Abstract
Hair follicle stem cells (HFSCs) regenerate hair in response to Wnt signalling. Here, we unfold genome-wide transcriptional and chromatin landscapes of β-catenin–TCF3/4–TLE circuitry, and genetically dissect their biological roles within the native HFSC niche. We show that during HFSC quiescence, TCF3, TCF4 and TLE (Groucho) bind coordinately and transcriptionally repress Wnt target genes. We also show that β-catenin is dispensable for HFSC viability, and that if TCF3/4 levels are sufficiently reduced, it is dispensable for proliferation. However, β-catenin is essential to activate genes that launch hair follicle fate and suppress sebocyte fate determination. TCF3/4 deficiency mimics Wnt–β-catenin-dependent activation of these hair follicle fate targets; TCF3 overexpression parallels their TLE4-dependent suppression. Our studies unveil TCF3/4–TLE histone deacetylases as a repressive rheostat, whose action can be relieved by Wnt–β-catenin signalling. When TCF3/4 and TLE levels are high, HFSCs can maintain stemness, but remain quiescent. When these levels drop or when Wnt–β-catenin levels rise, this balance is shifted and hair regeneration initiates.
Wnt signalling plays a role in many adult stem cells, but exactly how it functions and for what purpose is not clear1. The downstream effector of canonical Wnt signalling is β-catenin, which can act as a bipartite transcription factor for the lymphoid enhancer-binding factor 1 (LEF1) and/or T-cell factor (TCF) DNA-binding proteins1,2. Without TCF4, mice die at birth owing to failure to maintain developing intestinal crypts3. Conversely, intestinal stem cells maintain long-term organoid cultures in vitro when Wnt signalling is enhanced4. Wnt signalling can also stimulate stem cell maintenance in cultures of haematopoeitic stem cells5 and embryonic stem cells6 (ESCs). In these stem cells, Wnt–β-catenin and LEF1–TCF actions act cooperatively and positively. However, in the quiescent stem cell niche of the adult hair follicle, LEF1–TCF Wnt reporter (TOPGAL) activity has not been observed7,8, suggesting that if Wnt–β-catenin signalling is required universally to maintain stem cells, it acts through promoting activation rather than viability.
More recent evidence suggests that β-catenin is dispensable for ESC proliferation under some culture conditions, and ablation of Tcf7l1 (encoding TCF3) in these cells can even promote pluripotency9–11. In ESCs, TCF3 seems to dampen self-renewal while Wnt–β-catenin stimulates it by counteracting TCF3-mediated repression9,12–15. Similar antagonistic actions between Wnt signalling and LEF1–TCFs have been observed in developmental studies of both the epiblast and hair follicle8,15–18.
To some extent, the particular LEF1–TCF protein determines whether the outcome is transcriptional activation or repression. Thus, in the hair follicle, nuclear β-catenin and LEF1 occur concomitantly with Wnt reporter transactivation as transit-amplifying cells (TACs) commit to the hair lineage7, whereas β-catenin and TCF3/4 act antagonistically at earlier steps in the same lineage8,17,18. Similarly, in ESCs, TCF3 seems to function as a repressor, whereas TCF1 works in concert with β-catenin14,19. Compounding this problem further, the antagonistic effects of Wnt–β-catenin on TCF3 may even be outside the classic model of canonical Wnt–β-catenin signalling, such as influencing TCF3 stability13.
There are other cases where context and tissue rather than LEF1–TCF protein influence whether LEF1–TCFs and Wnt–β-catenin will act antagonistically or cooperatively. Thus, while TCF4 functions together with β-catenin as a transcriptional activator in intestine20,21, TCF4 acts as a repressor both in the hair follicle and also in colon and colorectal cancer17,22,23. Adding even greater complexity, although TCF3 represses some features of differentiation during early mouse development15, it is required for exit from pluripotency and in this regard acts positively on differentiation24.
Superimposed on these functional issues is how β-catenin and its LEF1–TCF DNA-binding partners act to recognize and regulate their target genes. Recent studies suggest that LEF1–TCF target genes differ across cell types. Thus, haematopoietic lineage regeneration following acute injury depends on Wnt-induced nuclear translocation and binding of TCF4 to key blood genes that are already bound by Gata2 but awaiting transactivation25. In contrast, TCF4 shows co-occupancy with a different transcription factor, CDX2, in colonic cells26, while it controls metabolic genes in neonatal and adult livers27.
