Abstract
Parkinson's disease (PD)-associated Pink1 and Parkin proteins are believed to function in a common pathway controlling mitochondrial clearance and trafficking. Glial cell line-derived neurotrophic factor (GDNF) and its signaling receptor Ret are neuroprotective in toxin-based animal models of PD. However, the mechanism by which GDNF/Ret protects cells from degenerating remains unclear. We investigated whether the Drosophila homolog of Ret can rescue Pink1 and park mutant phenotypes. We report that a signaling active version of Ret (RetMEN2B) rescues muscle degeneration, disintegration of mitochondria and ATP content of Pink1 mutants. Interestingly, corresponding phenotypes of park mutants were not rescued, suggesting that the phenotypes of Pink1 and park mutants have partially different origins. In human neuroblastoma cells, GDNF treatment rescues morphological defects of PINK1 knockdown, without inducing mitophagy or Parkin recruitment. GDNF also rescues bioenergetic deficits of PINK knockdown cells. Furthermore, overexpression of RetMEN2B significantly improves electron transport chain complex I function in Pink1 mutant Drosophila. These results provide a novel mechanism underlying Ret-mediated cell protection in a situation relevant for human PD.
Keywords: Drosophila, neurodegeneration, neurotrophic factors, OXPHOS, Parkinson's disease
Introduction
The etiology of Parkinson's Disease (PD) is highly complex and largely unknown, involving both environmental and genetic risk factors. Mitochondrial dysfunction, oxidative stress and protein aggregation are believed to be central events in the pathological process, but their interconnection remains unclear (Schapira & Jenner, 2011; Exner et al, 2012; McCoy & Cookson, 2012). The first indications of a role for mitochondria came with the discovery that the toxin 1-methyl-4-phenyl-1,2,3,4-tetrahydropyridine (MPTP) causes Parkinsonism in humans and animal models (Burns et al, 1983; Langston et al, 1983). Its active metabolite, 1-methyl-4-phenylpyridinium ion (MPP+), is selectively imported into dopaminergic neurons via the dopamine transporter, and inhibits complex I of the electron transport chain (ETC). Several other mitochondrial toxins, including paraquat and rotenone, generating either mitochondrial reactive oxygen species (ROS) or specifically inhibiting complex I, have been linked to PD in epidemiological studies and animal models (de Lau & Breteler, 2006). Furthermore, patients with sporadic PD can have decreased activity of complex I in brain and other tissues (Schapira et al, 1989; Parker & Swerdlow, 1998), or less complex I proteins in the substantia nigra (Mizuno et al, 1989).
Autosomal recessive PD-associated proteins Parkin, PINK1 and DJ-1 (OMIM #600116, 605909, 606324) have been shown to have functions related to mitochondrial integrity, (reviewed in Exner et al, 2012; Martin et al, 2011). In three seminal studies, Pink1 mutant Drosophila displayed mitochondrial abnormalities and muscle degeneration in a manner highly similar to park mutants, and Parkin overexpression largely rescued the phenotypes of Pink1 mutants, but not vice versa, suggesting that the two proteins act in a common linear pathway (Clark et al, 2006; Park et al, 2006; Yang et al, 2006). Manipulation of the mitochondrial remodeling machinery rescues some Pink1 and park mutant phenotypes in Drosophila and in mammalian cell lines. However, while increasing fission rescues the Drosophila phenotypes, shifting the fusion/fission balance in the opposite direction rescues mammalian cell lines, but the underlying mechanisms are not fully understood (Deng et al, 2008; Poole et al, 2008; Lutz et al, 2009). PINK1, a mitochondrial Ser/Thr kinase, and Parkin, an E3 Ubiquitin ligase, were found to regulate clearance of damaged mitochondria via mitophagy (Geisler et al, 2010; Narendra et al, 2010; Vives-Bauza et al, 2010), and microtubular transport (Weihofen et al, 2009; Wang et al, 2011). However, other studies have reported additional functions of Parkin in the regulation of stress response proteins and mitochondrial biogenesis (Bouman et al, 2011; Shin et al, 2011), in promoting NF-κB signaling (Henn et al, 2007; Muller-Rischart et al, 2013), and in controlling cytochrome-c release (Berger et al, 2009). PINK1 also has additional functions, unrelated to recruiting Parkin, such as regulating mitochondrial calcium buffering (Gandhi et al, 2009; Sandebring et al, 2009; Heeman et al, 2011). Furthermore, PINK1 mutant mitochondria have decreased activity of complex I of the ETC (Morais et al, 2009), and overexpression of a yeast substitute for complex I rescued many of the functional impairments of Pink1 mutant flies (Vilain et al, 2012). Additional studies are required to elucidate which of the functions reported for Parkin and PINK1 are critical for causing Parkinson pathology.
The neurotrophic factor Glial cell line-derived neurotrophic factor (GDNF) promotes the survival of dopamine neurons (Lin et al, 1993) and protects nigral dopamine neurons from cell death in rodent and primate toxin-models of PD such as 6-hydroxydopamine (6-OHDA) and MPTP (Kearns & Gash, 1995; Sauer et al, 1995; Tomac et al, 1995; Gash et al, 1996). Several clinical trials have been performed with mixed outcomes, but ongoing research and development aims at improving delivery methods of GDNF (Deierborg et al, 2008). GDNF signals via the GPI-anchored co-receptor GFR-α1 and the receptor tyrosine kinase Ret (Airaksinen & Saarma, 2002). Endogenous Ret expression is required for long-term survival of a fraction of nigral dopamine neurons in aged mice (Kramer et al, 2007). Conversely, mice that express a constitutively active Ret receptor in dopamine neurons (RetMEN2B) show increased numbers of dopamine neurons (Mijatovic et al, 2007). The mechanism by which GDNF/Ret protects dopamine neurons from cell death is not fully elucidated. We hypothesized that Ret-activated signaling pathways converge with functions of proteins associated with familial PD. We recently reported that Ret and DJ-1 double loss-of-function in aged mice exacerbates the neuron loss observed in Ret single mutants (Aron et al, 2010). Here, we investigated whether Ret interacts genetically with park and Pink1 in Drosophila. We found that constitutively active RetMEN2B specifically rescues phenotypes of Pink1 mutants, including muscle degeneration, mitochondrial morphology and function, whereas park mutants remained unaffected. Moreover, Ret signaling rescued mitochondrial morphological and functional defects of PINK1-deficient human SH-SY5Y cells, without activating mitophagy. Mechanistically, Ret signaling restored the activity of complex I of the ETC, which is reduced in Pink1, but not park mutant flies. Thus our study indicates that Ret signaling can specifically ameliorate Pink1 loss-of-function deficiencies that are relevant to human Parkinson's disease.
Results
Active Ret rescues Pink1 but not park mutant muscle degeneration
To study whether Ret can modify Pink1 and park phenotypes, we utilized the Drosophila indirect flight muscles (IFMs) as a model system. Here, Pink1 and park mutants undergo significant muscle degeneration, likely because of the high energy consumption of the IFMs, and display enlarged mitochondria with broken cristae. Late stage pupae display normal muscle morphology, but soon after eclosion, the muscle tissue degenerates (Greene et al, 2003; Clark et al, 2006; Park et al, 2006). In 3- to 5-day-old Pink1 and park mutant animals housed at 18°C, interrupted muscles were found, and one or several of the six muscles displayed degenerated, highly irregular myofibrils with abnormal sarcomere structure, hereafter referred to as “degenerated” (Fig1I and K) in approximately 65% of the animals as compared to controls, which never displayed this phenotype (Fig1A, B, E, F, L). To investigate whether Ret signaling could modify muscle degeneration, we utilized the constitutively active version, RetMEN2B, which has an activating point mutation in the kinase domain (M955T) (Read et al, 2005). In an expression analysis of endogenous Ret by reverse transcriptase PCR (RT-PCR), we detected high levels of Ret mRNA in larvae and pupae, and lower levels in the adult thorax and IFMs (Supplementary Fig S1). To achieve robust overexpression of activated Ret specifically in muscles, we used the UAS-GAL4 system and the Myocyte enhancer factor-2 (Mef2) GAL4 driver, which is active in all muscle tissues from the early embryo throughout larval and pupal stages and in the adult fly. Mef2>RetMEN2B overexpression caused lethality at 25°C, but at 18°C, viable progeny eclosed with lower frequency. Surviving transgenic flies displayed mild muscle abnormalities, including deposits of actin dispersed over the muscle tissue, and some abnormally thick and irregular myofibrils (Fig1C, G, J). A recent RNAi screen for modifiers of muscle development (Schnorrer et al, 2010) identified a large number of lines with a highly reminiscent phenotypic class and designated this “actin blobs”, we therefore refer to this by the same term. When RetMEN2B was overexpressed in the background of Pink1 mutants, the majority of flies showed significantly improved muscle morphology, with only 12% of flies displaying degenerated myofibrils (Fig1D and L). The frequency of flies with actin blobs also decreased markedly compared to RetMEN2B expressing controls, suggesting that Pink1 function may be required for this phenotype. However, in contrast to Pink1 mutants, park mutants overexpressing RetMEN2B showed no improvement as the frequency of degenerated myofibrils remained unchanged (Fig1H and L). Expression of the RetMEN2B protein was examined by Western Blot of thorax homogenates and levels were similar between the Pink1 and park mutants, indicating that differences in transgene expression were not a likely cause of the differential response (Fig1M). To determine if Ret protein expression or Ret signaling was required for the phenotypic rescue, we overexpressed wild-type (WT) Ret using the same GAL4 driver. We found that RetWT was unable to modify the phenotype probably because the putative Ret ligand was not present in the IFMs at significant levels at this stage (Supplementary Fig S2). Moreover, the effects of Ret on IFM morphology appeared rather specific, since overexpression of a constitutively active fibroblast growth factor receptor (FGFR), UAS-htlλ, caused a dramatic change in IFM fate (data not shown).
