Abstract
There is growing evidence for the involvement of Y-complex nucleoporins (Y-Nups) in cellular processes beyond the inner core of nuclear pores of eukaryotes. To comprehensively assess the range of possible functions of Y-Nups, we delimit their structural and functional properties by high-specificity sequence profiles and tissue-specific expression patterns. Our analysis establishes the presence of Y-Nups across eukaryotes with novel composite domain architectures, supporting new moonlighting functions in DNA repair, RNA processing, signaling and mitotic control. Y-Nups associated with a select subset of the discovered domains are found to be under tight coordinated regulation across diverse human and mouse cell types and tissues, strongly implying that they function in conjunction with the nuclear pore. Collectively, our results unearth an expanded network of Y-Nup interactions, thus supporting the emerging view of the Y-complex as a dynamic protein assembly with diverse functional roles in the cell.
Coat nucleoporins form the inner core of nuclear pores of eukaryotes, protein supercomplexes responsible for the regulated transport of macromolecules between the nucleus and the cytoplasm. The Y-shaped Nup84/Nup107-160 subcomplex (Y-complex) forms the outer ring scaffold, is evolutionarily conserved, and is composed of certain key proteins referred to as outer ring coat nucleoporins (Y-Nups – 9 in vertebrates and 7 in yeast)1,2, with common structural features yet elusive sequence similarities3,4. While the functional capacities of coat nucleoporins are primarily connected with the nuclear pore and, in fact, despite their key role in maintaining the integrity of the outer ring, there is growing evidence for their involvement in other processes5,6, including mitotic spindle assembly7 and transcription regulation8. Few other nucleoporins bind directly to the outer rings, rendering detection of their interacting partners experimentally highly challenging1.
In order to identify potential novel functional associations for coat nucleoporins, we thus set out to characterize the nine families of Y-Nups across eukaryotes. In particular, we examined in detail their multi-domain architectures and their membership in co-expression groups in human and mouse that further support functional interactions. By integrating information from these analyses with previous knowledge, we significantly extend the emerging evidence for Y-Nup roles outside the nuclear pore, defined as ‘moonlighting' roles in this broader context9.
Results
To augment the limited set of known protein associations for Y-Nups across eukaryotes10, we deploy computational and experimental sequence analysis involving extensive sequence comparisons, RNA-seq expression profiling across diverse human and mouse tissues, protein domain detection and inference of protein interactions11, using the Drosophila melanogaster Y-Nups as queries. Using established protocols for low-complexity masking, sensitive iterative sequence profile searches, consistent labeling and annotation of homologs, automated sequence clustering and visualization of sequence similarity, we unambiguously assign the initially detected homologies (Supplementary Fig. 1) into nine Y-Nup families (Figure 1). The resulting multiple sequence alignments share as low as <10% identity between certain members and their homologs (p<10−04, see Methods and Supplementary Fig. 2)12.
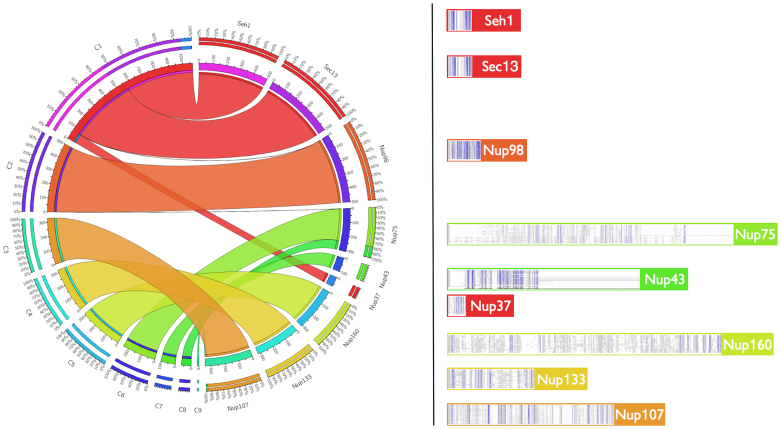
Figure 1. Automated clustering of validated Y-Nup family relationships.
Left: Circos Tableviewer (http://mkweb.bcgsc.ca/tableviewer/) representation (see Methods) of the nine automatically generated clusters (C1–C9) and the membership of the detected Y-Nup homologs from iterative sequence profile searches (named in alphabetical order, displayed in counterclockwise fashion). Stripes are color-coded according to clusters (for example, C1 is shown in red, spanning three Y-Nup homologs namely Nup37/SEH1/SEC13); there is a one-to-one correspondence between clusters and homologs except C1 (above), and Nup75 (C6, C7) and Nup107 (C3, C9). Outer circle values represent relative contributions, inner circle values correspond to absolute numbers. Right: Depiction of sequence alignments derived from profile searches (see Data Supplement DS03). Y-Nup families are assigned to the color of their highest-frequency cluster. Full-sized sample alignments are provided for Nup107 (minimum identity 4%, Supplementary Fig. 3) and Nup133 (minimum identity 9%, Supplementary Fig. 4). Note that Nup37 (in C1) and Nup43 (C8) detect fewer homologs due to their restricted phylogenetic distribution.