As important as these collective studies are, they do not explain at a molecular level how Wnt signalling can impact the switch from a repressive to an activated state and how stem cells change their transcriptional activity in response to Wnt signalling. Moreover, global chromatin immunoprecipitation (ChIP)-on-chip analysis on chromatin from cultured human ESCs shows that TCF3 binds not only to active pluripotency genes but also repressed differentiation genes28. A priori TCF3 might act as an activator for some genes and a repressor for others. Although a recent study shows that the β-catenin-binding domain of TCF3 is not required for gene activation in ESCs (ref. 13), TCF3 binding might still transactivate genes in particular if other transcription and/or chromatin modelling factors are involved. That said, if TCF3-expressing stem cells are in a Wnt-activated state it is hard to envision by a direct mechanism how β-catenin would influence this fate decision. On the flip side, TLE (transducin-like enhancer; human analogue of Drosophila melanogaster Groucho) repressor proteins, which can bind in vitro to LEF1–TCFs and histone deacetylases29–32 (HDACs), might be operative here in vivo. Additional possibilities come from reports that when either β-catenin or TCF3 is phosphorylated by certain kinases, their ability to bind DNA and/or function in transcription can be altered33–36. Possible heterogeneity in vitro and/or culture conditions might further add to the difficulty in assessing whether and how Wnts, β-catenin and LEF1–TCFs govern stem cell maintenance, proliferation and/or fate selection.
Here, we begin to address these issues, focusing on the adult stem cells that fuel cyclical bouts of active hair follicle regeneration and hair growth (anagen), followed by destruction (catagen) and then rest (telogen)37–41 (Supplementary Fig. 1a). HFSCs reside in a niche known as the bulge located at the base of the telogen-phase hair follicle. Using mice harbouring the highly sensitive Wnt reporter (Axin2LacZ/+; ref. 42), we first show that at the telogen-to-anagen transition, some HFSCs within the bulge begin to activate this reporter. Early HFSC progeny at the base of the bulge (hair germ) are molecularly similar to bulge HFSCs (ref. 43), but only hair germ cells exhibit detectable nuclear β-catenin, suggesting their enhanced sensitivity to Wnt signalling. We show that this difference is not merely rooted in proximity to the niche stimulus (dermal papilla), but is also critically linked to differential levels of TCF3/4. Our results resolve a number of existing paradoxes and provide several unexpected new insights into how Wnt signalling functions in orchestrating stem cell behaviour.