Figure 1.
- A–K Drosophila hemi-thoraces stained with phalloidin at low magnification (upper panels) showing overall indirect flight muscle (IFM) morphology, and at higher magnification (lower panels). High-magnification images of WT sarcomeres (I), sarcomeres with ‘actin blobs’ (J), and degenerated sarcomeres (K). Heterozygous controls (A, E) display normal IFM layout (upper panels), myofibril morphology (lower panels) and sarcomeres (I). Pink1 (B) and park mutants (F) display abnormal morphologies with truncated muscles (yellow arrow heads, upper panels) and disorganized myofibrils (lower panels) with degenerated sarcomere structure (K). Animals overexpressing RetMEN2B (C, G) display normal IFM layout (upper panels), fairly normal myofibril morphology with occasional deposits of mislocated actin filaments, and “actin blobs”, (red arrow heads, lower panels and J). RetMEN2B overexpression in Pink1 mutants largely rescues the mutant phenotypes, as the majority of animals display normal IFM morphology (D), while park mutants are not rescued (H).
- L Percentage of flies with phenotype “wild type” (blue), “actin blobs” (green), “degenerated” (red) or “actin blobs and degenerated” (yellow).
- M Western blot analysis of Ret expression in thorax homogenates from w1118 controls, and Pink1, or park mutants overexpressing RetMEN2B, indicating similar levels of Ret overexpression between the two mutant backgrounds. Tissue from three animals per sample. Tubulin was used as a loading control.
- N–U Overexpression of UAS-RetMEN2B under control of Mhc-GAL4 and Tub-GAL80ts, pupae were shifted from 18 to 30°C at pupal stage 11, activating expression after muscle formation is completed. Heterozygous controls (N, R) and RetMEN2B late overexpressing animals display normal muscle and myofibril morphologies (N, P, R, S, T). Pink1 (O) and park mutants (S) display abnormal morphologies with truncated muscles and disorganized myofibrils with degenerated sarcomere structure (lower panels). Late RetMEN2B overexpression in Pink1 mutants (Q) largely rescues the mutant phenotypes, while park mutants (U) are not rescued.
- V Percentage of flies with phenotype “wild type” (blue) or “degenerated” (red). Number of animals per genotype as depicted in figure.
Rescue of Pink1 mutants is not developmental
The partial embryonic lethality and appearance of actin blobs by Mef2>RetMEN2B overexpression indicated that high levels of Ret signaling interfered with normal muscle development. Other receptor tyrosine kinases such as epidermal growth factor receptor (EGFR) and FGFR are known to regulate embryonic myoblast specification via Ras/Erk signaling (Carmena et al, 1998; Halfon et al, 2000), and the insulin receptor controls muscle size (Demontis & Perrimon, 2009). Therefore, it is plausible that active RetMEN2B affects these, or similar developmental processes. To verify that the rescue of the Pink1 mutants is not a developmental interaction, we utilized the GAL80ts system which permits transgene expression in a defined time window regulated by temperature. To drive RetMEN2B expression, we chose the GAL4 driver, Myosin heavy chain (Mhc) GAL4, which expresses only in differentiated muscles, not in myoblasts, in difference to Mef-GAL4 and generates higher expression. Unlike Mef2-GAL4, it causes complete lethality when driving RetMEN2B from embryonic stages. Flies were crossed at 18°C (non-permissive temperature), after which pupae were shifted to 30°C (permissive temperature) at pharate adult stage P11 ± 3 h (equivalent of 75 h APF at 25°C) (Flybase FBdv:00005349), a time well after completion of IFM development, but before the onset of apoptotic degeneration in Pink1 and park mutants (Greene et al, 2003; Clark et al, 2006). Analyses were again performed at 3–5 days post-eclosion. Using this protocol, Pink1 and park mutants showed degenerated myofibrils with a frequency of approximately 90% and 80% respectively as compared to controls (Fig1N, O, R, S, V), the higher penetrance being likely due to the increased temperature. RetMEN2B-overexpressing flies eclosed with Mendelian frequencies and displayed fully normal muscle morphology, without the presence of actin blobs, confirming the hypothesis that the lethality and actin blob phenotypes have developmental origins (Fig1P, T, V). When RetMEN2B was expressed in Pink1 mutants from this late pupal stage and onwards, it again largely rescued muscle degeneration, indicating that the rescue is not due to a developmental interaction, but a direct protective effect of Ret signaling on degenerating tissue (Fig1Q and V). Interestingly, park mutants were again not rescued using this expression protocol (Fig1U and V).
Ret signaling rescues mitochondrial morphology in flight muscles
One possibility is that RetMEN2B inhibits muscle degeneration without directly targeting the primary cause of the Pink1 phenotype: mitochondrial impairments (Clark et al, 2006). To test this possibility, we analyzed the ultrastructure of mitochondria using transmission electron microscopy. IFMs from control flies showed regular organization of myofibrils and densely packed mitochondria with intact cristae (Fig2A, E, L, M). Pink1 and park mutants displayed a heterogeneous population of mitochondria with the majority having significantly enlarged sizes and mild or severe disruption of their cristae structure, when compared to control mitochondria (Fig2B, F, I–M). Mef2>RetMEN2B overexpression in control flies did not alter normal mitochondria morphology (Fig2C, G,L, M). However, in Pink1 mutants, RetMEN2B overexpression significantly reduced the fraction of severely impaired mitochondria and increased the fraction of mitochondria with WT-like cristae structure (Fig2D and L). In contrast, park mutants showed no improvement of structural impairments when RetMEN2B was overexpressed (Fig2H and M). These results demonstrate that RetMEN2B can rescue mitochondrial impairments of pink1 but not park mutants, suggesting that the mitochondrial deficiencies of the two mutant strains have partially different origins.
Figure 2.

- A–K Transmission electron microscopy images of indirect flight muscles. Heterozygous controls (A, E) and animals overexpressing RetMEN2B (B, F) display normal mitochondria of similar size with highly dense cristae structure. Pink1 and park mutants have enlarged mitochondria with broken cristae (C, G). Phenotype can vary from mild to severe. High-power images of mitochondria are shown for the categories “wild type” (I), “mild” (J), “severe” phenotype (K). RetMEN2B overexpression partially restores mitochondrial size and cristae structure in Pink1 (D), but not park mutants (H). Scale bar, 2 μm.
- L, M Percentages of mitochondria of the indicated categories, 500–800 mitochondria per animal, averages of 6 animals per genotype.
Ret rescues mitochondrial morphology in dopaminergic neurons
To address whether RetMEN2B also rescues the morphology of mitochondria in dopaminergic neurons, we overexpressed RetMEN2B using TH-GAL4 together with the mitochondrial marker mitoGFP (Pilling et al, 2006). Pink1 and park mutants displayed severely enlarged mitochondria as compared to controls (Fig3A, B, E, F, I, J). RetMEN2B overexpression in a control background did not significantly alter the normal mitochondrial background (Fig3C, G, I, J). However, when overexpressed in Pink1 mutants, mitochondrial size was significantly rescued (Fig3D and I). Quantification of mitochondrial volumes revealed that in the presence of RetMEN2B the abundance of normal mitochondria was increased, while the fraction of enlarged mitochondria decreased to levels similar to those of control flies. Merely, the 4% largest mitochondria were not rescued. In line with the analysis of mitochondria in muscle, mitochondrial morphology in neurons of park mutants was not rescued by RetMEN2B (Fig3H and J).
