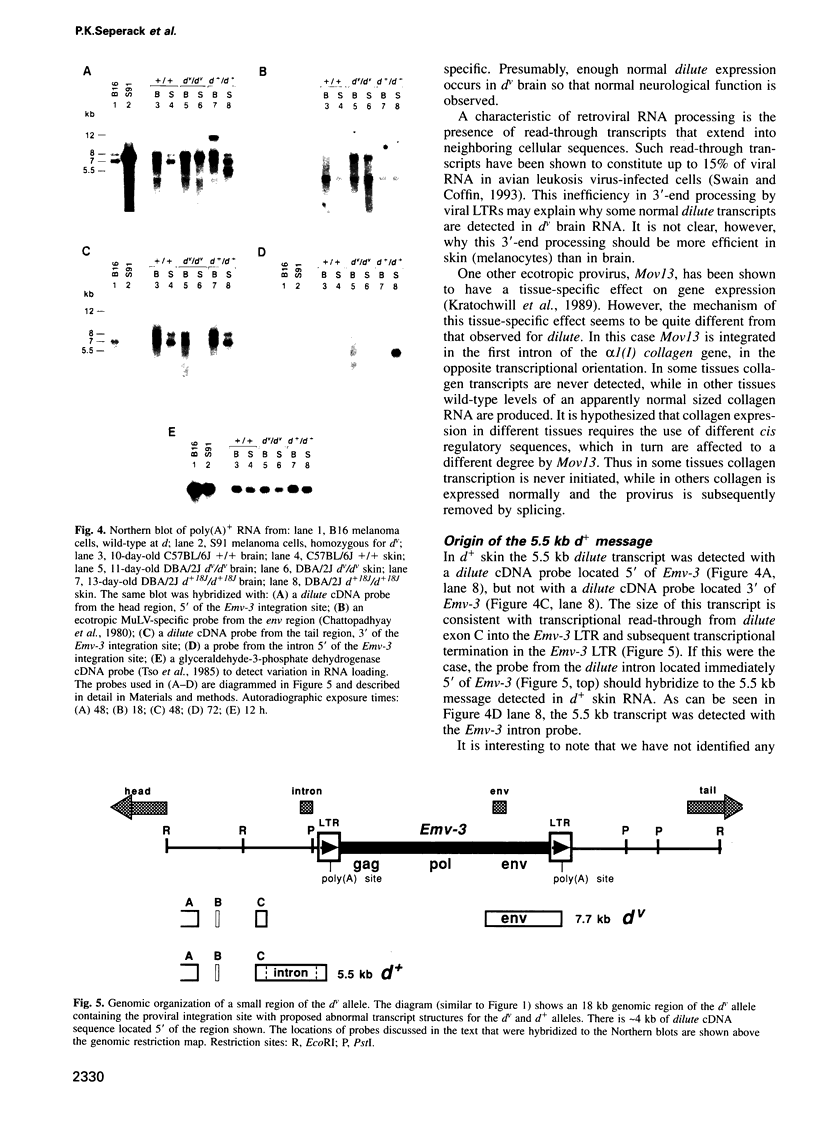


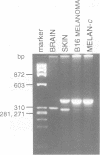
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi M., Watanabe-Fukunaga R., Nagata S. Aberrant transcription caused by the insertion of an early transposable element in an intron of the Fas antigen gene of lpr mice. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1756–1760. doi: 10.1073/pnas.90.5.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D. C., Cooper P. J., Dexter T. J., Devlin L. M., Heasman J., Nester B. Cloned mouse melanocyte lines carrying the germline mutations albino and brown: complementation in culture. Development. 1989 Feb;105(2):379–385. doi: 10.1242/dev.105.2.379. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Lander M. R., Rands E., Lowy D. R. Structure of endogenous murine leukemia virus DNA in mouse genomes. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5774–5778. doi: 10.1073/pnas.77.10.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheney R. E., Mooseker M. S. Unconventional myosins. Curr Opin Cell Biol. 1992 Feb;4(1):27–35. doi: 10.1016/0955-0674(92)90055-h. [DOI] [PubMed] [Google Scholar]
- Cheney R. E., O'Shea M. K., Heuser J. E., Coelho M. V., Wolenski J. S., Espreafico E. M., Forscher P., Larson R. E., Mooseker M. S. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 1993 Oct 8;75(1):13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland N. G., Hutchison K. W., Jenkins N. A. Excision of the DBA ecotropic provirus in dilute coat-color revertants of mice occurs by homologous recombination involving the viral LTRs. Cell. 1983 Jun;33(2):379–387. doi: 10.1016/0092-8674(83)90419-1. [DOI] [PubMed] [Google Scholar]
- Espreafico E. M., Cheney R. E., Matteoli M., Nascimento A. A., De Camilli P. V., Larson R. E., Mooseker M. S. Primary structure and cellular localization of chicken brain myosin-V (p190), an unconventional myosin with calmodulin light chains. J Cell Biol. 1992 Dec;119(6):1541–1557. doi: 10.1083/jcb.119.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fink G., Farabaugh P., Roeder G., Chaleff D. Transposable elements (Ty) in yeast. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):575–580. doi: 10.1101/sqb.1981.045.01.074. [DOI] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B. K., Petzold A., Lillie S. H., Brown S. S. Identification of MYO4, a second class V myosin gene in yeast. J Cell Sci. 1994 Apr;107(Pt 4):1055–1064. doi: 10.1242/jcs.107.4.1055. [DOI] [PubMed] [Google Scholar]
- Herman S. A., Coffin J. M. Differential transcription from the long terminal repeats of integrated avian leukosis virus DNA. J Virol. 1986 Nov;60(2):497–505. doi: 10.1128/jvi.60.2.497-505.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison K. W., Copeland N. G., Jenkins N. A. Dilute-coat-color locus of mice: nucleotide sequence analysis of the d+2J and d+Ha revertant alleles. Mol Cell Biol. 1984 Dec;4(12):2899–2904. doi: 10.1128/mcb.4.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N. A., Copeland N. G., Taylor B. A., Lee B. K. Dilute (d) coat colour mutation of DBA/2J mice is associated with the site of integration of an ecotropic MuLV genome. Nature. 1981 Oct 1;293(5831):370–374. doi: 10.1038/293370a0. [DOI] [PubMed] [Google Scholar]
- Jenkins N. A., Strobel M. C., Seperack P. K., Kingsley D. M., Moore K. J., Mercer J. A., Russell L. B., Copeland N. G. A retroviral insertion in the dilute (d) locus provides molecular access to this region of mouse chromosome 9. Prog Nucleic Acid Res Mol Biol. 1989;36:207–220. doi: 10.1016/s0079-6603(08)60172-5. [DOI] [PubMed] [Google Scholar]
- Johnston G. C., Prendergast J. A., Singer R. A. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991 May;113(3):539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil K., von der Mark K., Kollar E. J., Jaenisch R., Mooslehner K., Schwarz M., Haase K., Gmachl I., Harbers K. Retrovirus-induced insertional mutation in Mov13 mice affects collagen I expression in a tissue-specific manner. Cell. 1989 Jun 2;57(5):807–816. doi: 10.1016/0092-8674(89)90795-2. [DOI] [PubMed] [Google Scholar]
