Abstract
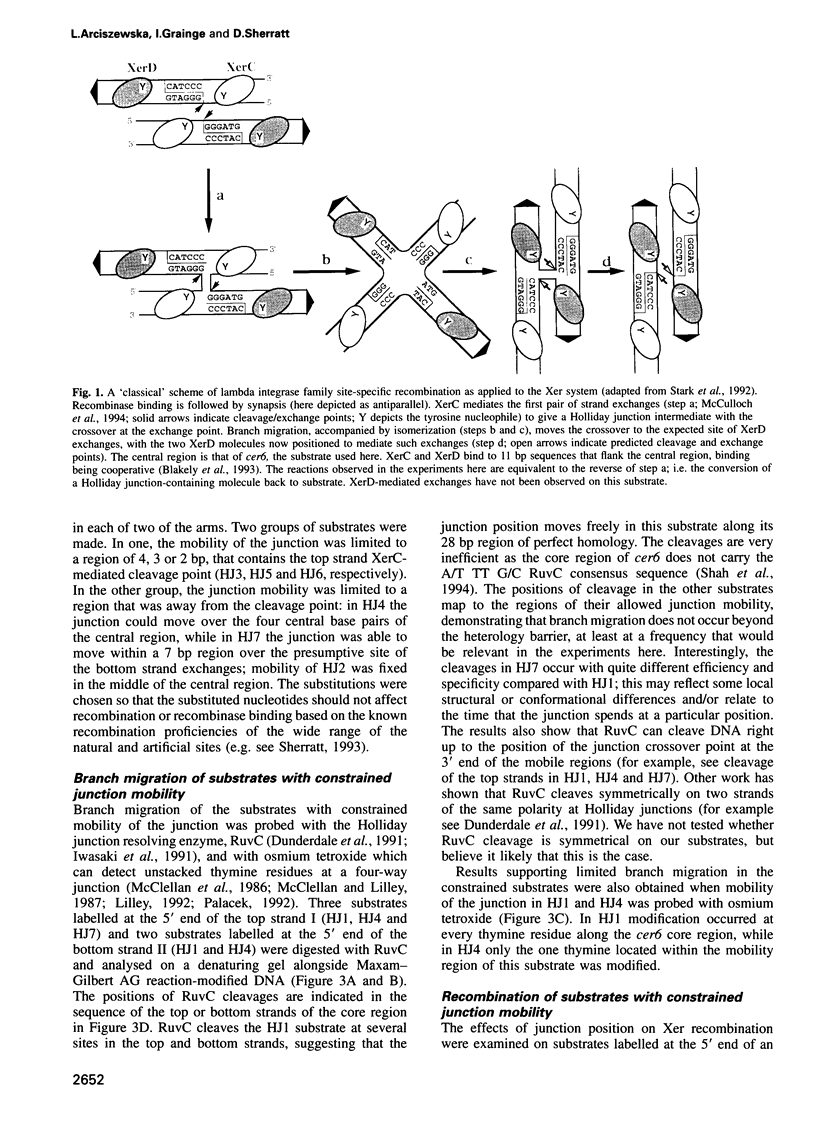
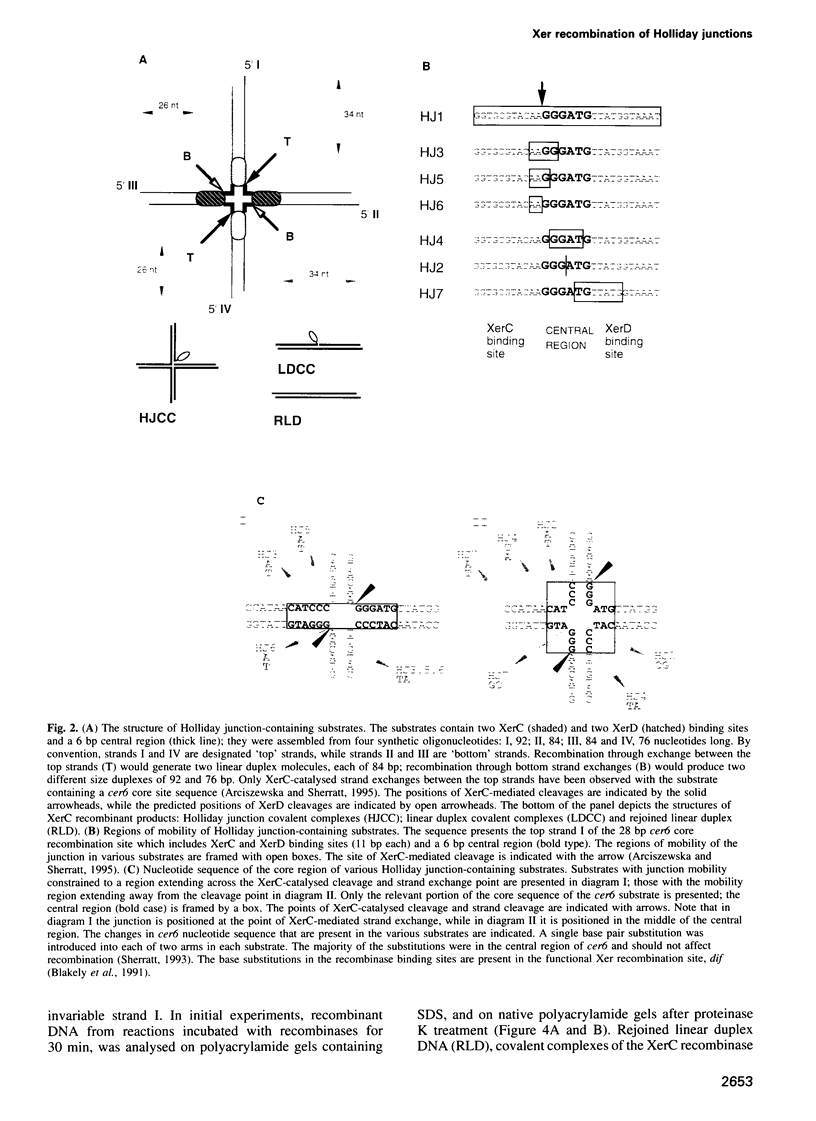
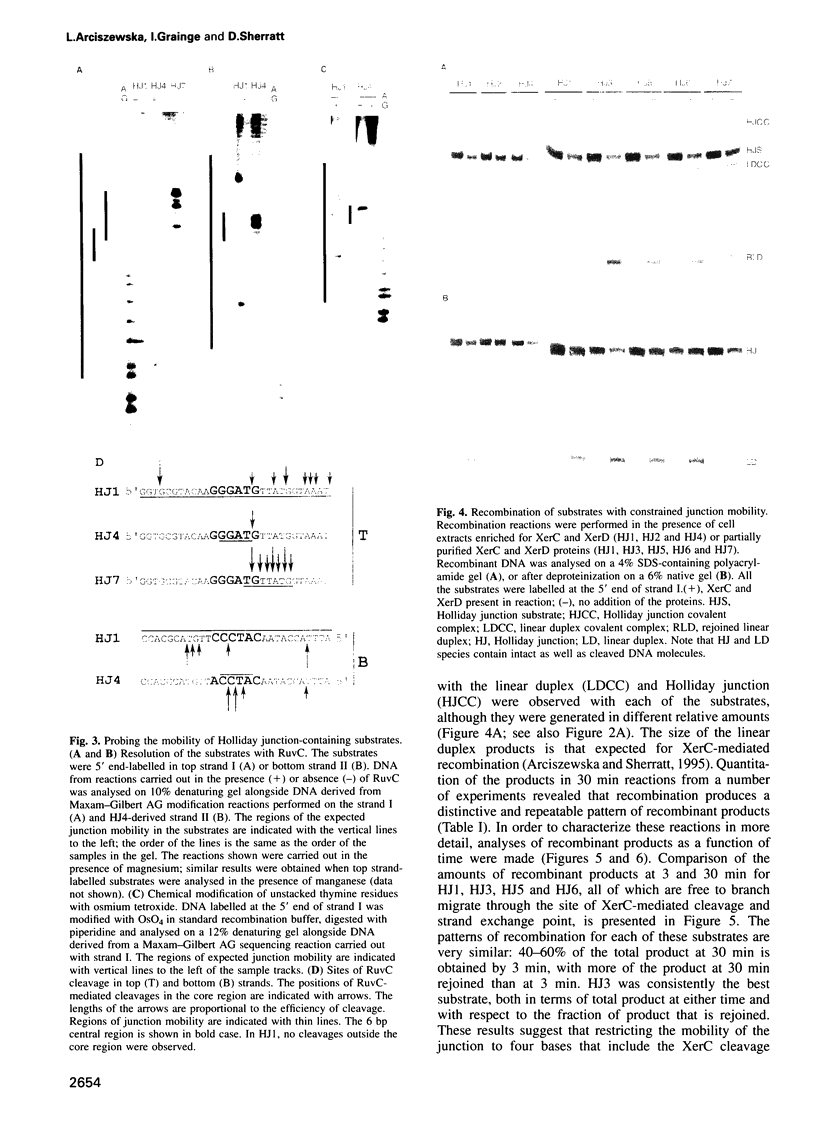
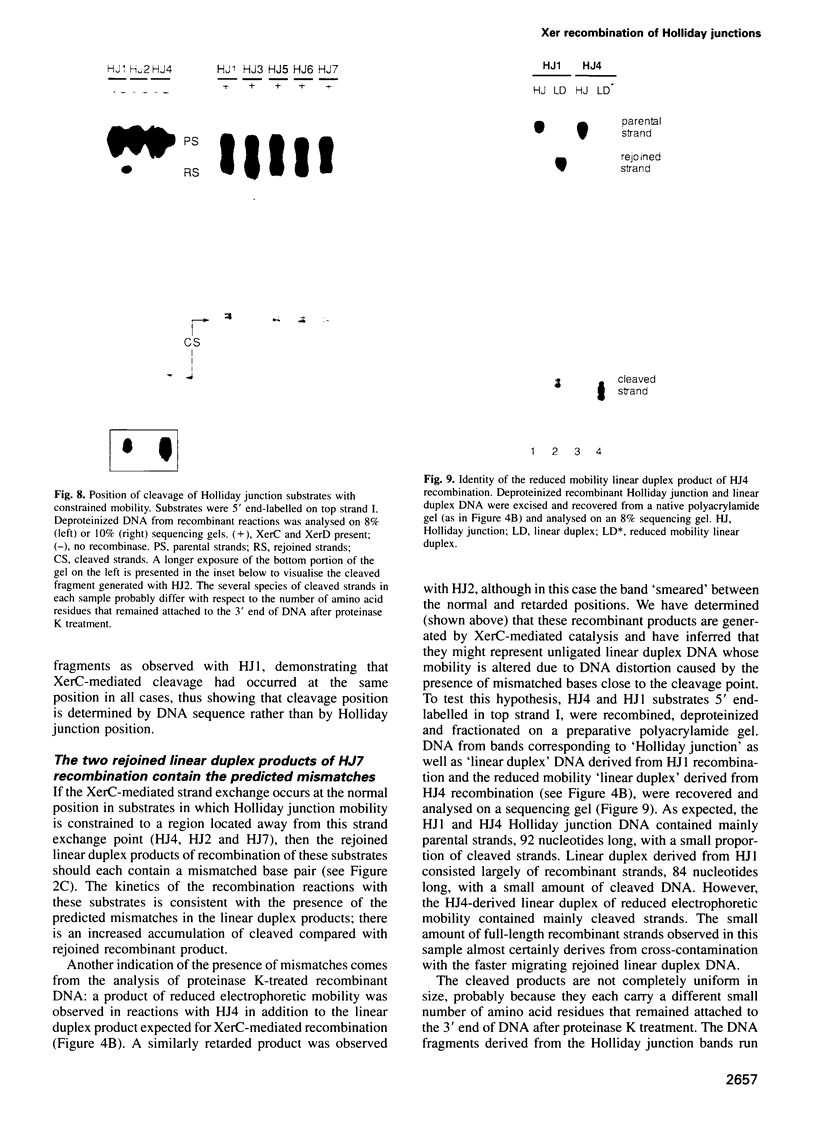
Site-specific recombination mediated by XerC and XerD functions in the segregation of circular replicons in Escherichia coli. A key feature of most models of recombination for the family of recombinases to which XerC and XerD belong is that a Holliday junction forms at the position of the first pair of recombinase-mediated strand exchanges and then branch migrates 6-8 bp to the position of the second pair of strand exchanges. We have tested this hypothesis for Xer recombination by studying the effects of junction position on XerC-mediated strand exchange in vitro. Recombination of synthetic Holliday junction substrates in which junction mobility was constrained to a region extending over or removed away from the normal cleavage and exchange point was analysed. All substrates undergo strand cleavage at the normal position. We infer that the Holliday junction need not be at this position during strand cleavage and exchange. With substrates in which the Holliday junction is constrained to a region away from the XerC-mediated cleavage point, strand exchange generates products with the predicted mispaired bases.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arciszewska L. K., Sherratt D. J. Xer site-specific recombination in vitro. EMBO J. 1995 May 1;14(9):2112–2120. doi: 10.1002/j.1460-2075.1995.tb07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C. E., Gardner J. F., Gumport R. I., Weisberg R. A. The effect of attachment site mutations on strand exchange in bacteriophage lambda site-specific recombination. Genetics. 