Figure 3.
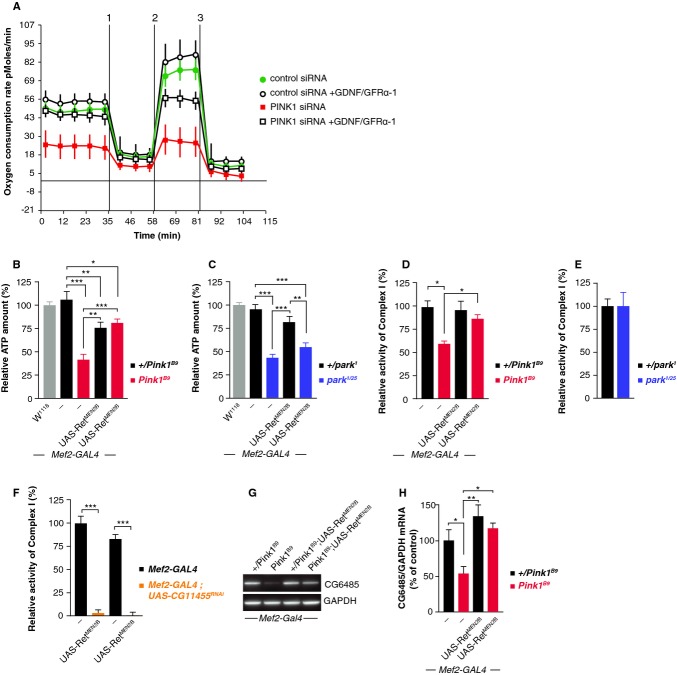
- A–H Confocal maximum projections (left panels) and isosurface renderings (right panels) of dopamine neuron mitochondria in the PPL1 cluster of dopaminergic neurons, visualized by mitoGFP and immunostainings against GFP and TH. Genotypes: All flies contain TH-GAL4 and UAS-mitoGFP and Pink1, park mutant alleles, as well as UAS-RetMEN2B as indicated. Isosurface renderings are color-coded according to volume from 0 to 3 μm3. RetMEN2B-overexpressing control animals (C, G) display normal mitochondrial morphology as compared to non-transgenic controls (A, E). Pink1 mutants (B) and park mutants (F) display severely enlarged mitochondria, and RetMEN2B partially rescues mitochondrial size in Pink1 mutants (D), but not in park mutants (H). Scale bar, 5 μm.
- I, J Mitochondrial volume distributions of (A–D) and (E–H) in categories as indicated. Due to differences in staining and imaging conditions, data between the Pink1 and park datasets cannot be directly compared. n = 8–20 animals per genotype.
GDNF/Ret signaling rescues mitochondrial defects in mammalian cells
In order to assess whether signaling from endogenous Ret can also rescue mitochondrial impairments caused by loss of PINK1 function, we used the human dopaminergic neuroblastoma cell line SH-SY5Y, which expresses endogenous Ret. Acute knock-down of PINK1 in this cell line was previously shown to cause fragmentation of the mitochondrial network (Lutz et al, 2009) (Fig4A, B, D). Stimulation of Ret by GDNF and soluble GFRα-1 rescued mitochondrial fragmentation, demonstrating that endogenous mammalian Ret can rescue mitochondrial impairments (Fig4C and D). A semi-quantitative RT-PCR analysis of PINK1 mRNA controlled that GDNF/GFRα-1 stimulation did not upregulate PINK1 levels (Fig4E).
Figure 4.
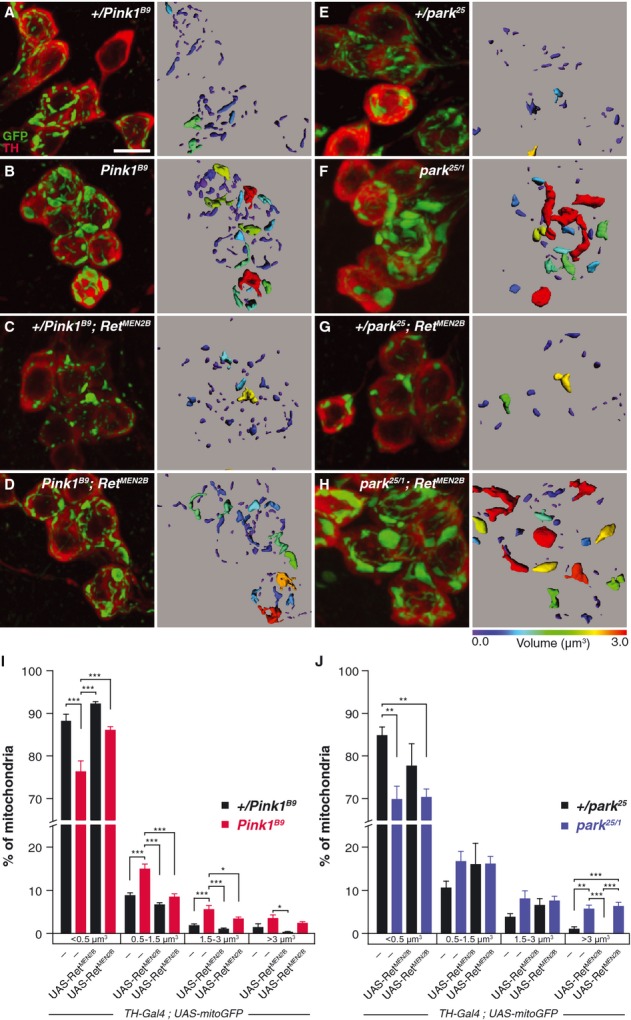
- A–C SH-SY5Y cells expressing endogenous Ret, transfected with scrambled control siRNA (A) display normal tubular mitochondrial morphology, visualized by immunostaining for TOM20 (white); DAPI (blue) indicates nuclei. Cells silenced for PINK1 expression display increased mitochondrial fragmentation (B). Stimulation of Ret signaling by treatment of cells with GDNF together with soluble GFRα-1 rescues mitochondrial fragmentation after PINK1 knockdown (C).
- D Quantification of cells with either tubular (gray) or fragmented (blue) mitochondria.
- E Quantification of PINK1 mRNA by quantitative RT-PCR indicates that GDNF/GFRα-1 treatment has no effect on PINK1 expression.
- F–M SH-SY5Y cells were treated with CCCP for 2 or 24 h to depolarize mitochondria, and then stained for HSP60 (red), DAPI (blue) and Parkin or Ret (green) as indicated. Cells with endogenous Parkin expression display low levels of mitophagy (F) and no cells fully cleared of mitochondria were detected 24 h after CCCP treatment. Cells overexpressing Parkin display translocation of Parkin to mitochondria 2 h after CCCP treatment and complete clearance of mitochondria by 24 h after adding CCCP (G). White arrowheads indicate cells without detectable mitochondria. Silencing of PINK1 by siRNA largely inhibits Parkin translocation and mitophagy (H), whereas silencing of Ret has no effect on mitophagy alone (I) or in cells overexpressing Parkin (J). Overexpression of constitutively active RetMEN2A does not activate mitophagy in control or PINK1-silenced cells (K, L), and does not modulate Parkin translocation or mitophagy in Parkin-overexpressing cells (M).
- N Quantification of the experiments described in (F–M).
- O Quantification of mRNA after PINK1 or Ret silencing by quantitative RT-PCR.
Ret rescues mitochondrial morphology independently of Parkin-induced mitophagy
Although the data so far suggested that Ret rescues Pink1-deficient mitochondria independently of Parkin, we cannot exclude that Ret signaling activates Parkin translocation to mitochondria, thus promoting their clearance through mitophagy. To test this hypothesis, we treated SH-SY5Y cells overexpressing Parkin with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) to depolarize mitochondria. CCCP treatment induced recruitment of Parkin to mitochondria (detected 2 h after adding CCCP) followed by the removal of depolarized mitochondria in about 50% of Parkin-expressing SH-SY5Y cells (monitored 24 h later) (Fig4G and N). Parkin-induced mitophagy required the presence of PINK1, as described previously (Geisler et al, 2010; Narendra et al, 2010; Vives-Bauza et al, 2010), but was not impaired in cells silenced for Ret expression (Fig4H, I, J, N, O). Moreover, the overexpression of constitutively active RetMEN2A did not induce Parkin translocation or mitophagy under any condition, including PINK1 knock-down with or without Parkin overexpression (Fig4K, L, M, N). Similar results were obtained when GDNF and soluble GFRα-1 was used to activate signaling via endogenous Ret (Fig4N). Furthermore, GDNF/GFRα-1 treatment also rescued mitochondrial fragmentation induced by PINK1 silencing HeLa cells, a cell type which does not express endogenous Parkin (Denison et al, 2003; Pawlyk et al, 2003), further indicating that Ret signaling rescues PINK1 loss-of-function phenotypes independently of Parkin (Supplementary Fig S3).