Clustering of Y-Nups into protein families
We identified ~3000 proteins as Y-Nups (Supplementary Table 1), many of which are reported here for the first time, especially for lower eukaryotes – including the previously undetected presence of Nup43 in fungal species (see Supplementary Text). These results confirm the universal distribution of the Y-Nup nuclear pore subcomplex in eukaryotes13. In particular, it is noteworthy that many of the protein sequences we detect here have not been reported previously in annotation efforts, due to the presence of subtle sequence similarities that are confounded by extensive low-complexity regions or repeats (e.g. WD40): of the 2962 entries in the resulting Y-Nup compendium, there are 1813 characterized and 1149 newly discovered Y-Nups, thus increasing the level of characterization by more than 63%. It should be pointed out that without low-complexity, compositionally biased region detection, the majority of these similarities are lost, mostly due to the presence of WD40 repeats, particularly for shorter query protein sequences. Based on our workflows (see Methods and Data Supplements DS01-09), we assigned detected homologs automatically into similarity clusters12 following detailed validation, essentially replicating our meticulous manual characterization in a highly consistent, reproducible manner. Of the 22,033 off-diagonal hits (i.e. excluding self-hits) in an all-against-all sequence comparison, 5,403 (24.5%) and 557 (2.5%) Y-Nups exhibit pairwise sequence identity <30% and ≥80% respectively (Supplementary Fig. 2). The nine independently derived clusters detected by the automated procedure correspond to all known classes of Y-Nups with the Nup37/SEH1/SEC13 families merging into the largest group (1077 members, minimum identity 7%), while the two smallest clusters represent distant sub-families of Nup75 (70 members from Ascomycota, minimum identity 8%) and Nup107 (10 members from Trypanosomatidae) (Figure 1).
Coordinated tissue-specific gene expression
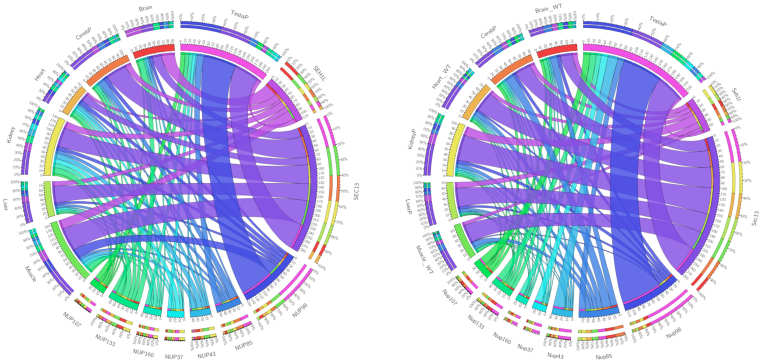
The rigorous delineation of Y-Nup structural features drawn from evolutionary relationships and multi-domain architecture is a prerequisite for the inference of genome-wide functional relationships both at the gene expression and protein interaction levels14. We thus examine Y-Nup gene expression tissue specificity patterns15, via RNA-seq data across a wide range of tissues and cell lines in human and mouse16 (Figure 2). There is a remarkable consistency of Y-Nup expression patterns across the two species17 (see Methods), with the most prominent feature a detected over-expression of Nup98 (Nup98-96) and SEC13 in testis. Also, SEC13 is more highly expressed in muscle, liver, kidney, heart and neural tissue than SEH1, Nup43 and Nup37 are significantly more expressed in mouse than in human testis, and mouse SEC13 has a higher expression in heart tissue compared to human (Figure 2). Exon skipping is found to be limited, with subtle tissue specificity patterns and minor alternative exon splicing events for Nup98 (Nup98-96) observed in both species (not shown), indicating a tight, evolutionarily conserved regulation at the transcriptional level (Figure 2).
Figure 2. Gene expression patterns of Y-Nups in human (left) and mouse (right) across seven representative tissues based on RNA-seq profiles.
Circos Tableviewer (http://mkweb.bcgsc.ca/tableviewer/) representation as in Figure 1. Y-Nups are labeled by their gene names in the corresponding species (as in Figure 1, except NUP85/Nup85 equivalent to Nup75). Tissue labels are self-explanatory (WT signifying wild-type for mouse, -P signifying a single tissue sample). Note that SEC13 exhibits a much higher expression level than SEH1 possibly due to its participation in other macromolecular complexes. The full RNA-seq patterns across a wide range of tissues and cell lines (see Methods) are provided (Data Supplement DS10).
Having established precise protein family relationships across Y-Nups and their coordinated gene expression patterns in two mammalian species, we then proceeded to the identification of domain associations and the extraction of their corresponding expression profiles. Domain associations can be used to infer the range of cellular functions that certain Y-Nup subunits might be performing, previously undetected by more traditional approaches18. These implied moonlighting functions19 for the homologous single-domain counterparts strongly point to the association of the Y-complex with other fundamental, yet transient processes at a given timepoint during the cell cycle20 and nuclear pore reorganization21,22. In fact, as mentioned above, the presence of common-repeat patterns in Y-Nups have occasionally confounded their detailed structural and functional characterization23, delineated with greater accuracy in this study.