1989 Aug;122(4):727–736. doi: 10.1093/genetics/122.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely G., Colloms S., May G., Burke M., Sherratt D. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991 Aug;3(8):789–798. [PubMed] [Google Scholar]
- Blakely G., May G., McCulloch R., Arciszewska L. K., Burke M., Lovett S. T., Sherratt D. J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993 Oct 22;75(2):351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- Colloms S. D., Sykora P., Szatmari G., Sherratt D. J. Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J Bacteriol. 1990 Dec;172(12):6973–6980. doi: 10.1128/jb.172.12.6973-6980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart M., Benkovic S. J., Nash H. A. Behavior of a cross-linked attachment site: testing the role of branch migration in site-specific recombination. J Mol Biol. 1991 Aug 5;220(3):621–629. doi: 10.1016/0022-2836(91)90105-f. [DOI] [PubMed] [Google Scholar]
- Craig N. L. The mechanism of conservative site-specific recombination. Annu Rev Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- Dixon J. E., Sadowski P. D. Resolution of immobile chi structures by the FLP recombinase of 2 microns plasmid. J Mol Biol. 1994 Oct 21;243(2):199–207. doi: 10.1006/jmbi.1994.1647. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Dunderdale H. J., Benson F. E., Parsons C. A., Sharples G. J., Lloyd R. G., West S. C. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991 Dec 19;354(6354):506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Shiba T., Nakata A., Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991 Dec;10(13):4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston B. H. Hydroxylamine and methoxylamine as probes of DNA structure. Methods Enzymol. 1992;212:180–194. doi: 10.1016/0076-6879(92)12012-f. [DOI] [PubMed] [Google Scholar]