Ret signaling rescues impaired bioenergetics of Pink1-deficient cells
It has been reported previously that PINK1 deficiency impairs mitochondrial respiration (Gautier et al, 2008, 2012; Gandhi et al, 2009; Lutz et al, 2009; Morais et al, 2009). We therefore investigated whether activation of Ret signaling via GDNF/GFRα-1 treatment could influence this phenotype. We measured mitochondrial function under basal and stress conditions in SH-SY5Y cells silenced for PINK1 expression by using an extracellular oxygen flux analyzer. In comparison to control siRNA-treated cells, PINK1-deficient cells were characterized by a decreased oxygen consumption rate even under basal conditions (Fig5A). Moreover, the spare respiratory capacity (difference between maximal and basal respiration) was markedly reduced, indicating that the ability of PINK1-deficient cells to respond to an increased energy demand under stress conditions is severely impaired. Remarkably, GDNF/GFRα-1 treatment fully rescued basal respiration and increased maximal respiration in PINK1-deficient cells, indicating that the beneficial effect of increased Ret signaling in PINK1-deficient models can be explained by influencing the bioenergetic capacity of mitochondria rather than mitophagy.
Figure 5.
- A Oxygen consumption rate in SH-SY5Y cells determined by an extracellular flux analyzer. 1: Injection of the F1FO–ATPase inhibitor oligomycin; 2: injection of the uncoupler FCCP; 3: injection of the complex I inhibitor rotenone and the and complex III inhibitor antimycin A. Under basal conditions, as well as FCCP-evoked maximum respiration, PINK1 knockdown cells (red squares) displayed markedly reduced oxygen consumption as compared to controls (green circles). Treatment of PINK1 knockdown cells with GDNF/GFRα-1 rescued basal respiration and increased FCCP-evoked respiration (open squares).
- B, C Relative ATP content in the thorax, normalized by total protein, expressed as percentage of w1118 controls. Pink1 and park mutants have reduced ATP amounts. RetMEN2B overexpression partially rescues ATP deficiency in Pink1 (B), but not park mutants (C). Averages of 6–12 animals per genotype.
- D–F Activity of Complex I (rotenone sensitive), normalized to citrate synthase activity, percentage of heterozygous controls. Pink1 mutants have impaired complex I function, which is rescued by RetMEN2B overexpression (D). park mutants have normal complex I activity as compared to controls (E). Inactivation of the complex I subunit CG11455 by RNAi driven by Mef2-GAL4 causes dramatically reduced complex I activity as compared to controls (F) and this was not rescued by RetMEN2B overexpression.
- G Semi quantitative RT-PCR analysis of complex I subunit CG6485 indicates upregulation by RetMEN2B overexpression in Pink1 mutants, GAPDH was used as a loading control.
- H Quantification of CG6485 mRNA normalized to GAPDH, averages of 3 experiments, RNA from 3 thoraces per sample.
Complex I deficiency of Pink1 mutants rescued by Ret signaling
To investigate whether Ret signaling also rescued mitochondrial functionality in Drosophila, we measured ATP content of thoracic homogenates. As previously shown (Clark et al, 2006; Park et al, 2006; Yang et al, 2006; Vos et al, 2012), Pink1 and park mutants showed reduced ATP content in the thorax to approximately 40% of controls, including flies carrying the Mef2-GAL4 driver (Fig5B and C). Mef2 > RetMEN2B overexpression in control flies caused a slight reduction in ATP as compared to controls, possibly as a result of their mild muscle phenotype. In line with the rescue of myofibril and mitochondrial structures, RetMEN2B overexpression largely rescued ATP levels in Pink1 mutants, while ATP levels of park mutants did not significantly improve (Fig5B and C). To unravel the underlying mechanism of the improved mitochondrial respiration, we turned our attention to complex I of the ETC. Recent reports had found that Pink1, in contrast to park mutants had decreased activity of the ETC, and specifically of complex I function (Morais et al, 2009; Vilain et al, 2012). For these reasons, we measured complex I activity in RetMEN2B-overexpressing Pink1 mutants, by monitoring rotenone-sensitive NADH oxidation by spectrophotometry, normalized to the activity of citrate synthase. As previously observed, Pink1 mutants displayed markedly reduced complex I activity (Fig5D). Interestingly, RetMEN2B significantly increased complex I activity to levels similar to controls (Fig5D). In accordance with previously reported data, park mutants showed no decreased complex I activity as compared to controls (Fig5E). Depleting the complex I subunit (CG11455) from muscles by RNAi abrogated most complex I activity (Fig5F), and RetMEN2B overexpression was not able to rescue this defect (Fig5F), suggesting that Ret signaling does not activate alternative means of NADH oxidation as previously shown for the yeast protein Ndi1p (Vilain et al, 2012). The mechanism by which Pink1 controls complex I function is still unknown. Drosophila complex I contains 48 subunits, six of which are mitochondrially encoded, the rest being nuclear. The supply of commercially available antibodies for Drosophila complex I is limited to the subunit NDUFS3, which has recently been shown to be reduced in Pink1 mutants (Liu et al, 2011). By Western blot, we could confirm the reduction of NDUFS3, but did not observe an upregulation by RetMEN2B (Supplementary Fig S4A and B). We performed a semi-quantitative RT-PCR screen of other complex I subunits in Pink1 mutants compared to RetMEN2B-overexpressing Pink1 mutants. Of 45 subunits analyzed, most were unchanged, but the transcript of CG6485, orthologous to human NDUFV2, was moderately elevated in RetMEN2B-overexpressing Pink1 mutants (Supplementary Fig S4C). Interestingly, when compared to controls, CG6485 mRNA was reduced by 46% in Pink1 mutants, and significantly increased to 117% of controls by RetMEN2B overexpression (Fig5G and H). This effect may at least in part be responsible for the Ret-mediated rescue of Pink1 deficiency.
Discussion
The receptor tyrosine kinase Ret is already known to be required for long-term survival of nigral dopamine neurons in mice, and stimulation with its ligand GDNF protects dopamine neurons from cell death in a variety of toxin-based rodent and primate models of PD. In the present work, we found that a signaling-active version of the Drosophila homolog of Ret suppresses degeneration of muscle tissue and mitochondrial abnormalities in Pink1 mutants. Interestingly, park mutants were not rescued. In human SH-SY5Y cells, stimulation of endogenous Ret by GDNF rescued both morphological and bioenergetic defects of mitochondria in PINK1-depleted cells. Pink1 and Parkin were previously shown to interact genetically in Drosophila in what was proposed to be a linear pathway, and a significant body of work has described how Pink1 and Parkin function to initiate mitophagy of impaired mitochondria, and arrest of mitochondrial trafficking. However, in our cell culture model, Ret signaling did not induce mitophagy or Parkin recruitment, arguing that Ret rescues PINK1 deficits independently of Parkin. A recent study demonstrated that Pink1 mutants in contrast to park mutants have decreased function of complex I of the electron transport chain, suggesting that Pink1 is required for maintaining efficient complex I enzymatic activity and that this function is upstream of mitochondrial remodeling. We found that Ret rescued both the impairment of complex I activity, and partially the mitochondrial morphology in Pink1 mutants, suggesting that complex I is a target of Ret signaling. Previous studies of complex I inhibition or genetic depletion have shown mild morphological impairments in Drosophila muscle, contrary to the stronger phenotype of Pink1 mutants. Therefore, it was somewhat unexpected that restoring complex I activity would be sufficient to rescue also morphological defects. One interpretation is that the Pink1 mutant morphological phenotype is more severe due to a synergistic effect of deficits in remodeling/mitophagy and complex I activity, which in this study was partially rescued. Another possibility is that Ret signaling not only targets complex I, but also morphology in a Parkin-independent manner.
