Functional proteomics is a rapidly evolving field that will have vast clinical implications in the near future. Therefore, it is important to understand its concept and role in the diagnosis, prognostication, and treatment in breast cancer. In this review, we present functional proteomic approaches mainly focusing on the recent clinical implications of utilizing the reverse-phase protein array platform in breast cancer research.
Keywords: Functional proteomics, Breast cancer, Prognostic marker, Predictive marker
Abstract
Breast cancer is one of the major public health problems of the Western world. Recent advances in genomics and gene expression-profiling approaches have enriched our understanding of this heterogeneous disease. However, progress in functional proteomics in breast cancer research has been relatively slow. Allied with genomics, the functional proteomics approach will be important in improving diagnosis through better classification of breast cancer and in predicting prognosis and response to different therapies, including chemotherapy, hormonal therapy, and targeted therapy. In this review, we will present functional proteomic approaches with a focus on the recent clinical implications of utilizing the reverse-phase protein array platform in breast cancer research.
Implications for Practice:
Functional proteomics is a rapidly evolving field that will have vast clinical implications in the near future. Therefore, it is important to understand its concept and role in the diagnosis, prognostication, and treatment in breast cancer. Mass spectrometry serves as a discovery platform, whereas reverse-phase protein array (RPPA) serves as a validation platform in breast cancer research. RPPA-based classification will complement gene expression-based classification. RPPA-based breast cancer subgroups are shown to be both prognostic of survival and predictive of response to chemotherapy. Prognostic or predictive protein panels in breast cancer will lead to discovery of novel therapeutic targets in breast cancer treatment.
Introduction
Breast cancer is one of the major health problems in the U.S. with 230,000 cases expected to occur in 2013 [1]. Recent advances in breast cancer classification, including human epidermal growth factor receptor 2 (HER2)-positive breast cancer, and its matched targeted therapy, including trastuzumab, have led to better outcomes of breast cancer [2]. In addition, the gene expression-profiling approach has identified at least six molecular subtypes that relate to different clinical presentations and outcome [3, 4]. For hormone-positive, node-negative breast cancer patients, the 21-gene profiling assay, Oncotype DX (Genomic Health, Redwood City, CA, http://www.genomichealth.com/), is currently being used to predict benefit from chemotherapy [5]. Despite the improvement in classification and recent development of targeted therapies in breast cancer that resulted in improved outcome, the survival gain has only been modest as a result of frequent relapses and failures to treatments.
The genomics approach has its limitations. It does not capture post-translational modification that affects protein function and stability [6, 7]. Proteins are the ultimate effector molecule of cellular functions, not genes or messenger RNAs. In addition, limited power of single gene or protein biomarkers for predicting clinical outcome has raised the need for integrated large-scale data analyses. Recent advances in functional proteomics have allowed high-throughput analysis of both basal and phosphorylated proteins active in carcinogenesis and tumor progression. Functional proteomic profiling has now enabled discovery of candidate proteins related to different pathophysiology or outcome in breast cancer [8]. In this review, we will examine the current technologies of functional proteomics and its clinical implications in breast cancer focusing on the reverse-phase protein array (RPPA) analysis.
Materials and Methods
The literature search was performed via online search engines including PubMed using “functional proteomics” and “breast cancer” as key words. Articles describing research using the “RPPA” platform were selected to be further reviewed. Articles without human breast cancer research were excluded. The level of evidence was not used in terms of evaluating the articles because none of the studies was performed in a randomized fashion. Unpublished data were not utilized in this review.
Protein Analysis Platforms
Proteomic platforms can be divided into antibody-based and nonantibody-based. The former includes Western blotting, enzyme-linked immunosorbent assay (ELISA), immunohistochemistry (IHC), and protein microarray. The latter includes protein mass spectrometry-based technology. Whereas the former requires prior knowledge of proteins to be tested and validated antibodies, the latter does not. Table 1 provides the overview and comparison between the aforementioned protein analysis platforms.
Table 1.
Overview and comparison of commonly used protein analysis platforms
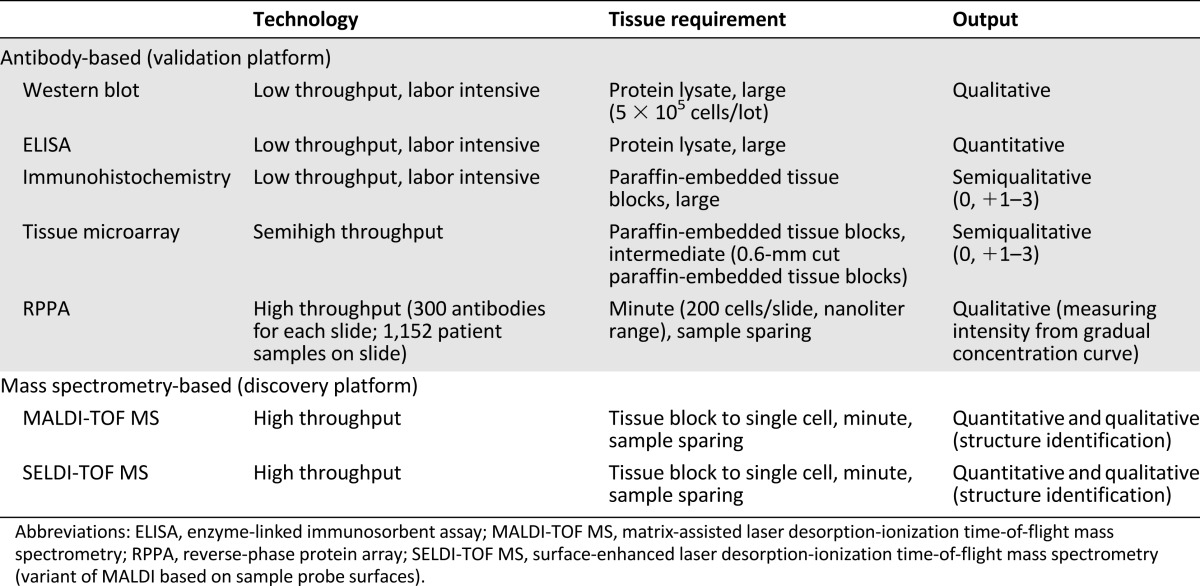
Western blotting is a widely used technique used to detect specific proteins in the given sample of tissue extract. It utilizes gel electrophoresis to separate proteins by the three-dimensional structure or the length of the polypeptide. The proteins are then transferred to a membrane, later stained with antibodies specific to the protein [9]. This is a low-throughput technology designed to survey one or a limited number of proteins at a time. However, the same concept of protein printing on the membrane is used in the protein microarray platform.