Multi-domain architectures of Y-Nups
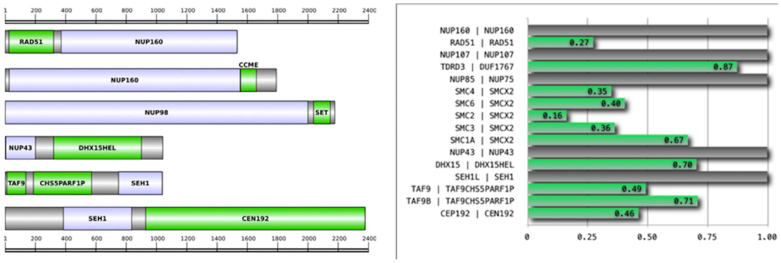
Following the above reasoning, we are thus able to detect 27 novel multi-domain architectures for Y-Nups (Supplementary Table 2), using an adaptive length threshold for the manual inspection of thousands of sequence alignments (see Methods), which in principle might involve genuine domain associations for Y-Nups11,18. These domains correspond to a wide range of functional categories, not directly related to nuclear pore formation, and thus warrant further investigation, using criteria for genome structure, gene expression and phylogenetic distribution. To validate the detected associations, we have first performed genomic sequence comparisons, using linker sequences of the corresponding multi-domain molecules as queries for genome and expression nucleotide sequence databases (see Methods and Data Supplements DS05-06): eight cases are supported by these exhaustive genomic searches (Supplementary Table 2, ‘by Genome'). Despite the fact that all homologs derive from complete genome sequences or assemblies (not shown) – represented by over 300,000 genes, there are quality issues that require independent experimental confirmation. We subsequently validate these architectures using the homology-based RNA-seq expression data from human and mouse (Supplementary Table 3): six cases are supported by this extensive genome-wide coverage (over 4 billion reads per species, Supplementary Table 4), across tissues and cell lines (Supplementary Table 2, ‘by Expression'). Genes that display coordinated expression across diverse cell and tissue types tend to share common functions, and the property of co-regulation has been used to predict gene function: herein, we use coordinated gene expression patterns as an additional level of validation for domain discovery associations. Remarkably, while there are three cases supported by both genomic and expression evidence, there are another three cases supported by either of the above, as well as presence in multiple species ('by Frequency') (Supplementary Table 2). While cases with variable support will require further experimental probing, six strongly supported cases (Table 1) can be unambiguously connected with coat nucleoporin function (Figure 3): five of these are found in more than one species.
Table 1. Moonlighting functions for Y-Nups indicated by functionally diverse domains. (column labels: Nucleoporin – name of Y-Nup; Y-Nup GI – representative composite protein identifier; Interaction partner – name of protein domain; Domain GI – representative single-domain protein identifier; Function – general function of associated domain; Biological process – Gene Ontology category identifier, representing functional association of corresponding domain).
| Nucleoporin | Y-Nup GI | Interaction partner | Domain GI | Function | Biological process |
|---|---|---|---|---|---|
| NUP160 | 322708659 | RAD51 | 322698012 | DNA repair | 0006281 |
| 166240053 | CcmE/CycJ domain¶ | 118587747 | cytochrome C biogenesis | 0017004 | |
| NUP98 | 342873147 | SET domain | 302917798 | chromatin regulation | 0016570 |
| NUP43 | 345482402 | DHX15 helicase¶ | 332019512 | mRNA processing | 0006397 |
| SEH1 | 320581285 | TAF9/Chs5p-Arf1p¶ | EFW95506.1 | transcription/protein export | 0006352/0015031 |
| 33086682 | Centrosomal protein 192¶§ | 351712025 | mitotic control | 0007051/0007098 |
Superscript symbols signify database records with a missing Y-Nup annotation¶ or the detection of the corresponding domain§ in the Y-Nup composite protein (Y-Nup GI) – all other cases can be regarded as correctly annotated database entries.
Figure 3.
A) Multi-domain architecture of representative Y-Nup homologs. Y-Nup domains identified by sequence searches (shown in light blue) found to be associated with protein domains with functions indicating moonlighting roles (shown in green). Grey boxes signify other protein regions. Scale is provided above and below the multi-domain diagram representations. Only the six cases with highest support (Supplementary Table 2) are shown (see also Table 1). A full characterization of the 27 validated domain associations is also provided (Supplementary Fig. 5). The phylogenetic distribution of all 27 reported domains is available in Data Supplement DS08. B) Coordinated RNA-seq expression patterns across twenty representative human tissues between Y-Nups and other domains supported by expression. Pearson correlation coefficient values across the twenty tissues are shown on the x-axis (see Methods). Gene names for human (left) and the encoded labels in this study (right) are shown on the y-axis (Supplementary Table 3), separated by a vertical bar. Full clustering analysis with Spearman rank correlation coefficients supports nine of the eleven cases (green bars) shown here, i.e. all listed genes except TDRD3 and TAF9B (middle block, in Figure 4).
Given the scarcity of known functional relationships for Y-Nups – partly due to technical limitations, the detection of novel genome-wide associations can expand their possible roles beyond the nuclear pore6, to include transient processes rarely detectable by targeted experiments. Thus, when validated by exhaustive functional genomics evidence, the inferred associations pointing to moonlighting roles of Y-Nups are highly consistent with the limited experimental evidence available both for gene expression (Figure 4) and protein interactions (Figure 5), in the broader context of biological processes as indicated by the associated domains (see also Data Supplement DS11). Beyond nuclear pore formation and maintenance5,6,7, the Y-Nups found associated with the strongly supported architectures (Figure 3, Table 1) can be linked to cellular processes – also previously reported, viz. cf. – involved in RNA processing and transport (cf. Rae124), DNA repair25 (cf. RAD5226), chromosome maintenance (cf. Sir4p27) and centrosome control28 (cf. Cenp-F29).