Extrapolated to mammalian models, our results suggest a novel mechanism by which the GDNF family of neurotrophic factors may promote survival of dopamine neurons in PD. Several of the mammalian models where the neuroprotective effects of GDNF treatment were initially discovered, were in fact models of mitochondrial dysfunction, either directly via complex I inhibition by MPTP treatment (Tomac et al, 1995; Gash et al, 1996), or the more general ROS toxicity of 6-OHDA (Kearns & Gash, 1995; Sauer et al, 1995), which also includes complex I impairments (Glinka et al, 1997). In light of our findings, it would be interesting to investigate whether or not GDNF improves complex I activity in these model systems. GDNF has been tested in models of alpha-synuclein overexpression, a pathology that is not known to cause complex I deficiency, but did not show any neuroprotective effects, fitting with our hypothesis (Lo Bianco et al, 2004; Decressac et al, 2011).
The current findings support recent evidence showing that Pink1 has an important function related to complex I activity, which is independent of its function in recruiting Parkin to the outer mitochondrial membrane upon loss of membrane potential. This model is consistent with a partial rescue of Pink1 deficiencies, e.g. by either overexpressing Parkin or the yeast complex I equivalent NADH dehydrogenase, or, in the current work, RetMEN2B (Clark et al, 2006; Park et al, 2006; Yang et al, 2006; Vilain et al, 2012). In addition, our findings are consistent with a recent study showing that Pink1-deficient flies but not Parkin-deficient flies can be rescued by TRAP1, which also seems to have beneficial effects on complex I activity (Zhang et al, 2013).
The pathways by which Ret signaling targets complex I and rescues Pink1 mutants requires further investigation. Also, the mechanism by which Pink1 regulates complex I remains elusive, it may regulate for example gene expression, phosphorylation status or assembly (Salvi et al, 2005; Pagliarini & Dixon, 2006) (Fig6). Our gene expression analysis showed that most subunits are unchanged by RetMEN2B, but interestingly one subunit was moderately downregulated in Pink1 mutants and upregulated by RetMEN2B, which may improve function. However, we do not exclude the possibility that Ret signaling targets complex I, and perhaps other metabolic components, by different means.
Figure 6.
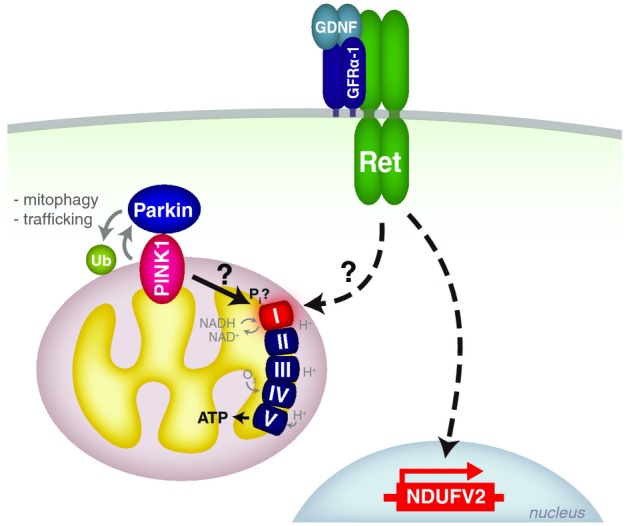
Model of Pink1 and RetMEN2B functions
Our results suggest a dual role for Pink1: One in recruiting Parkin to the mitochondria and initiating mitochondrial clearance or regulating mitochondrial trafficking, a second in regulating the activity of complex I via an as yet unclear pathway. This could be mediated, for example, via phosphorylation of the protein complex or by regulating expression of complex I components. Loss of Pink1 decreases complex I activity and respiratory function. Ret rescues specifically Pink1 mutants, by restoring complex I activity, respiration and ATP production, in part by upregulating the mRNA levels of the complex I subunit NDUFV2 (CG6485).
Brain-derived neurotrophic factor (BDNF) protects mouse cortical neurons against drug-induced excitotoxicity, an effect that was blocked by the complex I inhibitor Rotenone and a MEK1/2 inhibitor, suggesting that BDNF signaling via the Ras/Erk pathway can regulate complex I function (Markham et al, 2012). The signaling properties and functions of Drosophila Ret are not characterized in great detail, but it is structurally homologous to mammalian Ret and can, to some extent, activate the same signaling pathways (Abrescia et al, 2005). Mammalian Ret on the other hand, has been extensively characterized and is known to activate a number of downstream signaling pathways including Ras/ERK, phosphoinositol-3 kinase (PI3K)/Akt, phospholipase C-gamma (PLCγ), Janus kinase (JAK)/STAT, and ERK5, several of which have pro-survival effects, most notably the PI3K/Akt pathway (Sariola & Saarma, 2003; Pascual et al, 2011). Recent studies of Pink1 and park mutant Drosophila have indicated that PI3K/Akt signaling or components downstream of this pathway rather exacerbates Pink1 and park mutant phenotypes (Tain et al, 2009; Liu & Lu, 2010), making it an unlikely candidate for rescue.
Additional studies are required to elucidate the details by which Pink1 and Ret regulate complex I activity, and whether this finding is transferrable to mammalian models. In summary, this work shows that Ret signaling can rescue phenotypes of Pink1 mutants by restoring mitochondrial respiration and specifically complex I function, and thereby suggests a potential novel mechanism underlying GDNF-mediated protection in mammalian PD models. In the future, screening of PD patients for complex I deficiencies and subjecting specifically those individuals to GDNF treatment may provide a new therapeutic strategy.
Materials and Methods
Fly strains and procedures
Mef2-GAL4;UAS-RetMEN2B is lethal at 25°C, therefore all crosses were performed at 18°C. All analyses were performed with 2- to 5-day-old flies. In experiments with Mhc-GAL4;Tub-GAL80ts, pupae were shifted from 18 to 30°C at pharate adult stages P11-P12 (Flybase FBdv:00005349) and analyzed at 3–4 days post eclosion. park25 (Greene et al, 2003) was provided by Leo Pallanck, park1 (Cha et al, 2005) and Pink1B9 (Park et al, 2006) were provided by Jongkyeong Chung, Pink1B9::Mef2-GAL4 (Tain et al, 2009) was provided by Alex Whitworth, UAS-RetMEN2B (Read et al, 2005) was provided by Ross Cagan, TH-GAL4 (Friggi-Grelin et al, 2003), was provided by Hiromu Tanimoto, Mef2-GAL4 (Ranganayakulu et al, 1996), Tub-GAL80ts (McGuire et al, 2003), and UAS-mitoGFP (Pilling et al, 2006) were obtained from the Bloomington stock center, UAS-CG11455RNAi (#12838) was obtained from Vienna Drosophila RNAi Center. “+” controls depict Pink1 and park WT alleles from w1118 (Bloomington stock #5905). In For all histology experiments, flies were genotyped by PCR to assure correct genotypes and control for X-chromosome non-disjunction, for list of primers see Supplementary information.
Myosin heavy chain – GAL4 flies
A 2.5 kb Mhc enhancer was amplified from genomic DNA using primers FS124 (5′-tcaggtaccGGCCGCTCTAGAAATGATATGTG-3′) and FS125 (5′-tcacgcggccgcATTATCCTTGCTTAAATTTCGTTTAG-3′) and cloned with Asp718/NotI into a GAL4-containing Casper-based P-element transformation vector. Transgenic flies were generated using standard procedures. In contrast to the formerly published GAL4 line (Schuster et al, 1996), which shows a rather weak activity in embryos, larvae and adults, this new Mhc-GAL4 line is very strong and very specifically expressed in differentiated muscles from embryonic stages onwards (FS, unpublished).
Histology, transmission electron microscopy and analysis of mitochondrial morphology
Hemi-thoraces were prepared as described previously (Schnorrer et al, 2010), stained with Phalloidin-Alexa Fluor-568 (Molecular Probes), and single plane images were acquired on an Olympus FV1000 confocal scanning microscope. For transmission electron microscopy, hemi-thoraces were fixed in 2.5% Glutaraldehyde, from which semithin sections were prepared and stained with toluidine-blue, subsequently ultrathin serial sections were prepared using a Leica EM UC6 Ultramicrotome. Images at 5,000× magnification were acquired using a JEOL JEM-1230 transmission electron microscope at 80 kV, equipped with a Gatan Orius SC1000 digital Camera. Six TEM Images per animal were acquired from randomly selected regions of the indirect flight muscles. All mitochondria in these images (500–800 per animal) were grouped into three categories, based on the integrity of the cristae structure, with genotypes blinded to the experimenter, using the ImageJ software (NIH). Whole mount immunostaining of fly brains was performed according to standard procedures. The following antibodies were used: rabbit anti-tyrosine hydroxylase (ab152, Millipore; 1:200) and chick anti-GFP (Abcam ab13970; 1:500). The PPL1 cluster was imaged using an Olympus FV1000 confocal microscope with a 60× NA 1.3 objective with 4× zoom. 52 z-sections of 0.3 μm spacing were acquired and deconvolved by the nearest neighbor algorithm using Metamorph 7.5 (Molecular Devices). A volume corresponding to 26 × 26 × 15 μm was cropped, subjected to linear rescaling and analyzed in Imaris x64 6.4.2 (Bitplane Scientific Software). Mitochondrial volume was measured by 3D isosurface rendering using a fixed threshold.