ELISA is a highly specific assay mainly used for serum antigen or antibody. Although it is suitable for validating specific protein biomarkers, it is expensive and low throughput. It requires a large amount of protein lysates to have reasonable sensitivity. Western blotting and ELISA are currently not used in treatment guidelines [10]. ELISA technology is currently actively being utilized in the epidemiologic correlative studies assessing blood proteome-based biomarkers such as circulating tumor antigen [11].
Immunohistochemistry (IHC) is currently used to classify breast cancer into hormone receptor-positive (HER2-negative), HER2-positive, or triple-negative breast cancer. It is a conventional highly specific assay to assess protein expressions. It has the advantage of providing information of cellular and spatial localization of the protein. However, it is low-throughput and labor-intensive. When IHC is coupled with tissue microarray (TMA), it becomes possible to analyze a group of protein markers in multiple fragments of tumor tissues with a reasonable cost. The important limitation of IHC is that it is only semiquantitative. Thus, IHC can be observer-dependent. In breast cancer, estrogen receptor (ER), progesterone receptor (PR), and HER2 protein expression level by IHC are used for treatment algorithms [12, 13]. For example, tamoxifen or aromatase inhibitors can be used if ER or PR is measured equal to or greater than 1% by IHC depending on their menopausal status [13].
Definition of Functional Proteomics
Proteomics can be defined as a large-scale high-throughput study of proteins from a variety of different biological samples investigating their ontology, classification, expression levels, and properties. Functional proteomics, in contrast to conventional proteomics, indicates proteomics study that incorporates the examination of protein activation, protein-protein interactions, and activated pathway analyses. Serum, plasma, tissue, urine, cerebrospinal fluid, saliva, nipple fluid, ascites, pleural fluid, or any body fluid can be used by functional proteomics to classify breast cancer and predict survival and response to therapy in breast cancer by discovering proteomic biomarkers. Functional proteomics can also encompass classification of proteins into distinct groups such as exosomal proteins (exosome), secreted proteins (secretome), proteases (proteasome), kinases (kinome), and phosphorylated proteins (phosphoproteomics) and further characterization of them. Protein mass spectrometry and protein microarray are the commonly used high-throughput technologies that can utilize functional proteomics to its fullest potential in breast cancer research.
Protein Mass Spectrometry
Protein mass spectrometry is an analytical tool that generates spectra of the masses of the proteins consisting of a sample of material. It first ionizes compounds to generate charged molecules and then measures their mass-to-charge ratios. The spectra are examined to determine the elemental composition of a sample and the masses of proteins and to depict the chemical structures of proteins [14]. It has now become a major platform for discovery of unknown proteins associated with breast cancer pathogenesis. Recent advances in protein mass spectrometry have allowed high-sample throughput and near-complete coverage of whole-protein samples. The commonly used methodologies for whole-protein ionization are electrospray ionization, surface-enhanced laser desorption ionization, and matrix-assisted laser desorption ionization (MALDI). With the advent of MALDI-based imaging mass spectrometry method, it is now possible to simultaneously measure multiple analytes directly from intact tissue sections [15]. This is a major breakthrough because it does not require tissue consumption, in contrast to the IHC, which requires multiple fragments of tissue samples from TMA [16].
With the advent of MALDI-based imaging mass spectrometry method, it is now possible to simultaneously measure multiple analytes directly from intact tissue sections. This is a major breakthrough because it does not require tissue consumption, in contrast to the IHC, which requires requiring multiple fragments of tissue samples from TMA.
Mass spectrometry (MS)-based label-free proteomics provides an unbiased approach to screen biomarkers related to disease progression and therapy resistance of breast cancer on the global scale. It has shown promise in discovering new biomarkers related to early detection, prognostication, and prediction of response to different therapies in breast cancer [17–21]. However, this technology is only available in specialized centers with a large-scale mass spectrometer machine and specially trained staff with advanced bioinformatics skills. Therefore, the reproducibility and reliability of the findings from this platform have often been questioned, with some studies showing contradictory results [22]. Although the future of the protein mass spectrometry-based technologies is promising, it may not be cost-effective or clinically robust to be implemented into clinical practice. In addition, all reported biomarkers discovered are derived from retrospective patient samples. None of the markers is validated in the prospective clinical studies. This suggests both the limited validity of current mass spectrometry-based biomarkers and the importance of prospective validation to be applied in clinics in the near future.
Protein Microarray
Protein microarray is a high-throughput method used to assess the expression, interactions, and activities of proteins on a large scale. Its major advantage is that large numbers of proteins can be tracked in parallel. It is often compared with gene microarray, also known as gene chip.