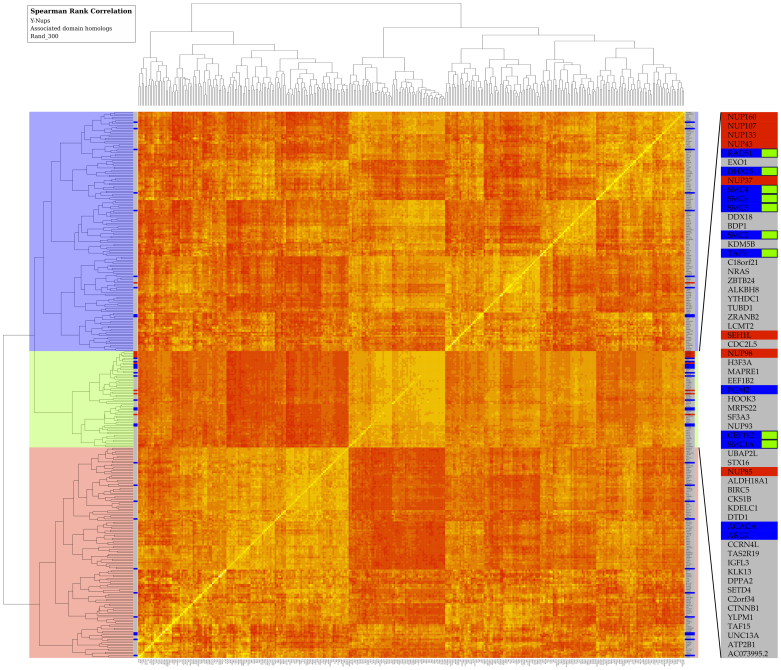
Figure 4. Clustering of human gene expression patterns across multiple tissues and cell lines.
Gene expression affinities are represented in a heat map by Spearman rank correlation coefficients as similarity measure across tissues and cell lines (see Methods). The narrow strip on both sides of the map indicates the entries corresponding to Y-Nups (red), homologs of associated domains (blue) – including human paralogs (Supplementary Table 3) found in other species (Supplementary Table 2), and a randomly selected set of 300 genes (grey); see also Methods. By splitting the clustering dendrogram at the second bifurcation, three clusters emerge depicted by three major blocks (color-coded, left side). The middle block (light green) is enriched in Y-Nups except SEC13, the upper cluster (light blue) contains SEC13, while the lower cluster (light red) does not contain any known Y-Nups. The inset (right side) magnifies entries in the middle block, where Y-Nups are included: gene names are shown, color scheme as for entire strip; green labels signify genes encoding for Y-Nup associated domains, supported by the bootstrap analysis (Supplementary Table 3). This figure is available at higher resolution as Supplementary Figure 6.
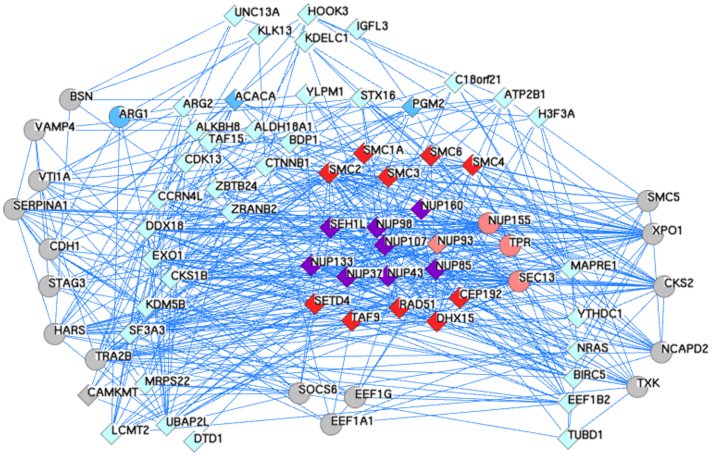
Figure 5. An expanded network of Y-Nup interactions from high-throughput experiments and the discovered multi-domain architectures.
Using as queries the human gene symbols of the molecules with coordinated gene expression patterns from the Y-Nup enriched middle block (see Figure 4), the resulting network is extracted by GeneMANIA with default parameters (only co-expression, physical and genetic interaction networks are retained). Queries were 60 (of which 3 are not found as interacting), depicted as diamonds (57 in total). An additional 20 genes are discovered by GeneMANIA, depicted as circles. The total genes in this network amount to 77, encompassing a number of known associations of Y-Nups (shown in purple, 8 in number) with other nucleoporins (shown in pink, 4 in number – including Sec13, not in query), e.g. TPR58 and related molecules (shown in light blue, 35 in number), e.g. EXO159. The reported molecules by GeneMANIA include interactions from large-scale experiments not further discussed (shown in grey, 17 in number). The three coordinated gene expression instances regarded as negatives in this study (shown in cyan, 3 in number) are ARG1 (curiously reported by GeneMANIA, thus shown as circle), ACACA and PGM2. The query molecule C2orf34 (synonym: CAMKMT, thus shown as diamond) is also reported by GeneMANIA. The six discovered novel domain associations (shown in purple, 10 in number), include the five of the six molecules with highest support (except CcmE/CycJ, Table 1) and five SMC paralogs (SMC1A, SMC2-4, SMC6), not previously found in association with Y-Nups. GeneMANIA reports no evidence for the association of NUP160-RAD51, NUP98-SET, NUP43-DHX15 and SEH1-TAF9, while providing strong evidence for SEH1-CEP19260,61,62. Common genes between PINA & GeneMANIA include other nucleoporins (e.g. NUP93) or others (e.g. EEF1G, Elongation factor 1-y) (see Methods). The annotated layout and GeneMANIA results with supporting literature are available in Data Supplement DS11.