RESULTS
HFSCs can remain for months in their niche without β-catenin but they do not undergo hair regeneration
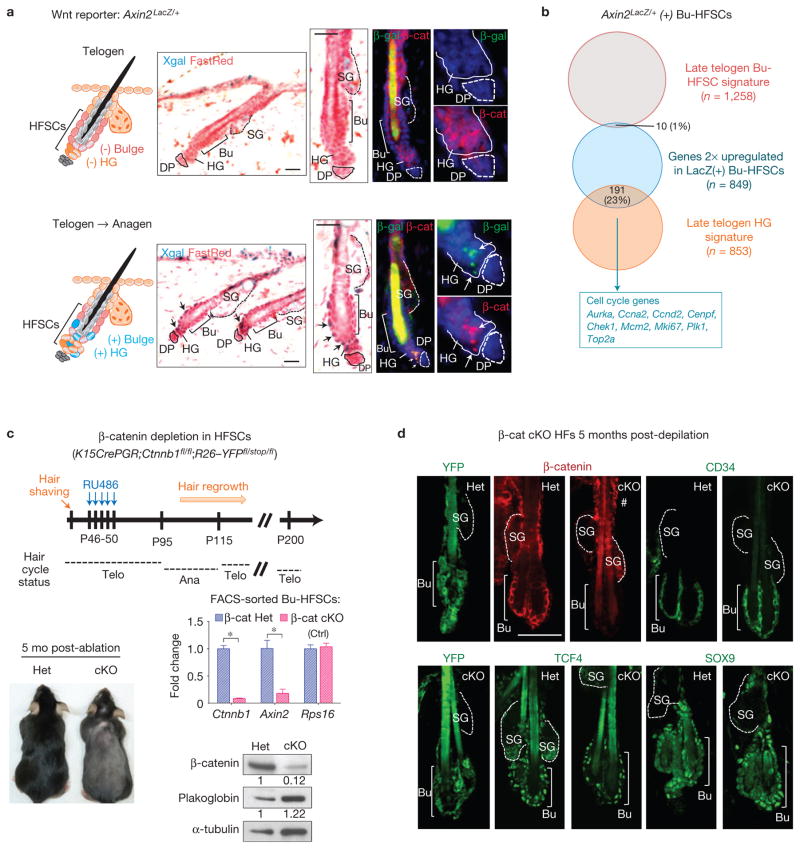
TOPGAL activity has not been observed in bulge HFSCs (ref. 7). We revisited this issue with the more sensitive Axin2LacZ/+ Wnt reporter mouse42 (Fig. 1a). Although sporadic Axin2LacZ/+ activity was constitutive in interfollicular epidermis (IFE), it was hair cycle dependent in hair follicles. The lack of Axin2LacZ/+ activity in resting-phase HFSCs confirmed their Wnt-silent, quiescent state. However, at the telogen-to-anagen transition, Axin2LacZ/+ activity was induced both in bulge and underlying hair germ.
Figure 1.
HFSCs are Wnt-responsive, but do not require β-catenin to exist in a quiescent, undifferentiated state. (a) X-gal staining of adult Axin2LacZ/+ hair follicles shows that β-galactosidase (β-gal; Wnt-responsive) activity is not detected in quiescent (Telogen) HFSCs, but can be seen in bulge (Bu) and hair germ (HG) cells at anagen onset (Telogen → Anagen). β-Galactosidase expression at anagen onset coincides with nuclear β-catenin most clearly in the hair germ (bottom right), consistent with its enhanced sensitivity to this signal. DP, dermal papilla; SG, sebaceous gland. (b) Venn diagram showing significant overlap between the genes upregulated in Axin2–LacZ(+) versus Axin2–LacZ(−) Bu-HFSCs and the signature from late telogen (Wnt-activated) hair germ43. Overlapping genes include those involved in cell cycle progression. (c) β-catenin conditional-knockout (β-cat cKO) HFSCs fail to activate Axin2 expression and launch a new hair cycle. Top: schematic showing regimen for Ctnnb1 ablation in adult HFSCs during second telogen. K15CrePGR;Ctnnb13/3;Rosa26–YFP3/stop/3 mice were treated with RU486 to activate Cre recombinase in HFSCs. Lower left: K15CrePGR;Ctnnb1+/3;Rosa26–YFP3/stop/3 (Het) and K15CrePGR;Ctnnb13/3;Rosa26–YFP3/stop/3 (cKO) mice at 5 months (5 mo) after hair-shaving and RU486 treatment. Note that the β-cat cKO mouse fails to regrow a new hair coat. Lower right: qPCR and immunoblotting analyses of FACS-isolated β-cat cKO HFSCs. (d) 5 mo after ablation of β-catenin, YFP(+) Bu-HFSCs exist in a prolonged resting state, but their expression levels of stem cell markers appear unchanged. # denotes upper hair follicle, which is not targeted by K15-Cre and remains β-catenin-positive. Scale bars, 50 μm (a,d). Data in c are reported as average + s.d. n = 3; *P < 0.01. Error bars for qPCR calculated from technical triplicates. Similar results were reproduced in five independent experiments. Uncropped images of blots are shown in Supplementary Fig. 8.