Protein microarrays can be divided into two distinct formats: forward-phase protein array (FFPA) and reverse-phase protein array (RPPA) [23, 24]. The former is also termed analytic capture array, whereas the latter is termed lysate array. In FFPA, antibodies are immobilized in each spot and each array is queried with a tumor sample that contains multiple protein lysates. Therefore, multiple protein expression and phosphorylation levels will be measured at the same time in a single array with a set of antibodies. In contrast, in RPPA, a set of tumor sample lysates is immobilized in each array spot and each array is queried with one antibody and later with affinity reagent. More than one thousand patient samples with multiple protein lysates can be investigated in parallel in a single array for each protein of interest using a validated antibody per each array.
Protein microarray is an automated, rapid, cost-effective, and highly sensitive technology consuming only small quantities of samples and reagents. Because RPPA technology can be used to compare multiple protein samples in parallel, enabling high-throughput analyses in a large number of patient cohorts, it is more commonly used and a feasible platform for biomarker discovery in breast cancer [25].
Use of Reverse-Phase Protein Array (RPPA) in Breast Cancer
The feasibility and validity of the RPPA platform to map functional proteomics of breast cancer have been repeatedly demonstrated with fine-needle aspiration (FNA) samples, core biopsies, resected tissue blocks, and in laser-capture microdissected tissue samples of both primary and metastatic breast cancer lesions [26–28]. Of note, one pilot study suggested the feasibility of using the RPPA with limited random periareolar FNA samples obtained from high-risk women in a prospective setting [27].
Laser-capture microdissection (LCM) is now applied to accurately capture tumor from stroma or the surrounding normal tissue [29]. In other words, it is used to procure specific cell subpopulations under direct microscopic visualization of a standard-stained frozen or formalin-fixed tissue section on a glass slide. Its feasibility and the proof of concept have been shown with the mass spectrometry-based technology as well as the RPPA in breast cancer [29–31]. The combination of the LCM and the RPPA technologies enables the analysis of various key cellular signaling proteins from pure tumor cell populations. Currently, different protein pathways are being monitored with the LCM and the RPPAs in clinical trials [32].
For example, a phase II neoadjuvant randomized clinical trial of trastuzumab and/or lapatinib with chemotherapy in HER2-positive breast cancer is still ongoing (NCT15585078). It was designed to monitor changes in the HER2 and other related protein pathways between the pre- and post-treatment tumor samples using the LCM and the RPPA. One study found a high concordance rate between the HER2 amplification by fluorescent in-situ hybridization (FISH) and HER2 level measured by the RPPA [33]. Intriguingly, a group of patients with low HER2 expression (by IHC) and amplification (by FISH) and high HER2 phosphorylation was identified [33, 34]. This group had HER2 pathway and its downstream pathway activation independent of total HER2 levels and functional signaling through HER3 and epidermal growth factor receptor (EGFR). These patients, currently excluded from HER2-directed therapy, may potentially benefit from it, leading to improved survival outcome [34]. Validation of this hypothesis in a prospective clinical trial setting will likely provide a proof of concept supporting the clinical utility of functional proteomics.
Nonmicrodissected tumor samples are also commonly used for the RPPA analyses. Variability in multistep tissue-handling process and apparent intratumoral heterogeneity is thought to potentially hamper the clinical utility of the nonmicrodissected tumor samples in proteomics. However, the RPPA technology demonstrated intraslide and interslide coefficients of variability of less than 15% [25]. In one study, although modest time-dependent instability of phosphoproteins at room temperature was evident, protein functional proteomic signature (finger print) was robust in most tumors even when it was maintained at room temperature for 24 hours before freezing. Intratumoral protein levels were remarkably less variable than intertumoral protein levels. Even between different tumor samples, functional proteomic signatures of prognostic implications were robust and reproducible. Moreover, significant correlation between IHC and RPPA was found among breast cancer samples [25].
Functional Proteomics in Classification
Breast cancer being a clinically heterogeneous disease, an effort to accurately classify breast cancer at a molecular level is ongoing. Gene expression-profiling approaches have initially separated breast cancer into distinct subgroups, such as luminal, HER2-enriched, basal-like, and normal breast-like [35]. With further studies, the normal breast-like subtype was overlooked as a possible artifact because of low tumor cellularity, and the luminal subtype was subdivided into luminal A, luminal B/HER2-negative, and luminal B/HER2-positive. These heterogeneous molecular subtypes were distinct in biological and clinical characteristics [4, 36]. They demonstrated correlation with progression-free survival, overall survival, and chemotherapy response [37–39].
Because gene expression microarrays are relatively expensive and not readily available, in practice, IHC is widely used to molecularly classify breast cancer [40]. It has been demonstrated that molecular classification by gene expression microarrays has a high concordance rate with the IHC classification used in the clinic [41]. In fact, PAM50, the 50-gene reverse transcription-polymerase chain reaction test, is not currently used in the clinic to make any treatment decisions.