Immunoblot analysis
Thoraces from three animals per sample were homogenized in Triton-lysis buffer, protein concentration was determined using the BCA method (BioRad), equal amounts of protein were separated using SDS-PAGE and blotted according to standard procedures. Antibodies used were: panRet (provided by C. Ibanez) and alpha-Tubulin (clone DM1A, Sigma).
Cell culture, treatments and RNA Interference
SH-SY5Y (DSMZ number ACC 209) cells were cultivated as described previously (Henn et al, 2005; Schlehe et al, 2008). For acute stimulation of Ret, cells were incubated for 3–4 h with recombinant hGDNF (Shenandoah Biotechnology Inc.) and hGFRα-1 (R&D Systems) at a final concentration of 100 ng/mL. PINK1 and Ret gene silencing was performed with the following stealth siRNA oligos (Invitrogen) using Lipofectamine RNAiMAX (Invitrogen): PINK1 human HSS127945 (SH-SY5Y), Ret human HSS109181.
Assessment of mitochondrial morphology
SH-SY5Y: Cells grown on 15-mm glass coverslips were fixed with 3.7% PFA in PBS for 10 min. Cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and blocked with 5% BSA in PBS at room temperature. Fixed cells were sequentially incubated with primary antibody diluted in blocking solution (TOM20 pAb, overnight at 4°C) and secondary antibody diluted in blocking buffer (goat anti rabbit Alexa555- coniugated, 2 h at room temperature). Nuclei were counterstained with DAPI. Coverslips were mounted on glass slides and images were acquired with a Zeiss LSM710 confocal microscope equipped with a 63× oil objective (NA 1.4). Cells displaying an intact network of tubular mitochondria were classified as tubular. When this network was disrupted and mitochondria appeared either globular or rod-like they were classified as fragmented. The mitochondrial morphology of the cells was determined in a blinded manner. Quantifications were based on 150 cells from at least 3 independent experiments.
Assessment of mitophagy
SH-SY5Y cells were plated on glass coverslips and reversely transfected with siRNA and 24 h later with the indicated DNA plasmid. Human GDNF and GFRalpha were added to the cells 24 h after siRNA transfection and 3 h before CCCP treatment. The next day, cells were treated with 10 μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP, Sigma) for 2 or 24 h. Recruitment of parkin to mitochondria (after 2 h CCCP) and removal of mitochondria (after 24 h CCCP) was detected by indirect immunofluorescence using a monoclonal anti-Parkin antibody (PRK8, Santa Cruz Biotechnology) and a polyclonal antibody against HSP60 (Santa Cruz Biotechnology). Nuclei were stained by DAPI. Cells were analysed by fluorescence microscopy using a Leica DMRB microscope and confocal images were taken using a Zeiss LSM710 confocal microscope equipped with a 63× oil objective (NA 1.4). Quantifications are based on three independent experiments. At least 1,500 cells were analysed for each condition.
Real-time RT-PCR, cultured cells
Knock-down efficiency of PINK1 and Ret was evaluated by real-time RT-PCR with the 7500 Fast Real Time System (Applied Biosystems) as previosly described (Bouman et al, 2011). Statistical analysis of RT-PCR data is based on at least four independent experiments with triplicate samples. For list of primers, see Supplementary information.
Measurement of mitochondrial oxygen consumption
The oxygen consumption rate was determined using a Seahorse XF 96 analyzer (Seahorse Biosciences). SH-SY5Y cells were reversely transfected and plated in a XF 96 cell culture microplate. The next day, fresh medium containing human GDNF/GFRα-1 was added to the cells where indicated. The cells were incubated with low-glucose (1 mM) medium overnight and the sensor cartridge was hydrated overnight according to the manufacturers' instructions. Measurements were performed 48 h after transfection. The measured values were normalized to protein levels. PINK1 knockdown did not induce apoptosis under these conditions. The cells were washed using the XF Prep Station three times with Seahorse Medium containing 10 mM galactose and 1 mM pyruvate. Mitochondrial function was analyzed using the XF Cell Mito Stress Test Kit (Seahorse Biosciences) and all measurements were carried out at 37°C. The following drugs were diluted in Seahorse Medium and loaded on the sensor cartridge: oligomycin (injection port A), carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone (FCCP; injection port B), rotenone and antimycin A (both injection port C). The drugs were diluted in Seahorse Medium and loaded on the sensor cartridge. Measured values were normalized to protein levels.
ATP measurement
Measurements of thoracic ATP were performed using a luciferase assay as described previously (Park et al, 2006) with some modifications: Briefly, single thoraces from 3-day-old flies with heads and wings removed were homogenized in 50 μl of extraction buffer (100 mM Tris-HCl, 4 mM EDTA pH 7.8) with 6 M Guanidine-HCl using a teflon-on-glass dounce homogenizer. The lysate was boiled for 3 min and cleared by centrifugation at 20,000 g for 1 min. The samples were diluted 1:100 in extraction buffer before analyzing using the ATP determination kit (Invitrogen), according to the manufacturer's instruction. Values were normalized to total protein content, measured by absorbance at 280 nm using a NanoDrop spectrophotometer. All measurements were performed in triplicate.
Enzymatic measurements
Activity of complex I (NADH: ubiquinone oxidoreductase) was assessed by monitoring the oxidation of NADH as previously described (Fischer et al, 1986). Briefly, thoraces from 20 animals were homogenized in 250 mM sucrose, 10 mM Tris pH 7.4, 0.15 mM MgCl2, after which mitochondria were isolated as described previously (Walker et al, 2006). Enzymatic activity of complex I was assessed by NADH oxidation, monitored at A340 nm as described (Bugiani et al, 2004), and rotenone insensitive activity was subtracted. The activity of complex I was normalized to Citrate Synthase activity, which was measured indirectly by AcCoA-SH formation, as described (Ferguson & Williams, 1966).
RT-PCR, Drosophila complex I subunits
Thoraces were dissected and snap-frozen, homogenized in RLT buffer (Qiagen) using a rotor-stator homogenizer. Total RNA was prepared using the RNeasy mini kit according to instructions. Samples were treated with DNase1 on-column for 15 min (RNase-free DNase set, Qiagen). RT-PCR analysis was performed using the OneStep RT-PCR kit (Qiagen) using 20 ng of template RNA and 35–40 cycles of PCR amplification depending on signal strength of the primer pair. Primers were designed using the primerBLAST tool (NCBI), and when possible exon-junction spanning primers were used, for list of primers, see Supplementary information. As some of the analyzed transcripts are single-exon, control reactions omitting the reverse transcriptase amplification step were performed to assure that samples were free of contaminating genomic DNA, despite DNase1 treatment.
Statistical analysis
Data represent mean ± SEM. Statistical analysis was carried out using analysis of variance (ANOVA) or Student's t-test; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Acknowledgments
We would like to thank Marianne Braun and Ursula Weber for transmission electron microcopy and Pilar Alcalá for molecular biology assistance, the Bloomington stock center for fly strains, Liviu Aron for discussions at early stages of the project and Louise Gaitanos for critically reading the manuscript. This work was in part supported by the Max-Planck Society, and grants from the German Research Foundation (DFG), the European Union (MOLPARK) and the European Research Council (TOPAG).