- Johnston B. H., Rich A. Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell. 1985 Oct;42(3):713–724. doi: 10.1016/0092-8674(85)90268-5. [DOI] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Homology-dependent interactions in phage lambda site-specific recombination. Nature. 1987 Sep 24;329(6137):346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994 Sep;58(3):401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Landy A. Mechanistic and structural complexity in the site-specific recombination pathways of Int and FLP. Curr Opin Genet Dev. 1993 Oct;3(5):699–707. doi: 10.1016/s0959-437x(05)80086-3. [DOI] [PubMed] [Google Scholar]
- Leslie N. R., Sherratt D. J. Site-specific recombination in the replication terminus region of Escherichia coli: functional replacement of dif. EMBO J. 1995 Apr 3;14(7):1561–1570. doi: 10.1002/j.1460-2075.1995.tb07142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of branched DNA species. Q Rev Biophys. 1993 May;26(2):131–175. doi: 10.1017/s0033583500004054. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Palecek E. The supercoil-stabilised cruciform of ColE1 is hyper-reactive to osmium tetroxide. EMBO J. 1984 May;3(5):1187–1192. doi: 10.1002/j.1460-2075.1984.tb01949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. Probes of DNA structure. Methods Enzymol. 1992;212:133–139. doi: 10.1016/0076-6879(92)12009-f. [DOI] [PubMed] [Google Scholar]
- McClellan J. A., Lilley D. M. A two-state conformational equilibrium for alternating (A-T)n sequences in negatively supercoiled DNA. J Mol Biol. 1987 Oct 20;197(4):707–721. doi: 10.1016/0022-2836(87)90477-3. [DOI] [PubMed] [Google Scholar]