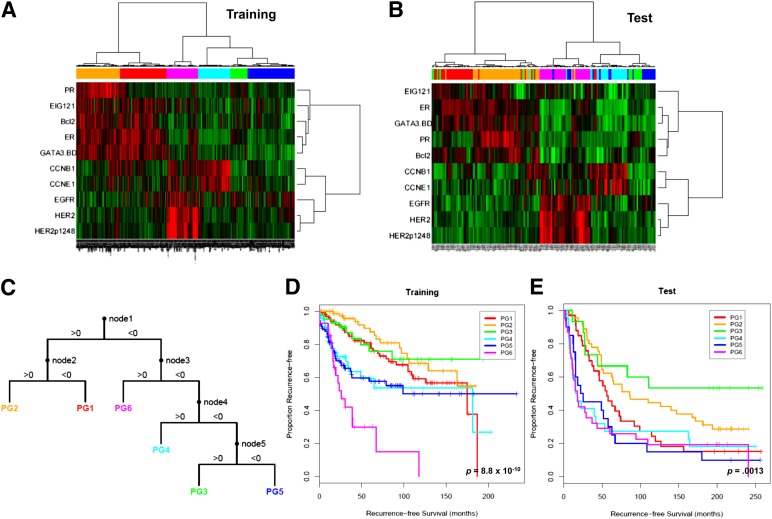
We have previously demonstrated for the first time that functional proteomic classification of breast cancer is feasible and is of clinical relevance [42]. Using the RPPA platform with 146 validated antibodies relevant to breast cancer in three independent tumor sets, including surgical resection samples, we were able to identify six breast cancer subgroups from 10 protein panels (Fig. 1). Ten proteins were selected based on their contribution to the differences among the new subgroups and relapse-free survival (RFS) in patients who received neoadjuvant systemic therapy (NST). They were ER, PR, Bcl2, GATA3, CCNB1, CCNE1, EGFR, HER2, HER2p1248, and EIG121. In addition to validating the importance of ER, PR, HER2, and proliferation-related markers emphasized in previous gene expression-profiling approaches, including the Oncotype DX, these findings support the vital role of other protein markers that define the 10 protein panels and the novel concept of using protein panels to define subgroups of prognostic implications.
Figure 1.
Supervised clustering of breast cancers with quantification data for 10 proteins derived using reverse-phase protein arrays. The 712 breast tumor samples (training set) (A) were clustered with the 10 markers using an “uncentered correlation” distance metric along with the Ward linkage rule. This analysis yielded six subgroups (BG1–6). The 168 breast tumor samples (test set) (B) were subgrouped into one of six groups (PG1–6) using the decision tree (C) that was derived from the training set. Patients in the six subgroups differed significantly in their recurrence-free survival in both training (D) and test (E) sets. Reprinted from Gonzalez-Angulo AM, Hennessy BT, Meric-Bernstam F et al. Functional proteomics can define prognosis and predict pathologic complete response in patients with breast cancer. Clinical Proteomics, 2011;8:11.
Recently, a pivotal multiplatform study of breast cancer by The Cancer Genome Atlas (TCGA) confirmed the four heterogenous subgroups: luminal A, luminal B, HER2-enriched, and basal-like [43]. This study also used the RPPA platform in addition to genomic DNA copy number arrays, DNA methylation, exome sequencing, messenger RNA arrays, and microRNA sequencing. Each group exhibited significant molecular heterogeneity. The RPPA analysis identified two novel protein expression-defined subgroups within the luminal group, possibly produced by stromal/microenvironmental elements, defined as reactive I and reactive II. In fact, seven RPPA clusters were identified in total. They were termed HER2, luminal A, luminal A/B, X, reactive I, and reactive II.
Furthermore, the integrated analyses identified specific signaling pathways dominant in each molecular subtype, including a HER2/phosphorylated HER2/EGFR/phosphorylated EGFR signature within the HER2-enriched expression subtype. RPPA and mRNA microarray demonstrated a high correlation between HER2 clinical status, HER2 protein by RPPA, pHER2, EGFR, and pEGFR. These multiple signatures were able to identify two subgroups within clinically HER2-positive tumors. These signatures may represent predictive biomarkers for response to anti-HER2-targeted therapies.
Gujral et al. [44] reported another RPPA-based breast cancer classification using 56 breast cancers and matched normal tissues. They classified breast cancers into 12 distinct clusters with an average cluster size of 6 samples: HER2 coclustered in group II with prosurvival signaling molecules such as pAkt1, pPI3K, pRSK3, pGSK3β, PDGFRβ, and insulin receptor; progesterone receptor coclustered in group X with Vav1, peIF4G, and pPLA2; and ER clustered by itself (group XI). The groups in general reflected the known biology of receptor tyrosine kinase cascade. For instance, phosphorylated forms of extracellular signal-regulated kinase (pT202/pY204), mitogen-activated protein kinase kinase (pS217/pS221), and S6K (pS240/pS244) kinases made up group VIII, consistent with the known topology of the mitogen-activated protein kinase signaling and its significance in breast cancer.
Functional Proteomics in Prognostication
Six breast cancer subgroups identified by the RPPA platform developed by our group were associated with different RFS (log rank p = 8.8 × 10−10) [42]. This was first found in a tumor training set (n = 712) and then validated on a test set (n = 168) in two cohorts of patients with primary breast cancer. A score was created via ordinal logistic regression to quantify the probability of recurrence. With the strong bioinformatics support, this was the first study to validate the utility of the RPPA platform in capturing prognostic differences in different breast cancer subgroups. Six proteomic signatures obtained from resected breast cancer tissues in the nonmetastatic setting accurately and reproducibly classified breast cancer patients into six groups of patients with distinctly different 5-year RFS.
NST provides an excellent opportunity to assess in vivo chemosensitivity by evaluating the resected surgical breast cancer tissue. Attaining a pathological complete response (pCR) following NST is a validated surrogate marker for improved survival outcome [45–47]. Conversely, patients with residual viable tumors after NST portend poor prognosis, suggesting possible chemoresistance with the future therapy. These patients have heterogeneous tumors with different biology. Therefore, the use of the residual tumor tissues to identify subgroups of patients with higher risk of recurrence presents an exciting opportunity for functional proteomics.