Author contributions
PK designed, performed and analyzed the majority of the experiments. CS and FS contributed to the design of the fly genetics and analysis of muscle morphology, and FS generated the Mhc-GAL4 line. EM and AKM-R designed, performed and analyzed the SH-SY5Y experiments. KFW supervised the cell culture work and contributed to the analysis of the fly data. RK supervised the project, designed experiments and co-wrote the manuscript with PK.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Methods
Review Process File
Source Data for Figure 1M
References
- Abrescia C, Sjostrand D, Kjaer S, Ibanez CF. Drosophila RET contains an active tyrosine kinase and elicits neurotrophic activities in mammalian cells. FEBS Lett. 2005;579:3789–3796. doi: 10.1016/j.febslet.2005.05.075. [DOI] [PubMed] [Google Scholar]
- Airaksinen MS, Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–394. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- Aron L, Klein P, Pham TT, Kramer ER, Wurst W, Klein R. Pro-survival role for Parkinson's associated gene DJ-1 revealed in trophically impaired dopaminergic neurons. PLoS Biol. 2010;8:e1000349. doi: 10.1371/journal.pbio.1000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AK, Cortese GP, Amodeo KD, Weihofen A, Letai A, LaVoie MJ. Parkin selectively alters the intrinsic threshold for mitochondrial cytochrome c release. Hum Mol Genet. 2009;18:4317–4328. doi: 10.1093/hmg/ddp384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouman L, Schlierf A, Lutz AK, Shan J, Deinlein A, Kast J, Galehdar Z, Palmisano V, Patenge N, Berg D, Gasser T, Augustin R, Trumbach D, Irrcher I, Park DS, Wurst W, Kilberg MS, Tatzelt J, Winklhofer KF. Parkin is transcriptionally regulated by ATF4: evidence for an interconnection between mitochondrial stress and ER stress. Cell Death Differ. 2011;18:769–782. doi: 10.1038/cdd.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugiani M, Invernizzi F, Alberio S, Briem E, Lamantea E, Carrara F, Moroni I, Farina L, Spada M, Donati MA, Uziel G, Zeviani M. Clinical and molecular findings in children with complex I deficiency. Biochim Biophys Acta. 2004;1659:136–147. doi: 10.1016/j.bbabio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Burns RS, Chiueh CC, Markey SP, Ebert MH, Jacobowitz DM, Kopin IJ. A primate model of parkinsonism: selective destruction of dopaminergic neurons in the pars compacta of the substantia nigra by N-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Proc Natl Acad Sci USA. 1983;80:4546–4550. doi: 10.1073/pnas.80.14.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena A, Gisselbrecht S, Harrison J, Jimenez F, Michelson AM. Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes Dev. 1998;12:3910–3922. doi: 10.1101/gad.12.24.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha GH, Kim S, Park J, Lee E, Kim M, Lee SB, Kim JM, Chung J, Cho KS. Parkin negatively regulates JNK pathway in the dopaminergic neurons of Drosophila. Proc Natl Acad Sci USA. 2005;102:10345–10350. doi: 10.1073/pnas.0500346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Dagda RK, Gusdon AM, Pien I, Strack S, Green S, Li C, Van Houten B, Cherra SJ, 3rd, Chu CT. Mitochondrially localized PKA reverses mitochondrial pathology and dysfunction in a cellular model of Parkinson's disease. Cell Death Differ. 2011;18:1914–1923. doi: 10.1038/cdd.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decressac M, Ulusoy A, Mattsson B, Georgievska B, Romero-Ramos M, Kirik D, Bjorklund A. GDNF fails to exert neuroprotection in a rat alpha-synuclein model of Parkinson's disease. Brain. 2011;134:2302–2311. doi: 10.1093/brain/awr149. [DOI] [PubMed] [Google Scholar]
- Deierborg T, Soulet D, Roybon L, Hall V, Brundin P. Emerging restorative treatments for Parkinson's disease. Prog Neurobiol. 2008;85:407–432. doi: 10.1016/j.pneurobio.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Demontis F, Perrimon N. Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development. 2009;136:983–993. doi: 10.1242/dev.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison SR, Wang F, Becker NA, Schule B, Kock N, Phillips LA, Klein C, Smith DI. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene. 2003;22:8370–8378. doi: 10.1038/sj.onc.1207072. [DOI] [PubMed] [Google Scholar]
- Exner N, Lutz AK, Haass C, Winklhofer KF. Mitochondrial dysfunction in Parkinson's disease: molecular mechanisms and pathophysiological consequences. EMBO J. 2012;31:3038–3062. doi: 10.1038/emboj.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, Williams GR. The effect of malate and other dicarboxylic acids on mitochondrial isocitrate metabolism. J Biol Chem. 1966;241:3696–3700. [PubMed] [Google Scholar]
- Fischer JC, Ruitenbeek W, Trijbels JM, Veerkamp JH, Stadhouders AM, Sengers RC, Janssen AJ. Estimation of NADH oxidation in human skeletal muscle mitochondria. Clin Chim Acta. 1986;155:263–273. doi: 10.1016/0009-8981(86)90246-9. [DOI] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- Gandhi S, Wood-Kaczmar A, Yao Z, Plun-Favreau H, Deas E, Klupsch K, Downward J, Latchman DS, Tabrizi SJ, Wood NW, Duchen MR, Abramov AY. PINK1-associated Parkinson's disease is caused by neuronal vulnerability to calcium-induced cell death. Mol Cell. 2009;33:627–638. doi: 10.1016/j.molcel.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gash DM, Zhang Z, Ovadia A, Cass WA, Yi A, Simmerman L, Russell D, Martin D, Lapchak PA, Collins F, Hoffer BJ, Gerhardt GA. Functional recovery in parkinsonian monkeys treated with GDNF. Nature. 1996;380:252–255. doi: 10.1038/380252a0. [DOI] [PubMed] [Google Scholar]
- Gautier CA, Giaime E, Caballero E, Nunez L, Song Z, Chan D, Villalobos C, Shen J. Regulation of mitochondrial permeability transition pore by PINK1. Mol Neurodegener. 2012;7:22. doi: 10.1186/1750-1326-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci USA. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Glinka Y, Gassen M, Youdim MB. Mechanism of 6-hydroxydopamine neurotoxicity. J Neural Transm Suppl. 1997;50:55–66. doi: 10.1007/978-3-7091-6842-4_7. [DOI] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jimenez F, Baylies MK, Michelson AM. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- Heeman B, Van den Haute C, Aelvoet SA, Valsecchi F, Rodenburg RJ, Reumers V, Debyser Z, Callewaert G, Koopman WJ, Willems PH, Baekelandt V. Depletion of PINK1 affects mitochondrial metabolism, calcium homeostasis and energy maintenance. J Cell Sci. 2011;124:1115–1125. doi: 10.1242/jcs.078303. [DOI] [PubMed] [Google Scholar]
- Henn IH, Bouman L, Schlehe JS, Schlierf A, Schramm JE, Wegener E, Nakaso K, Culmsee C, Berninger B, Krappmann D, Tatzelt J, Winklhofer KF. Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling. J Neurosci. 2007;27:1868–1878. doi: 10.1523/JNEUROSCI.5537-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn IH, Gostner JM, Lackner P, Tatzelt J, Winklhofer KF. Pathogenic mutations inactivate parkin by distinct mechanisms. J Neurochem. 2005;92:114–122. doi: 10.1111/j.1471-4159.2004.02854.x. [DOI] [PubMed] [Google Scholar]
- Kearns CM, Gash DM. GDNF protects nigral dopamine neurons against 6-hydroxydopamine in vivo. Brain Res. 1995;672:104–111. doi: 10.1016/0006-8993(94)01366-p. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Aron L, Ramakers GM, Seitz S, Zhuang X, Beyer K, Smidt MP, Klein R. Absence of Ret signaling in mice causes progressive and late degeneration of the nigrostriatal system. PLoS Biol. 2007;5:e39. doi: 10.1371/journal.pbio.0050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–980. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- de Lau LM, Breteler MM. Epidemiology of Parkinson's disease. Lancet Neurol. 2006;5:525–535. doi: 10.1016/S1474-4422(06)70471-9. [DOI] [PubMed] [Google Scholar]