- McClellan J. A., Palecek E., Lilley D. M. (A-T)n tracts embedded in random sequence DNA--formation of a structure which is chemically reactive and torsionally deformable. Nucleic Acids Res. 1986 Dec 9;14(23):9291–9309. doi: 10.1093/nar/14.23.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch R., Coggins L. W., Colloms S. D., Sherratt D. J. Xer-mediated site-specific recombination at cer generates Holliday junctions in vivo. EMBO J. 1994 Apr 15;13(8):1844–1855. doi: 10.1002/j.1460-2075.1994.tb06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Leon L., Inman R. B., Cox M. M. Characterization of Holliday structures in FLP protein-promoted site-specific recombination. Mol Cell Biol. 1990 Jan;10(1):235–242. doi: 10.1128/mcb.10.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J. E., Kemper B., Cunningham R. P., Kallenbach N. R., Seeman N. C. T4 endonuclease VII cleaves the crossover strands of Holliday junction analogs. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9441–9445. doi: 10.1073/pnas.85.24.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Azaro M. A., Landy A. Swapping DNA strands and sensing homology without branch migration in lambda site-specific recombination. Curr Biol. 1995 Feb 1;5(2):139–148. doi: 10.1016/s0960-9822(95)00035-2. [DOI] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Matsumoto L., Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987 Aug 28;50(5):779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- Palecek E. Probing DNA structure with osmium tetroxide complexes in vitro. Methods Enzymol. 1992;212:139–155. doi: 10.1016/0076-6879(92)12010-n. [DOI] [PubMed] [Google Scholar]