Although nuclear β-catenin was also hair cycle dependent, it was detected only in the hair germ, and at anagen onset43. We therefore wondered whether Axin2–LacZ(+) bulge (Bu)-HFSCs might share similar characteristics with activated hair germ cells, albeit at a reduced threshold. Indeed, by performing fluorescence-activated cell sorting (FACS)-Gal to purify β-gal(+) Bu-HFSCs (Supplementary Fig. 2a) and conducting RNA-seq analysis, we learned that transcripts that were ≥2× upregulated in LacZ(+) versus LacZ(−) Bu-HFSCs overlapped with those upregulated in the activated hair germ43 (Fig. 1b). These data suggest that Bu-HFSCs are also Wnt-responsive even though they are not as closely situated as the hair germ to the dermal papilla (Supplementary Fig. 2b). Directly beneath the hair germ, dermal papilla is known to instigate the Wnt-inhibitory BMP crosstalk involved in the telogen-to-anagen transition. To further explore β-catenin’s role in HFSCs, we engineered K15CrePGR;Ctnnb1fl /fl;Rosa26-YFPfl/stop/fl mice, and used RU486 to ablate Ctnnb1 in the stem cell niche during their second telogen (Fig. 1c). Just before targeting, we shaved the mice to remove their prior hair coat, so we could monitor entry into the next hair cycle. In contrast to their heterozygous littermates, YFP(+) β-catenin conditional knockout (β-cat cKO) HFSCs remained quiescent and showed no signs of anagen re-entry, that is, the point at which Wnt reporter activity is activated in the wild-type bulge and hair germ (Fig. 1c and Supplementary Fig. 3a). Remarkably however, the β-cat cKO Bu-HFSC pool remained comparable in size to their control counterparts (Supplementary Fig. 3b). Even 5 months after Ctnnb1 ablation, YFP(+) β-cat cKO Bu-HFSCs continued to express stemness genes at normal levels (Fig. 1d and Supplementary Fig. 3c). These findings were surprising because HFSCs are lost when tamoxifen is topically applied to ablate Ctnnb1 in K14CreER;Ctnnb1fl/fl mice44. Later, we address the reasons underlying this difference, and show that it depends on whether β-cat cKO HFSCs are exposed to a wound-like situation (for example, induced by topical tamoxifen) or allowed to exist in their normal homeostatic environment.
Bulge HFSCs can proliferate long-term without β-catenin
We next addressed whether the failure of β-cat cKO HFSCs to initiate hair cycling reflects an inability to proliferate and/or a block in activating cell fate commitment. For this purpose, we used fluorescence-activated cell sorting (FACS) to isolate CD34+α6+YFP+ Bu-HFSCs from our β-cat cKO and control littermates. When cultured under conditions that promote HFSC proliferation, β-cat cKO Bu-HFSCs exhibited relatively normal colony numbers and sizes (Fig. 2a). Even on long-term passaging, no major differences were seen (Fig. 2b). Moreover, as confirmed by quantitative PCR (qPCR) and immunoblotting, colonies forming in β-cat cKO HFSC cultures were not simply ‘escapers’. Although β-catenin’s dispensability for proliferation was expected for cultured IFE keratinocytes45, it was surprising for HFSCs, where Wnt signalling is required for their specification.
Figure 2.
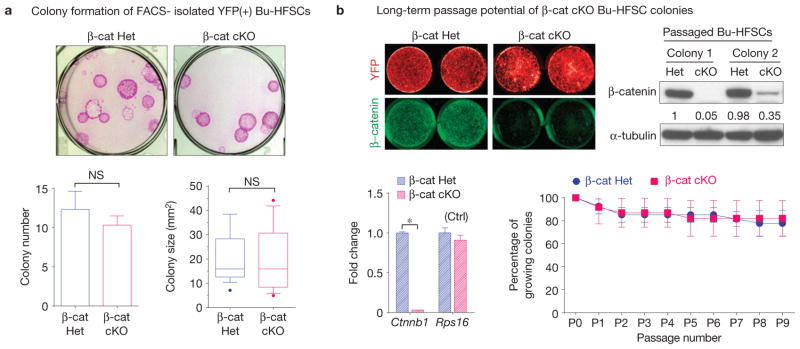
Bulge HFSCs can proliferate in vitro without β-catenin. (a) When cultured two weeks in medium rich in serum and growth factors, β-cat cKO Bu-HFSCs formed colonies of comparable numbers and sizes to control Bu-HFSCs. Rhodamine stained cultures at top; quantifications, below. (b) RosaYFP(+) Bu-HFSC colonies null or heterozygous for Ctnnb1 grow at comparable rates and can be passaged long-term. β-catenin depletion was verified for individual colonies, as shown by in-cell immunostaining (top left), immunoblotting (top right), and qPCR of mRNAs (bottom left) from passaged colonies. As measured by passaging in vitro, the long-term potential is similar for β-cat cKO and control Bu-HFSCs (bottom right). Data in a (left) and b are reported as average+ s.d. n = 3; *P < 0.01; NS, not significant. Error bars for qPCR in b calculated from technical triplicates. Similar results were reproduced in three independent experiments. Uncropped images of blots are shown in Supplementary Fig. 8.