Certain domain configurations with limited support might be due to sequencing artifacts, gene prediction or short-read assembly errors. Of those, four cases deserve further discussion although they are not admitted in our final list. The association of Nup75 (Nup153) from Naegleria gruberi (GI:290983204) with FG-repeats30 might represent a genuine case (see Supplementary Text). Another intriguing, low-support architecture is an association of SMC domains6 (condensins) with Nup75 of Chlorella variabilis (GI:307108886): both pairwise correlations (Figure 3B) and rank correlation clustering (Figure 4) indicate a co-expression of human paralogs with Nup75, SMC1A being the most Nup75-coordinated paralog across human tissues (Supplementary Table 3); independent observations from stem cell Oct4 interactions provide additional evidence31, although the particular Chlorella instance will need to be further validated. The third case with no counterpart elsewhere is the co-occurence of Nup107 with DUF1767 (domain of unknown function) in the flatworm Clonorchis sinensis (GI:358337287); moreover, DUF1767 is found in Rmi1, a protein controlling genome stability in yeast32 and exhibits the highest pairwise correlation of coordinated gene expression with Nup107 (Figure 3B). Finally, while not adequately supported, the co-occurrence of acetyl-CoA carboxylase with Nup75 in Rhodotorula glutinis (GI:342319109) provides clues for a suspected role of lipids in nuclear pore formation33,34. While all other cases are indeed tantalizing (including, e.g. aminopeptidase35), we conclude that more experimental and phylogenetic evidence is required and thus might not deem them as strong candidates for functional association with Y-Nups.
Validation and discovery of Y-Nup moonlighting functions
Strong functional genomics evidence for association with Y-Nups is detected for six domains (Table 1). Using the enriched Y-Nup group discovered by tissue-specific expression (middle block in light green, Figure 4), further substantial support for the six novel discoveries is obtained from high-throughput experiments (Figure 5), via a composite query to GeneMANIA36 (see Methods). By partitioning this network into two sub-networks, with the known cases and discovered multi-domain architectures deemed as positives (25 in number, average network connectivity 18) and all other nodes as negatives (depicted in light blue and grey, 52 in number, average network connectivity 12), the inferred nucleoporin-induced network exhibits a striking difference in topological complexity, thus placing the newly discovered multi-domain architectures pointing to moonlighting roles into a functionally coherent context.
The RAD51-Nup160 composite protein found in two fungal species, Metarhizium anisopliae ARSEF 23 (GI:322708659) and Phaeosphaeria nodorum SN15 (GI:169623440), annotated automatically in the corresponding sequence records, is strongly supported by gene expression data for human tissues, tightly co-ordinated not only with Nup160 but also Nup107, Nup133 and Nup43 (Figure 4). Interestingly, this association is also observed as a tandem gene cluster in Fusarium oxysporum lycopersici supercontig 2.1 (genes Foxg00234/5: https://img.jgi.doe.gov/cgi-bin/imgm_hmp/main.cgi?section=ScaffoldGraph&page=alignment&scaffold_id=2507525031,supercontig_2.1&coord1=779427&coord2=779510), further detected as a conserved pattern in multiple species where Nup160 remains unidentified (see https://img.jgi.doe.gov/cgi-bin/imgm_hmp/main.cgi?section=GeneNeighborhood&page=geneOrthologNeighborhood&gene_oid=2508346114&show_checkbox=1&cog_color=yes&use_bbh_lite=1). Additional experimental evidence is provided by Oct4 interactions (e.g. RAD50)31 as well as DNA damage response (DDR) studies, e.g. a si-RNA-based microscopy screening of ionizing radiation responses, pointing out the critical role that nucleoporins might play in genome maintenance37. The adjacent configuration of Nup160-CcmE in four uncharacterized proteins of Amoebozoa (Figure 3A) Dictyostelium discoideum AX4 (GI:166240053), D. purpureum (GI:330796511), D. fasciculatum (GI:328873820) and Polysphondylium pallidum PN500 (GI:281210825) provides strong comparative genomics evidence, in the absence of solid expression data: the role of CcmE in this context remains unknown at present38. NUP98 (Nup98-96) is found in association with a SET domain in three fungal species, namely in annotated Fusarium oxysporum Fo5176 (GI:342873147), and two uncharacterized proteins in Metarhizium acridum CQMa 102 (GI:322698664) and M. anisopliae ARSEF 23 (GI:322711125). Curiously, a similar configuration is found in patients with acute myeloblastic leukemia, with the fusion protein N-terminal NUP98-MLL acquiring a H3K4 methyltransferase ability through the SET domain present in MLL39. Similar observations support the association of SET with Nup9840, for instance the fusion of Nup98 to NSD1 (another SET-containing histone methyltransferase)41. The Nup43-DHX15 helicase association found in uncharacterized proteins of multiple insect species, for instance Nasonia vitripennis (GI:345482402), is consistent with the presence of a Werner helicase interacting protein in the Y-complex42 and DDX10 in leukemia43, while it is also detected in Oct4 interactions along with Nup4331 and very strong correlations with multiple Y-Nups (Supplementary Table 3, Figure 4). Most importantly, the interaction of DEAD-box helicases with other nucleoporins, for instance Ddx19 with Nup159, has been reported at the molecular level44. The association of TAF9 domain linked to Chs5p-Arf1p-binding domain and SEH1 in Ogataea parapolymorpha (GI:320581285) is supported by coordinated expression in human (TAF9, Figure 4) and the known involvement of TAF9 in the SAGA complex for chromatin remodelling6. Finally, centrosomal protein 192 (CEP192 in human) with a role in both centrosome maturation and spindle assembly45 is detected at the C-terminus of SEH1-specific WD40 repeats in multiple vertebrate species including rodents and marsupials, with high support by correlation clustering (Figure 4) and the presence of CEP192 with other centrosomal proteins and – curiously – Nup160 (figure 2 of cited work)28. Remarkably, this gene pair is also conserved in tandem organization across several vertebrate genomes (not shown). A set of complex patterns of variable functions is thus suggested by domain association analysis and validation (Table 1).