Using the RPPA platform, we have recently found models that independently predicted RFS in residual breast cancer after NST [48]. This was the first study to analyze the residual breast tumor after NST using functional proteomics. RPPA of 76 proteins in test (n = 99) and validation (n = 79) sets with the CoxBoost method identified a three-protein panel that was asso-ciated with RFS in all residual breast cancers. Three proteins were CHK1pS345, Caveolin1, and RAB25. There was unsupervised clustering of separated patients into two groups with different 3-year RFS (p < .001) representing high and low relapse-risk groups.
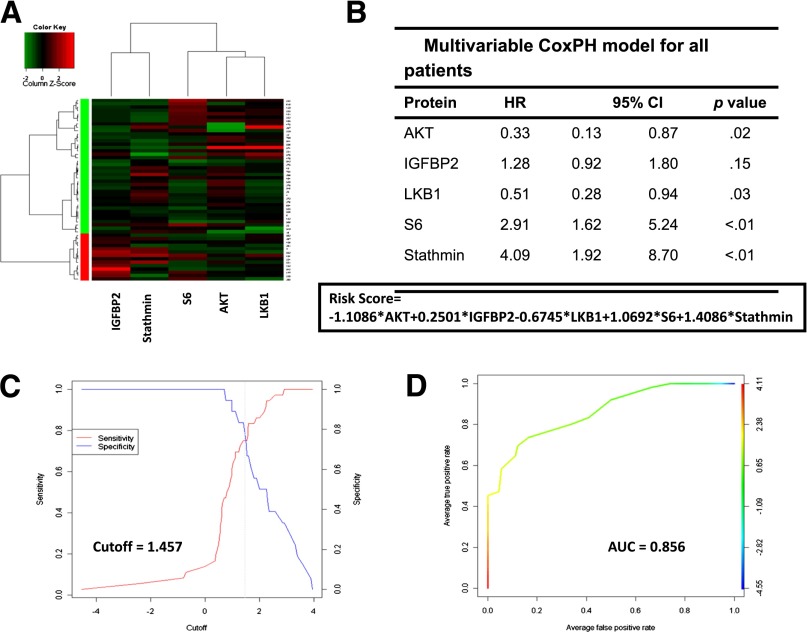
Because breast cancer is a heterogeneous disease, the same analysis was applied to ER-positive disease and triple-negative breast cancer (TNBC) [48, 49]. A two-protein panel model consisting of CD31 and cyclin E1 demonstrated correlation with RFS in HR-positive tumors [48]. In a study analyzing 54 residual TNBCs, multivariate analysis using the top 25 proteins from univariable analysis at the false discovery rate (FDR) of 0.3 revealed a five-protein panel model consisting of AKT, IGFBP2, LKB1, S6, and Stathmin accurately predicted RFS [49] (Fig. 2). A risk score (RS) was calculated with the sum of the coefficients from the final Cox proportional hazard model for all tumors, HR-positive tumors, and TNBCs. RS remained as a robust independent predictor of RFS after adjustment for other clinic-pathological characteristic such as tumor grade or stage (all p < .001).
Figure 2.
Clustering into green and red groups depending on the expression levels of AKT, IGFBP2, LKB1, S6, and stathmin 54 residual triple-negative breast cancers (A). Multivariable Cox proportional hazard model and calculated risk score (RS) (B). Optimal cutoff point at 1.457 (sensitivity versus 1; specificity for the RS in all 54 cases) (C). Receiving operating curve of the RS model (area under the curve = 0.856) (D). Reprinted from Sohn J, Do KA, Liu S et al. Functional proteomics characterization of residual triple-negative breast cancer after standard neoadjuvant chemotherapy. Annals of Oncology 2013;24:2522–2526.
Abbreviations: AUC, area under the curve; CI, confidence interval; HR, hazard ratio.
Functional Proteomics in Prediction of Response to Therapy
Biomarkers of clinical implications can be divided into ones with prognostic or predictive relevance. Whereas most gene expression profiling and proteomic studies describe subgroups of different prognosis, few studies identified predictive biomarkers for different cancer therapies. For example, clinically HER2-positive breast cancers defined by IHC and/or FISH is a compelling predictive biomarker for HER2-directed therapy such as trastuzumab or lapatinb [50, 51]. However, there is still a group of HER2-positive tumors that do not respond to HER2-directed therapies. Also, some groups of HER2-negative tumors with HER2 pathway activation depicted from the TCGA data [43] may show benefit from HER2-directed therapies. With the use of RPPA that can actually capture the HER2 and its downstream pathway activation, we may be able to better identify functional proteomic biomarkers that accurately predict response for HER2-directed therapies. In fact, HER2 phosphorylation was reported to be associated with HER2 therapy resistance [52] and may be used for preselection of HER2-positive breast cancer patients displaying resistance to the first-line trastuzumab treatment. The same approach can be applied to different novel targeted therapies. For example, phosphatidylinositol 3-kinase (PI3K) pathway activation defined from the RPPA could be a candidate biomarker to predict response to PI3K pathway inhibitors in addition to the use of genomic information such as PIK3CA mutation or PTEN loss.
With the use of RPPA that can actually capture the HER2 and its downstream pathway activation, we may be able to better identify functional proteomic biomarkers that accurately predict response for HER2-directed therapies. In fact, HER2 phosphorylation was reported to be associated with HER2 therapy resistance and may be used for preselection of HER2-positive breast cancer patients displaying resistance to the first-line trastuzumab treatment.