β-cat cKO HFSCs differentiate into sebocytes when activated in their niche
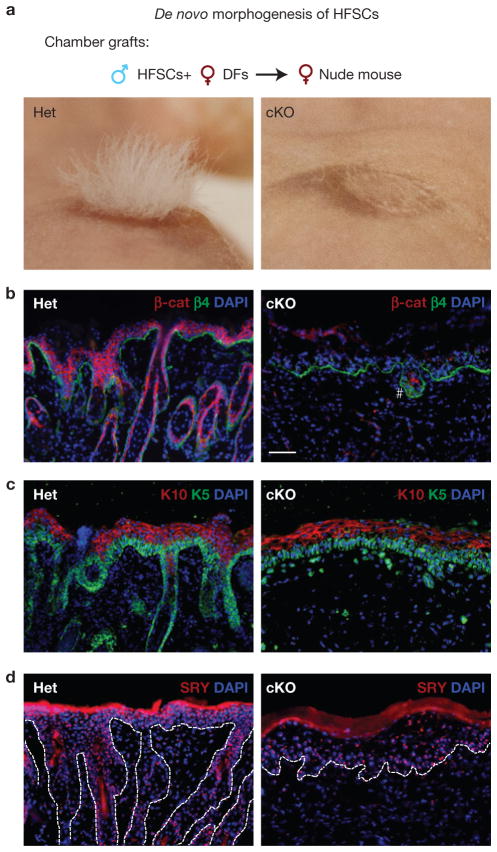
We next wondered what would happen if β-cat cKO Bu-HFSCs were forced to become activated in vivo. We approached this problem in two ways. In the first experiment, we engrafted our cultured and long-term passaged male (SRY+) Bu-HFSCs onto immunodeficient (nude) female mice (Fig. 3a). Immunofluorescence microscopy revealed that while control (Het) HFSCs generated IFE and hair follicles, β-cat cKO HFSCs produced IFE but not hair follicles (Fig. 3b–d).
Figure 3.
Engrafted β-catenin-deficient bulge HFSCs make only epidermis and not hair. When engrafted, control Bu-HFSCs generate epidermis and hair follicles, but β-cat cKO Bu-HFSCs produce only epidermis. (a) Experiments were performed with cultured male (Het or β-cat cKO) Bu-HFSCs and freshly purified female dermal fibroblasts, engrafted in chambers onto female nude mice. (b,c) Grafts were assayed visually and by immunostaining for β-catenin (β-cat), intergrin β4 (β4), keratin 5 (K5), and 10 (K10). # denotes a rare hair-follicle-like structure that contains a few residual β-catenin-positive cells. Scale bar, 50 μm. (d) Y-chromosome (SRY) fluorescence in situ hybridization (FISH) confirms the formation of epidermis from engrafted β-cat cKO Bu-HFSCs.