Discussion
We have demonstrated the presence of particular domains with a wide range of functional roles in four Y-Nup instances (Table 1), indicating the association of those domains with the nuclear pore as unraveled by functional genomics evidence and evolutionary conservation. While issues of sequencing or assembly artifacts remain a possibility and will pose a continuing challenge for whole-genome analysis of this kind, there is strong evidence supporting our findings in recent experimental studies5,27. In this work, we encountered those issues arising from short-read genome assemblies which required the use of independently derived information to support domain association analysis: our approach can thus be regarded as a proposed framework for function prediction, which could be further automated and made available for the wider community. In particular, comparative genomics reveals the extent to which the discovered domain relationships are conserved and can pinpoint towards species-specific adaptations rather than artifacts. These instances can be assessed experimentally in situ, with advances in novel imaging and molecular technologies46: indeed, further experimental analysis will shed light into these multi-domain associations.
Our analysis exemplifies how genome sequence and functional genomics data can be coupled to unravel intricate associations of key supramolecular complexes known to defy biochemical characterization at present. Although our results cannot prove the discovered associations definitively, they direct future experimental efforts. As recently articulated, domain association inference (if properly executed) can yield low-coverage yet high-precision functional relationships and might supplement interaction proteomics18. Herein, we augment substantially the set of known interactions for Y-Nups, contributing evidence for new instances of functionally diverse molecules that are omnipresent in different taxonomic categories. Our results indicate that the structural and functional characterization of Y-Nups thus obtained represents a step towards a better understanding of the functional versatility of this key nuclear pore subcomplex. In summary, our results are consistent with the emerging view that Y-Nups, rather than serving as inert components of the nuclear pore, are actually functionally diverse and possess unexpected moonlighting functions5,46,47.
Methods
Data collection
Proteins from the D. melanogaster nuclear pore complex considered as stoichiometrically assembled Y-Nups (i.e. explicitly excluding ELYS) were collected and tabulated (Supplementary Table 1). We maintain the order according to previous reports10.
Sequence filtering & searching
All sequences were masked using CAST48 with score ≥15 and otherwise default parameters, to exclude subtle compositional bias, including well-known repeats found in these proteins (Data Supplement DS01). In total, 160 regions were filtered out for such elements. These low-complexity, compositionally biased regions are provided separately, for further study (Data Supplement DS02).
The masked sequences were used as queries against the non-redundant protein sequence database (NRDB) at NCBI (15,052,178 entries)49 with BLAST (e-value cut-off threshold 10−06)50. Furthermore, these searches were manually executed with PSI-BLAST with a variable number of iterations until convergence (PSI-BLAST parameters: e-value cut-off threshold 10−04, 500 alignments, CAST score ≥15) (Supplementary Table 1), in particular to delineate possible anomalies such as multi-domain structure (Data Supplement DS03). Results from the above searches were evaluated (and confirmed with reverse sequence searches, not shown) and multi-domain similarities were extracted for subsequent analysis (for similarity distributions see Data Supplement DS04). Validity of domain associations was assessed by searching with linker sequences (Data Supplement DS05) against nucleotide databases – as a proxy for visual inspection of genome browser tracks; linkers were extracted and searched against these data collections within boundaries of ±20 amino acid residues where possible (Data Supplement DS06) and associated domains were separately extracted (Data Supplement DS07) and examined for taxonomic distribution (Data Supplement DS08). Multiple alignments were extracted and visualized by JalView51 – using redundancy elimination interactively until the production of visually appealing multiple alignments (Data Supplement DS03).
Clustering & annotation
All detected homologs labeled accordingly were compared using BLAST in an all-against-all mode (e-value cut-off threshold 0.01), following CAST masking as above. The similarity pairwise list was submitted to MCL sequence clustering using an inflation value of 1.2; clusters were incrementally assigned to an integer identifier12. Clusters are sorted by their size (number of members in a cluster, Data Supplement DS09); thus, the largest clusters have smallest integer identifiers (groups with 2 or less members are omitted, namely 12 instances). These cases (12/2962 or 0.4%) yield a sensitivity level of 99.6%. Conversely, two ‘false' positives in clusters C1 (Nup98, GI:262118708) and C2 (Nup75, GI:307191801) yield a specificity level of 99.9% (under further investigation – Promponas et al., in preparation).
Expression profiles and protein interactions
Next-generation sequencing (NGS) data for a wide range of human and mouse tissues and cell lines were extracted from multiple available sources (Supplementary Table 4 – for other species, data are not as rich). Expression data for each instance were measured using cRPKM units [corrected (form mappability) Reads Per Kilobase per Million mapped reads], calculated as previously described16,52. The orthologs from human and mouse were analyzed for tissue-specific gene expression across all samples16,17 (Data Supplement DS10). Identification of cassette alternative exons and quantification of their transcript inclusion levels across samples was performed as previously described53 (see also: Hon et al., submitted). Both sequence clusters and gene expression profiles (Figures 1 and 2) were visualized with Circos54.