- Mercer J. A., Seperack P. K., Strobel M. C., Copeland N. G., Jenkins N. A. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 1991 Feb 21;349(6311):709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- Moore K. J., Seperack P. K., Strobel M. C., Swing D. A., Copeland N. G., Jenkins N. A. Dilute suppressor dsu acts semidominantly to suppress the coat color phenotype of a deletion mutation, dl20J, of the murine dilute locus. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8131–8135. doi: 10.1073/pnas.85.21.8131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascolo S., Ghazvini M., Boyer J., Colleaux L., Thierry A., Dujon B. The sequence of a 9.3 kb segment located on the left arm of the yeast chromosome XI reveals five open reading frames including the CCE1 gene and putative products related to MYO2 and to the ribosomal protein L10. Yeast. 1992 Nov;8(11):987–995. doi: 10.1002/yea.320081109. [DOI] [PubMed] [Google Scholar]
- RUSSELL W. L. X-ray-induced mutations in mice. Cold Spring Harb Symp Quant Biol. 1951;16:327–336. doi: 10.1101/sqb.1951.016.01.024. [DOI] [PubMed] [Google Scholar]
- Rinchik E. M., Russell L. B., Copeland N. G., Jenkins N. A. Molecular genetic analysis of the dilute-short ear (d-se) region of the mouse. Genetics. 1986 Feb;112(2):321–342. doi: 10.1093/genetics/112.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell E S. A Quantitative Histological Study of the Pigment Found in the Coat-Color Mutants of the House Mouse. IV. the Nature of the Effects of Genic Substitution in Five Major Allelic Series. Genetics. 1949 Mar;34(2):146–166. doi: 10.1093/genetics/34.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L. B. Definition of functional units in a small chromosomal segment of the mouse and its use in interpreting the nature of radiation-induced mutations. Mutat Res. 1971 Jan;11(1):107–123. doi: 10.1016/0027-5107(71)90036-4. [DOI] [PubMed] [Google Scholar]
- Sanders G., Lichte B., Meyer H. E., Kilimann M. W. cDNA encoding the chicken ortholog of the mouse dilute gene product. Sequence comparison reveals a myosin I subfamily with conserved C-terminal domains. FEBS Lett. 1992 Oct 26;311(3):295–298. doi: 10.1016/0014-5793(92)81123-4. [DOI] [PubMed] [Google Scholar]
- Schlager G., Dickie M. M. Natural mutation rates in the house mouse. Estimates for five specific loci and dominant mutations. Mutat Res. 1971 Jan;11(1):89–96. doi: 10.1016/0027-5107(71)90034-0. [DOI] [PubMed] [Google Scholar]
- Seperack P. K., Strobel M. C., Corrow D. J., Jenkins N. A., Copeland N. G. Somatic and germ-line reverse mutation rates of the retrovirus-induced dilute coat-color mutation of DBA mice. Proc Natl Acad Sci U S A. 1988 Jan;85(1):189–192. doi: 10.1073/pnas.85.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M. C., Seperack P. K., Copeland N. G., Jenkins N. A. Molecular analysis of two mouse dilute locus deletion mutations: spontaneous dilute lethal20J and radiation-induced dilute prenatal lethal Aa2 alleles. Mol Cell Biol. 1990 Feb;10(2):501–509. doi: 10.1128/mcb.10.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain A., Coffin J. M. Influence of sequences in the long terminal repeat and flanking cell DNA on polyadenylation of retroviral transcripts. J Virol. 1993 Oct;67(10):6265–6269. doi: 10.1128/jvi.67.10.6265-6269.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso J. Y., Sun X. H., Kao T. H., Reece K. S., Wu R. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res. 1985 Apr 11;13(7):2485–2502. doi: 10.1093/nar/13.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar Z., Davison D., Garza D., Bingham P. M. A detailed developmental and structural study of the transcriptional effects of insertion of the Copia transposon into the white locus of Drosophila melanogaster. Genetics. 1985 Nov;111(3):495–515. doi: 10.1093/genetics/111.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]