Reliably predicting response to standard chemotherapy is also an important unmet need. If we know in advance that there is low likelihood of downsizing tumor with NST using the conventional anthracycline and/or taxane regimen, NST approach will be discouraged. Rather, upfront surgery followed by investigational therapy approach may be considered. Our group used the FNA samples from 132 patients who later received NST to investigate the predictive value of the six subgroups derived from the RPPA analyses [42]. Because pCR after NST is a surrogate marker for survival outcome in breast cancer [45–47], biomarkers predictive of pCR are of clinical significance. The prognostic score constructed using the 10-protein panel that defined six subgroups demonstrated a significant association with likelihood of pCR to NST in the FNA set (p = .002). This study provided the first proof of concept that functional proteomics can be used to generate robust predictive biomarkers in breast cancer.
Functional Proteomics as a Discovery Tool for Targeted Therapy
One of the important roles of functional proteomics in clinical breast cancer research is to discover potential targets for therapy. The fact that RPPA enables more than 1,000 patient samples (protein lysates in nanoliters) to be printed on one slide makes it an ideal high-throughput discovery platform. Our group has used RPPA to determine the molecular characteristics of residual breast cancer after NST, which can provide significant insight into the biology behind chemoresistance and lead to identification of novel targets for future therapy [48, 49].
RPPA analysis of residual TNBC after NST found that a protein panel model including Stathmin, AKT, and ribosomal protein S6 was associated with RFS. These proteins are known to play a role in PTEN/PI3K/AKT/mTOR pathway [53, 54], suggesting that activation of this PI3K pathway characterizes residual TNBC and that suppression of this pathway may represent a novel strategy to overcome resistance to the current standard NST.
Likewise, RPPA analysis of residual HR-positive breast cancer after NST identified cyclin E to be part of the final two-protein panel model that correlated with RFS. Cyclin E is a cyclin family protein that forms a complex with cyclin-dependent kinase (CDK2) to facilitate the G1 to S phase transition. Cyclin E/CDK2 phosphorylates retinoblastoma protein to promote G1 progression [55]. Of note, cyclin E is a known prognostic marker in breast cancer, its expression being increased with the increasing stage and grade of the tumor [56]. Our group has previously reported that breast cancer patients with overexpression of low molecular weight cyclin E failed to respond to aromatase inhibitors [57]. Recently, the use of CDK inhibitors in combination with aromatase inhibitors has shown promising results in HR-positive breast cancers. In the randomized phase II interim analysis, addition of palbociclib (PD-0332991; Pfizer, New York, NY, http://www.pfizer.com), a CDK inhibitor, to letrozole compared with letrozole alone prolonged median progression-free survival from 7.5 months to 26.1 months (hazard ratio = 0.37, p < .001) [58]. In fact, there is an ongoing multicenter randomized phase III trial with palbociclib with Food and Drug Administration breakthrough therapy designation (NCT01740427). All of the above findings corroborate the utility of functional proteomics as a major discovery tool for identifying novel targets for therapy.
Recently, interaction between cMET and Axl was suggested based on the RPPA breast cancer clustering and network analysis [44]. Both cMET and Axl have been implicated in the pathogenesis of breast cancer and in resistance to anticancer therapies. However, the two receptor tyrosine kinases are not known to be functionally associated. Interestingly, the cross-talk between the two was further verified with in vitro experiments suggesting that dual tyrosine kinases targeting cMET and Axl may play a role in breast cancer therapy [44]. It can be speculated that, among cMET pathway inhibitors, bispecific receptor inhibitors such as small kinase inhibitors including GSK1363089, BMS-777607, or MP470 may be more efficacious than the monospecific therapeutic antibodies currently in development, such as MetMAb.
Limitations of the Functional Proteomics Approach Using RPPA
One of the major limitations of RPPA is that it requires a set of validated antibodies. The performance of RPPA is dependent on the quality of the antibody. Therefore, rigorous antibody validation for RPPA is critical to ensure that the detected signals are representative of the proteins of interest. The other limitation of RPPA is that it requires prior knowledge of proteins being tested. Validated antibodies are selected for a specific RPPA analysis usually based on their known relevance to breast pathogenesis, possibly introducing a bias to the study. Therefore, mass spectrometry is generally recommended when the aim of a study is to discover proteins previously not known to be associated with the disease process. Also, developing a model that correctly predicts clinical outcome requires meticulous application of relevant bioinformatics algorithms. Therefore, developing reliable biomarkers from the RPPA platform will require strong skill sets in bioinformatics.
Conclusion
We have shown that functional proteomics research in breast cancer is feasible and is of utmost clinical relevance. Allied with genomics, the functional proteomics approach will be essential to improving diagnosis through better classification of breast cancer and in predicting prognosis and response to different therapies, including chemotherapy, hormonal therapy, and targeted therapy. Furthermore, it will identify hub proteins or protein-protein interactions and driver pathways, leading to the implementation of rational biomarker-based and individualized clinical trials that increase the success rate, bringing the latest bench discoveries to bedside.
There is no doubt that the field of functional proteomics is evolving quickly. In the next 5 years, we believe that functional proteomics will play an important role in reclassifying breast cancer subtypes. We may have to redefine responders to the HER2-directed therapy with the use of functional proteomics. With emergence of novel targeted therapies in breast cancer, there is increasing need to develop predictive functional proteomic biomarkers that are pathway-based. Proteomic platforms will become more prevalent in early clinical trials in the next 5 years.
Acknowledgments
This work was supported in part by National Cancer Institute 1K23CA121994 (to A.M.G.-A.), American Society of Clinical Oncology Career Development Award (to A.M.G.-A.), Komen for the Cure SAC100004 (to A.M.G.-A.), and American Cancer Society Research Scholar Grant 121329-RSG-11-187-01-TBG (to A.M.G.-A.).