Gene expression data for Y-Nups and associated domain homologs in human (Supplementary Table 4) were subject to bootstrap rank correlation statistics (Supplementary Table 3). Expression patterns from 300 randomly selected human genes were systematically sampled 500 times with replacement for bootstrapping, in subsets of 100 expression patterns. Each subset was merged with Y-Nup and associated domain homolog expression profiles for Spearman rank correlation analysis, and average ranks were recorded (Supplementary Table 3). The complete gene expression dataset (human Y-Nups, human homologs of the 27 associated domains, random sample of 300 human genes) was clustered based on Spearman rank correlation coefficients (Figure 4).
Known protein interactions were extracted from the PINA database55,56 and annotated appropriately; these data were augmented by the discovered domain associations (Data Supplement DS11a), and are made available in BioLayout57 format for visual exploration. Coordinated tissue-specific gene expression data enriched in Y-Nups were used as a composite query to GeneMANIA36 resulting in supporting evidence from high-throughput experiments (Data Supplement DS11b). Note that the PINA results are used only to reflect the current status of knowledge for Y-Nup interactions while the GeneMANIA results are used to discover and provide support for the novel findings reported here.
Entire Y-Nup sequence compendium
All 2962 Y-Nups +27 external domain = 2989 sequences detected by the above analysis are labeled by property and domain association and provided in FASTA format for further study and a possible basis for a more consistent nomenclature (Data Supplement DS12).
Data availability
All results (in 12 Data Supplements) are available as a ZIP archive (58.3 MBytes) on http://dx.doi.org/10.6084/m9.figshare.840452
Author Contributions
K.R.K. was involved in study design, literature searches, data analysis and annotation; M.I. and B.J.B. provided expression data and assisted in their interpretation; C.K. assisted with visualisation and analysis; Z.G.S. was involved in study design, data interpretation and project coordination; V.J.P. and C.A.O. designed the study, performed computational analysis, analysed data, coordinated collaborative project and wrote the paper. All authors analysed data, discussed the results and commented on the manuscript.
Supplementary Material
SREP-13-05332A SI
Supplementary Figure 6
Acknowledgments
Parts of this work have been supported by the FP7 Collaborative Projects MICROME (grant agreement # 222886-2) and CEREBRAD (grant agreement # 295552), both funded by the European Commission. C.A.O. thanks the Department of Biological Sciences at the University of Cyprus for their kind hospitality during 2012. All authors wish to thank Dr. Valérie Doye (Université Paris Diderot, Sorbonne Paris) for critical comments on the manuscript and for suggesting the term Y-Nups. M.I. is the recipient of a HFSP Long Term Fellowship, and B.J.B. gratefully acknowledges funding from the Canadian Institutes for Health Research.
References
- Grossman E., Medalia O. & Zwerger M. Functional architecture of the nuclear pore complex. Annu Rev Biophys 41, 557–584 (2012). [DOI] [PubMed] [Google Scholar]
- Hoelz A., Debler E. W. & Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem 80, 613–643 (2011). [DOI] [PubMed] [Google Scholar]
- Devos D. et al. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci U S A 103, 2172–2177 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Mitchell J. M., Wozniak R. W., Blobel G. & Fan J. Structural evolution of the membrane-coating module of the nuclear pore complex. Proc Natl Acad Sci U S A 109, 16498–16503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M. & D'Angelo M. A. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol 13, 687–699 (2012). [DOI] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M. & Rout M. P. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol 11, 490–501 (2010). [DOI] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C. & Askjaer P. Dissecting the NUP107 complex: multiple components and even more functions. Nucleus 3, 340–348 (2012). [DOI] [PubMed] [Google Scholar]
- Sarma N. J. & Willis K. The new nucleoporin: Regulator of transcriptional repression and beyond. Nucleus 3, 508–515 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle S. J. Protein adaptation: mitotic functions for membrane trafficking proteins. Nat Rev Mol Cell Biol 14, 592–599 (2013). [DOI] [PubMed] [Google Scholar]
- Neumann N., Lundin D. & Poole A. M. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One 5, e13241 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A. J., Iliopoulos I., Kyrpides N. C. & Ouzounis C. A. Protein interaction maps for complete genomes based on gene fusion events. Nature 402, 86–90 (1999). [DOI] [PubMed] [Google Scholar]
- Enright A. J., Van Dongen S. & Ouzounis C. A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30, 1575–1584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste E., Charlebois R. L., MacLeod D. & Brochier C. The two tempos of nuclear pore complex evolution: highly adapting proteins in an ancient frozen structure. Genome Biol 6, R85 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounis C. A., Coulson R. M., Enright A. J., Kunin V. & Pereira-Leal J. B. Classification schemes for protein structure and function. Nat Rev Genet 4, 508–519 (2003). [DOI] [PubMed] [Google Scholar]
- Lupu F., Alves A., Anderson K., Doye V. & Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell 14, 831–842 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais N. L. et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 338, 1587–1593 (2012). [DOI] [PubMed] [Google Scholar]
- Lopez-Bigas N., Blencowe B. J. & Ouzounis C. A. Highly consistent patterns for inherited human diseases at the molecular level. Bioinformatics 22, 269–277 (2006). [DOI] [PubMed] [Google Scholar]
- Promponas V. J., Ouzounis C. A. & Iliopoulos I. Experimental evidence validating the computational inference of functional associations from gene fusion events: a critical survey. Brief Bioinform (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley S. D. Moonlighting is mainstream: paradigm adjustment required. Bioessays 34, 578–588 (2012). [DOI] [PubMed] [Google Scholar]