- Panyutin I. G., Hsieh P. Formation of a single base mismatch impedes spontaneous DNA branch migration. J Mol Biol. 1993 Mar 20;230(2):413–424. doi: 10.1006/jmbi.1993.1159. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980 Oct 24;8(20):4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R., Bennett R. J., West S. C. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell. 1994 Dec 2;79(5):853–864. doi: 10.1016/0092-8674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Sherratt D. J., Arciszewska L. K., Blakely G., Colloms S., Grant K., Leslie N., McCulloch R. Site-specific recombination and circular chromosome segregation. Philos Trans R Soc Lond B Biol Sci. 1995 Jan 30;347(1319):37–42. doi: 10.1098/rstb.1995.0006. [DOI] [PubMed] [Google Scholar]
- Stark W. M., Boocock M. R., Sherratt D. J. Catalysis by site-specific recombinases. Trends Genet. 1992 Dec;8(12):432–439. [PubMed] [Google Scholar]
- Stark W. M., Sherratt D. J., Boocock M. R. Site-specific recombination by Tn3 resolvase: topological changes in the forward and reverse reactions. Cell. 1989 Aug 25;58(4):779–790. doi: 10.1016/0092-8674(89)90111-6. [DOI] [PubMed] [Google Scholar]
- Summers D. K., Sherratt D. J. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984 Apr;36(4):1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- West S. C. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- de Massy B., Dorgai L., Weisberg R. A. Mutations of the phage lambda attachment site alter the directionality of resolution of Holliday structures. EMBO J. 1989 May;8(5):1591–1599. doi: 10.1002/j.1460-2075.1989.tb03543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]