Author Contributions
Conception/Design: Ana Maria Gonzalez-Angulo, Young Kwang Chae
Provision of study material or patients: Ana Maria Gonzalez-Angulo, Young Kwang Chae
Collection and/or assembly of data: Ana Maria Gonzalez-Angulo, Young Kwang Chae
Data analysis and interpretation: Ana Maria Gonzalez-Angulo, Young Kwang Chae
Manuscript writing: Ana Maria Gonzalez-Angulo, Young Kwang Chae
Final approval of manuscript: Ana Maria Gonzalez-Angulo, Young Kwang Chae
Disclosures
The authors indicated no financial relationships.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Jelovac D, Emens LA. HER2-directed therapy for metastatic breast cancer. Oncology (Williston Park) 2013;27:166–175. [PubMed] [Google Scholar]
- 3.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 4.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 6.Yeh E, Cunningham M, Arnold H, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 7.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Lam SW, Jimenez CR, Boven E. Breast cancer classification by proteomic technologies: Current state of knowledge. Cancer Treat Rev. 2014;40:129–138. doi: 10.1016/j.ctrv.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Burnette WN. “Western blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 10.Theriault RL, Carlson RW, Allred C, et al. Breast cancer, version 3.2013: Featured updates to the NCCN guidelines. J Natl Compr Canc Netw. 2013;11:753–760; quiz 761. doi: 10.6004/jnccn.2013.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanash S. A call for a fresh new look at the plasma proteome. Proteomics Clin Appl. 2012;6:443–446. doi: 10.1002/prca.201200052. [DOI] [PubMed] [Google Scholar]
- 12.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 13.Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez P, Müller M, Appel RD. Automated protein identification by tandem mass spectrometry: Issues and strategies. Mass Spectrom Rev. 2006;25:235–254. doi: 10.1002/mas.20068. [DOI] [PubMed] [Google Scholar]
- 15.Seeley EH, Caprioli RM. Molecular imaging of proteins in tissues by mass spectrometry. Proc Natl Acad Sci USA. 2008;105:18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schöne C, Höfler H, Walch A. MALDI imaging mass spectrometry in cancer research: Combining proteomic profiling and histological evaluation. Clin Biochem. 2013;46:539–545. doi: 10.1016/j.clinbiochem.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 17.Bauer JA, Chakravarthy AB, Rosenbluth JM, et al. Identification of markers of taxane sensitivity using proteomic and genomic analyses of breast tumors from patients receiving neoadjuvant paclitaxel and radiation. Clin Cancer Res. 2010;16:681–690. doi: 10.1158/1078-0432.CCR-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K, Yuan K, Wu H, et al. Identification of potential markers related to neoadjuvant chemotherapy sensitivity of breast cancer by SELDI-TOF MS. Appl Biochem Biotechnol. 2012;166:753–763. doi: 10.1007/s12010-011-9464-z. [DOI] [PubMed] [Google Scholar]
- 19.Yang WS, Moon HG, Kim HS, et al. Proteomic approach reveals FKBP4 and S100A9 as potential prediction markers of therapeutic response to neoadjuvant chemotherapy in patients with breast cancer. J Proteome Res. 2012;11:1078–1088. doi: 10.1021/pr2008187. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa T, Huang SK, Martinez SR, et al. Proteomic profiling of primary breast cancer predicts axillary lymph node metastasis. Cancer Res. 2006;66:11825–11830. doi: 10.1158/0008-5472.CAN-06-2337. [DOI] [PubMed] [Google Scholar]
- 21.Kabbage M, Chahed K, Hamrita B, et al. Protein alterations in infiltrating ductal carcinomas of the breast as detected by nonequilibrium pH gradient electrophoresis and mass spectrometry. J Biomed Biotechnol. 2008;2008:564127. doi: 10.1155/2008/564127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gast MC, Schellens JH, Beijnen JH. Clinical proteomics in breast cancer: A review. Breast Cancer Res Treat. 2009;116:17–29. doi: 10.1007/s10549-008-0263-3. [DOI] [PubMed] [Google Scholar]
- 23.Cahill DJ. Protein and antibody arrays and their medical applications. J Immunol Methods. 2001;250:81–91. doi: 10.1016/s0022-1759(01)00325-8. [DOI] [PubMed] [Google Scholar]
- 24.Wingren C, Borrebaeck CA. Antibody-based microarrays. Methods Mol Biol. 2009;509:57–84. doi: 10.1007/978-1-59745-372-1_5. [DOI] [PubMed] [Google Scholar]
- 25.Hennessy BT, Lu Y, Gonzalez-Angulo AM, et al. A technical assessment of the utility of reverse phase protein arrays for the study of the functional proteome in non-microdissected human breast cancers. Clin Proteomics. 2010;6:129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rapkiewicz A, Espina V, Zujewski JA, et al. The needle in the haystack: Application of breast fine-needle aspirate samples to quantitative protein microarray technology. Cancer. 2007;111:173–184. doi: 10.1002/cncr.22686. [DOI] [PubMed] [Google Scholar]
- 27.Ibarra-Drendall C, Troch MM, Barry WT, et al. Pilot and feasibility study: Prospective proteomic profiling of mammary epithelial cells from high-risk women provides evidence of activation of pro-survival pathways. Breast Cancer Res Treat. 2012;132:487–498. doi: 10.1007/s10549-011-1609-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wulfkuhle JD, Speer R, Pierobon M, et al. Multiplexed cell signaling analysis of human breast cancer applications for personalized therapy. J Proteome Res. 2008;7:1508–1517. doi: 10.1021/pr7008127. [DOI] [PubMed] [Google Scholar]