- Lin LF, Doherty DH, Lile JD, Bektesh S, Collins F. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- Liu S, Lu B. Reduction of protein translation and activation of autophagy protect against PINK1 pathogenesis in Drosophila melanogaster. PLoS Genet. 2010;6:e1001237. doi: 10.1371/journal.pgen.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Acin-Perez R, Geghman KD, Manfredi G, Lu B, Li C. Pink1 regulates the oxidative phosphorylation machinery via mitochondrial fission. Proc Natl Acad Sci USA. 2011;108:12920–12924. doi: 10.1073/pnas.1107332108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Bianco C, Deglon N, Pralong W, Aebischer P. Lentiviral nigral delivery of GDNF does not prevent neurodegeneration in a genetic rat model of Parkinson's disease. Neurobiol Dis. 2004;17:283–289. doi: 10.1016/j.nbd.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Lutz AK, Exner N, Fett ME, Schlehe JS, Kloos K, Lammermann K, Brunner B, Kurz-Drexler A, Vogel F, Reichert AS, Bouman L, Vogt-Weisenhorn D, Wurst W, Tatzelt J, Haass C, Winklhofer KF. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J Biol Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham A, Cameron I, Bains R, Franklin P, Kiss JP, Schwendimann L, Gressens P, Spedding M. Brain-derived neurotrophic factor-mediated effects on mitochondrial respiratory coupling and neuroprotection share the same molecular signalling pathways. Eur J Neurosci. 2012;35:366–374. doi: 10.1111/j.1460-9568.2011.07965.x. [DOI] [PubMed] [Google Scholar]
- Martin I, Dawson VL, Dawson TM. Recent advances in the genetics of Parkinson's disease. Annu Rev Genomics Hum Genet. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MK, Cookson MR. Mitochondrial quality control and dynamics in Parkinson's disease. Antioxid Redox Signal. 2012;16:869–882. doi: 10.1089/ars.2011.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Mijatovic J, Airavaara M, Planken A, Auvinen P, Raasmaja A, Piepponen TP, Costantini F, Ahtee L, Saarma M. Constitutive Ret activity in knock-in multiple endocrine neoplasia type B mice induces profound elevation of brain dopamine concentration via enhanced synthesis and increases the number of TH-positive cells in the substantia nigra. J Neurosci. 2007;27:4799–4809. doi: 10.1523/JNEUROSCI.5647-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno Y, Ohta S, Tanaka M, Takamiya S, Suzuki K, Sato T, Oya H, Ozawa T, Kagawa Y. Deficiencies in complex I subunits of the respiratory chain in Parkinson's disease. Biochem Biophys Res Commun. 1989;163:1450–1455. doi: 10.1016/0006-291x(89)91141-8. [DOI] [PubMed] [Google Scholar]
- Morais VA, Verstreken P, Roethig A, Smet J, Snellinx A, Vanbrabant M, Haddad D, Frezza C, Mandemakers W, Vogt-Weisenhorn D, Van Coster R, Wurst W, Scorrano L, De Strooper B. Parkinson's disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol Med. 2009;1:99–111. doi: 10.1002/emmm.200900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Rischart AK, Pilsl A, Beaudette P, Patra M, Hadian K, Funke M, Peis R, Deinlein A, Schweimer C, Kuhn PH, Lichtenthaler SF, Motori E, Hrelia S, Wurst W, Trumbach D, Langer T, Krappmann D, Dittmar G, Tatzelt J, Winklhofer KF. The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol Cell. 2013;49:908–921. doi: 10.1016/j.molcel.2013.01.036. [DOI] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Dixon JE. Mitochondrial modulation: reversible phosphorylation takes center stage? Trends Biochem Sci. 2006;31:26–34. doi: 10.1016/j.tibs.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Parker WD, Jr, Swerdlow RH. Mitochondrial dysfunction in idiopathic Parkinson disease. Am J Hum Genet. 1998;62:758–762. doi: 10.1086/301812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual A, Hidalgo-Figueroa M, Gomez-Diaz R, Lopez-Barneo J. GDNF and protection of adult central catecholaminergic neurons. J Mol Endocrinol. 2011;46:R83–R92. doi: 10.1530/JME-10-0125. [DOI] [PubMed] [Google Scholar]
- Pawlyk AC, Giasson BI, Sampathu DM, Perez FA, Lim KL, Dawson VL, Dawson TM, Palmiter RD, Trojanowski JQ, Lee VM. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J Biol Chem. 2003;278:48120–48128. doi: 10.1074/jbc.M306889200. [DOI] [PubMed] [Google Scholar]
- Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell. 2006;17:2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganayakulu G, Schulz RA, Olson EN. Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev Biol. 1996;176:143–148. doi: 10.1006/dbio.1996.9987. [DOI] [PubMed] [Google Scholar]
- Read RD, Goodfellow PJ, Mardis ER, Novak N, Armstrong JR, Cagan RL. A Drosophila model of multiple endocrine neoplasia type 2. Genetics. 2005;171:1057–1081. doi: 10.1534/genetics.104.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi M, Brunati AM, Toninello A. Tyrosine phosphorylation in mitochondria: a new frontier in mitochondrial signaling. Free Radical Biol Med. 2005;38:1267–1277. doi: 10.1016/j.freeradbiomed.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Sandebring A, Thomas KJ, Beilina A, van der Brug M, Cleland MM, Ahmad R, Miller DW, Zambrano I, Cowburn RF, Behbahani H, Cedazo-Minguez A, Cookson MR. Mitochondrial alterations in PINK1 deficient cells are influenced by calcineurin-dependent dephosphorylation of dynamin-related protein 1. PLoS ONE. 2009;4:e5701. doi: 10.1371/journal.pone.0005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariola H, Saarma M. Novel functions and signalling pathways for GDNF. J Cell Sci. 2003;116:3855–3862. doi: 10.1242/jcs.00786. [DOI] [PubMed] [Google Scholar]
- Sauer H, Rosenblad C, Bjorklund A. Glial cell line-derived neurotrophic factor but not transforming growth factor beta 3 prevents delayed degeneration of nigral dopaminergic neurons following striatal 6-hydroxydopamine lesion. Proc Natl Acad Sci USA. 1995;92:8935–8939. doi: 10.1073/pnas.92.19.8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD. Mitochondrial complex I deficiency in Parkinson's disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- Schapira AH, Jenner P. Etiology and pathogenesis of Parkinson's disease. Mov Disord. 2011;26:1049–1055. doi: 10.1002/mds.23732. [DOI] [PubMed] [Google Scholar]
- Schlehe JS, Lutz AK, Pilsl A, Lämmermann K, Grgur K, Henn IH, Tatzelt J, Winklhofer KF. Aberrant folding of pathogenic parkin mutants: aggregation versus degradation. J Biol Chem. 2008;283:13771–13779. doi: 10.1074/jbc.M707494200. [DOI] [PubMed] [Google Scholar]
- Schnorrer F, Schonbauer C, Langer CC, Dietzl G, Novatchkova M, Schernhuber K, Fellner M, Azaryan A, Radolf M, Stark A, Keleman K, Dickson BJ. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 2010;464:287–291. doi: 10.1038/nature08799. [DOI] [PubMed] [Google Scholar]
- Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron. 1996;17:641–654. doi: 10.1016/s0896-6273(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci. 2009;12:1129–1135. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomac A, Lindqvist E, Lin LF, Ogren SO, Young D, Hoffer BJ, Olson L. Protection and repair of the nigrostriatal dopaminergic system by GDNF in vivo. Nature. 1995;373:335–339. doi: 10.1038/373335a0. [DOI] [PubMed] [Google Scholar]
- Vilain S, Esposito G, Haddad D, Schaap O, Dobreva MP, Vos M, Van Meensel S, Morais VA, De Strooper B, Verstreken P. The yeast complex I equivalent NADH dehydrogenase rescues pink1 mutants. PLoS Genet. 2012;8:e1002456. doi: 10.1371/journal.pgen.1002456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrane J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos M, Esposito G, Edirisinghe JN, Vilain S, Haddad DM, Slabbaert JR, Van Meensel S, Schaap O, De Strooper B, Meganathan R, Morais VA, Verstreken P. Vitamin K2 is a mitochondrial electron carrier that rescues pink1 deficiency. Science. 2012;336:1306–1310. doi: 10.1126/science.1218632. [DOI] [PubMed] [Google Scholar]
- Walker DW, Hajek P, Muffat J, Knoepfle D, Cornelison S, Attardi G, Benzer S. Hypersensitivity to oxygen and shortened lifespan in a Drosophila mitochondrial complex II mutant. Proc Natl Acad Sci USA. 2006;103:16382–16387. doi: 10.1073/pnas.0607918103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen A, Thomas KJ, Ostaszewski BL, Cookson MR, Selkoe DJ. Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry. 2009;48:2045–2052. doi: 10.1021/bi8019178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci USA. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Karsten P, Hamm S, Pogson JH, Muller-Rischart AK, Exner N, Haass C, Whitworth AJ, Winklhofer KF, Schulz JB, Voigt A. TRAP1 rescues PINK1 loss-of-function phenotypes. Hum Mol Genet. 2013;22:2829–2841. doi: 10.1093/hmg/ddt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Methods
Review Process File
Source Data for Figure 1M