- Chakraborty P. et al. Nucleoporin levels regulate cell cycle progression and phase-specific gene expression. Dev Cell 15, 657–667 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo M. A., Raices M., Panowski S. H. & Hetzer M. W. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136, 284–295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger S., Laurell E. & Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol 10, 178–191 (2009). [DOI] [PubMed] [Google Scholar]
- Tamura K., Fukao Y., Iwamoto M., Haraguchi T. & Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22, 4084–4097 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Seo H. S., Blobel G. & Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc Natl Acad Sci U S A 107, 10406–10411 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B. et al. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol Biol Cell 18, 2912–2923 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen R. D. et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35, 228–239 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vosse D. W. et al. A Role for the Nucleoporin Nup170p in Chromatin Structure and Gene Silencing. Cell 152, 969–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins J. R. et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhy S. et al. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol 192, 855–871 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn L. A., Shen T., Shulga N., Goldfarb D. S. & Wente S. R. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol 6, 197–206 (2004). [DOI] [PubMed] [Google Scholar]
- Cheong C. Y. et al. In silico tandem affinity purification refines an Oct4 interaction list. Stem Cell Res Ther 2, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Nallaseth F. S., Lan Y. Q., Slagle C. E. & Brill S. J. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol 25, 4476–4487 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E. & Weis K. Nuclear pore complex-a coat specifically tailored for the nuclear envelope. Curr Opin Cell Biol 23, 293–301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R. et al. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol 16, 7161–7172 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A. & Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8, 761–773 (2007). [DOI] [PubMed] [Google Scholar]
- Warde-Farley D. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38, W214–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudry P. et al. Nucleoporin NUP153 guards genome integrity by promoting nuclear import of 53BP1. Cell Death Differ 19, 798–807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer E. B. & Barker P. D. Continued surprises in the cytochrome c biogenesis story. Protein Cell 3, 405–409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach S. et al. NUP98-MLL fusion in human acute myeloblastic leukemia. Blood 116, 2332–2335 (2010). [DOI] [PubMed] [Google Scholar]
- Light W. H. et al. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol 11, e1001524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. G., Cai L., Pasillas M. P. & Kamps M. P. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 9, 804–812 (2007). [DOI] [PubMed] [Google Scholar]
- Kaur S., White T. E., DiGuilio A. L. & Glavy J. S. The discovery of a Werner Helicase Interacting Protein (WHIP) association with the nuclear pore complex. Cell Cycle 9, 3106–3111 (2010). [PubMed] [Google Scholar]
- Yassin E. R., Abdul-Nabi A. M., Takeda A. & Yaseen N. R. Effects of the NUP98-DDX10 oncogene on primary human CD34+ cells: role of a conserved helicase motif. Leukemia 24, 1001–1011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit B. et al. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472, 238–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ferreria M. A. & Sharp D. J. Cep192 and the generation of the mitotic spindle. Cell Cycle 7, 1507–1510 (2008). [DOI] [PubMed] [Google Scholar]
- Adams R. L. & Wente S. R. Uncovering Nuclear Pore Complexity with Innovation. Cell 152, 1218–1221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B., Pickersgill H., Shloma V. V. & Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140, 360–371 (2010). [DOI] [PubMed] [Google Scholar]
- Promponas V. J. et al. CAST: an iterative algorithm for the complexity analysis of sequence tracts. PubMed artifact. Bioinformatics 16, 915–922 (2000). [DOI] [PubMed] [Google Scholar]
- Sayers E. W. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 40, D13–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer A. A. et al. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res 29, 2994–3005 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M., Clamp M. & Barton G. J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe R. M. et al. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells 30, 1734–1745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare T. et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat Struct Mol Biol 19, 1037–1043 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. et al. Integrated network analysis platform for protein-protein interactions. Nat Methods 6, 75–77 (2009). [DOI] [PubMed] [Google Scholar]
- Cowley M. J. et al. PINA v2.0: mining interactome modules. Nucleic Acids Res 40, D862–865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldovsky L., Cases I., Enright A. J. & Ouzounis C. A. BioLayout(Java): versatile network visualisation of structural and functional relationships. Appl Bioinformatics 4, 71–74 (2005). [DOI] [PubMed] [Google Scholar]
- Snow C. J., Dar A., Dutta A., Kehlenbach R. H. & Paschal B. M. Defective nuclear import of Tpr in Progeria reflects the Ran sensitivity of large cargo transport. J Cell Biol 201, 541–557 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R. et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146, 233–246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res 11, 5730–5739 (2005). [DOI] [PubMed] [Google Scholar]
- Hummel M. et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med 354, 2419–2430 (2006). [DOI] [PubMed] [Google Scholar]
- Nakayama R. et al. Gene expression analysis of soft tissue sarcomas: characterization and reclassification of malignant fibrous histiocytoma. Mod Pathol 20, 749–759 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SREP-13-05332A SI
Supplementary Figure 6
Data Availability Statement
All results (in 12 Data Supplements) are available as a ZIP archive (58.3 MBytes) on http://dx.doi.org/10.6084/m9.figshare.840452