- 29.Espina V, Wulfkuhle J, Liotta LA. Application of laser microdissection and reverse-phase protein microarrays to the molecular profiling of cancer signal pathway networks in the tissue microenvironment. Clin Lab Med. 2009;29:1–13. doi: 10.1016/j.cll.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Liu NQ, Braakman RB, Stingl C, et al. Proteomics pipeline for biomarker discovery of laser capture microdissected breast cancer tissue. J Mammary Gland Biol Neoplasia. 2012;17:155–164. doi: 10.1007/s10911-012-9252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Improta G, Zupa A, Fillmore H, et al. Protein pathway activation mapping of brain metastasis from lung and breast cancers reveals organ type specific drug target activation. J Proteome Res. 2011;10:3089–3097. doi: 10.1021/pr200065t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowherd SM, Espina VA, Petricoin EF, III, et al. Proteomic analysis of human breast cancer tissue with laser-capture microdissection and reverse-phase protein microarrays. Clin Breast Cancer. 2004;5:385–392. doi: 10.3816/cbc.2004.n.046. [DOI] [PubMed] [Google Scholar]
- 33.Wulfkuhle JD, Berg D, Wolff C, et al. Molecular analysis of HER2 signaling in human breast cancer by functional protein pathway activation mapping. Clin Cancer Res. 2012;18:6426–6435. doi: 10.1158/1078-0432.CCR-12-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tessitore A, Zazzeroni F, Alesse E. Reverse-phase protein microarray highlights HER2 signaling activation in immunohistochemistry/FISH/HER2-negative breast cancers. Expert Rev Proteomics. 2013;10:223–226. doi: 10.1586/epr.13.18. [DOI] [PubMed] [Google Scholar]
- 35.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 36.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blows FM, Driver KE, Schmidt MK, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: A collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fountzilas G, Dafni U, Bobos M, et al. Differential response of immunohistochemically defined breast cancer subtypes to anthracycline-based adjuvant chemotherapy with or without paclitaxel. PLoS One. 2012;7:e37946. doi: 10.1371/journal.pone.0037946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 40.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: When will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 41.Guiu S, Michiels S, André F, et al. Molecular subclasses of breast cancer: How do we define them? The IMPAKT 2012 Working Group Statement. Ann Oncol. 2012;23:2997–3006. doi: 10.1093/annonc/mds586. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez-Angulo AM, Hennessy BT, Meric-Bernstam F, et al. Functional proteomics can define prognosis and predict pathologic complete response in patients with breast cancer. Clin Proteomics. 2011;8:11. doi: 10.1186/1559-0275-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gujral TS, Karp RL, Finski A, et al. Profiling phospho-signaling networks in breast cancer using reverse-phase protein arrays. Oncogene. 2013;32:3470–3476. doi: 10.1038/onc.2012.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rastogi P, Anderson SJ, Bear HD, et al. Preoperative chemotherapy: Updates of National Surgical Adjuvant Breast and Bowel Project Protocols B-18 and B-27. J Clin Oncol. 2008;26:778–785. doi: 10.1200/JCO.2007.15.0235. [DOI] [PubMed] [Google Scholar]
- 46.Guarneri V, Broglio K, Kau SW, et al. Prognostic value of pathologic complete response after primary chemotherapy in relation to hormone receptor status and other factors. J Clin Oncol. 2006;24:1037–1044. doi: 10.1200/JCO.2005.02.6914. [DOI] [PubMed] [Google Scholar]
- 47.Symmans WF, Peintinger F, Hatzis C, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25:4414–4422. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez-Angulo AM, Liu S, Chen H, et al. Functional proteomics characterization of residual breast cancer after neoadjuvant systemic chemotherapy. Ann Oncol. 2013;24:909–916. doi: 10.1093/annonc/mds530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sohn J, Do KA, Liu S, et al. Functional proteomics characterization of residual triple-negative breast cancer after standard neoadjuvant chemotherapy. Ann Oncol. 2013;24:2522–2526. doi: 10.1093/annonc/mdt248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konecny G, Slamon DJ. Her2 testing and correlation with efficacy of trastuzumab therapy. Oncology. 2002;16:1576–1578. [PubMed] [Google Scholar]
- 51.Giampaglia M, Chiuri VE, Tinelli A, et al. Lapatinib in breast cancer: Clinical experiences and future perspectives. Cancer Treat Rev. 2010;36(suppl 3):S72–S79. doi: 10.1016/S0305-7372(10)70024-4. [DOI] [PubMed] [Google Scholar]
- 52.Ramić S, Asić K, Balja MP, et al. Correlation of phosphorylated HER2 with clinicopathological characteristics and efficacy of trastuzumab treatment for breast cancer. Anticancer Res. 2013;33:2509–2515. [PubMed] [Google Scholar]
- 53.Saal LH, Johansson P, Holm K, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci USA. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun SY, Rosenberg LM, Wang X, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 55.Hinds PW, Mittnacht S, Dulic V, et al. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 56.Keyomarsi K, O’Leary N, Molnar G, et al. Cyclin E, a potential prognostic marker for breast cancer. Cancer Res. 1994;54:380–385. [PubMed] [Google Scholar]
- 57.Agarwal R, Gonzalez-Angulo AM, Myhre S, et al. Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin Cancer Res. 2009;15:3654–3662. doi: 10.1158/1078-0432.CCR-08-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn R. Results of a randomized phase 2 study of pd 0332991, a cyclin-dependent kinase (cdk) 4/6 inhibitor, in combination with letrozole vs letrozole alone for first-line treatment of er+/her2- advanced breast cancer (bc). San Antonio Breast Cancer Symposium (SABCS) 4–8 December 2012, San Antonio, TX. [Google Scholar]