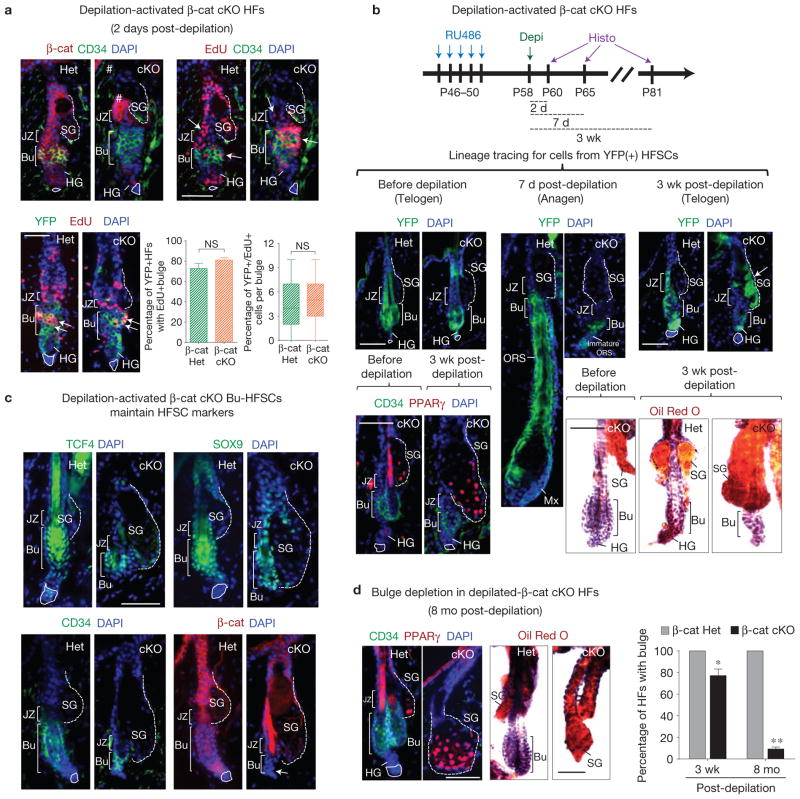
To examine the consequences of activating β-cat cKO HFSCs in their native niche, we used hair plucking, typically inducing a wound response in the bulge and junctional zone46,47 (JZ). Hair plucking performed in mid-telogen resulted in proliferation of both β-cat-negative and control β-cat-positive hair follicles throughout these regions (Fig. 4a, top). Lineage tracing with YFP confirmed that proliferating Bu-HFSCs were indeed derived from Cre-activated Rosa26–YFP(+) HFSCs (Fig. 4a, bottom). Consistent with our in vitro studies, this finding demonstrated that β-cat cKO Bu-HFSCs can be also stimulated to proliferate in vivo.
Figure 4.
β-catenin-deficient bulge HFSCs can become activated in their native niche, but progress to make the wrong fate choice. (a) Depilation-activated β-cat cKO HFSCs can proliferate in vivo. Two days post-waxing and 24-h post-EdU labelling, depilated skins were sectioned, immunostained with the antibodies shown and quantified. Note proliferation within hair germ, bulge and junctional zone (JZ) of both β-cat cKO and control hair follicles in vivo. Those proliferating Bu-HFSCs were YFP(+). (b) β-cat cKO HFSCs differentiate into sebocytes when activated in their native niche. Lineage tracing reveals that by 3-week post-depilation, control HFSC progeny (YFP+) had repaired the junctional zone, initiated and completed one hair cycle, and were in their next telogen. In contrast, β-cat cKO HFSCs failed to generate hair follicle, but their YFP+ progeny moved upwards and differentiated into sebocytes (arrow). PPARγstaining on β-cat cKO skin section (bottom left) and Oil Red O staining of sebocytes on whole mount skins (bottom right) before and 3 weeks post depilation (c) Depilation-activated β-cat cKO HFSCs are diminished, but still maintain HFSC markers. Immunolabelling with HFSC markers shows that 3 weeks post-depilation, the β-cat cKO bulge still maintains HFSC markers although it has shrunk in size concomitant with enlargement of the sebaceous gland. (d) Depilation-activated β-cat cKO HFSCs progressively differentiate and eventually deplete the niche of its stem cells. PPARγ staining on skin sections and Oil Red O staining of sebocytes on whole-mount skins at 8 months after depilation show enlarged sebaceous glands and loss of bulge in β-cat cKO skin. Quantification of hair follicles with bulge is shown at the right. Scale bars, 50 μm (a–d). Data in a,d are reported as average+ s.d. n = 3; *P < 0.05; **P < 0.01; NS, not significant.
Hair plucking also promotes entry of telogen-phase hair follicles into anagen. Indeed, within 2 d post-depilation, control hair follicles entered a new hair cycle, and by three weeks had entered their next telogen (Fig. 4b). Lineage tracing further revealed that control Rosa26–YFP(+) HFSC progeny not only regenerate the hair follicle but also move upward and repair the JZ after depilation (shown). In contrast, β-cat cKO HFSCs not only failed to initiate hair cycling, but in addition, they showed signs of massive sebocyte differentiation (PPARγ+; Oil-red-O+) within their ‘repaired’ junctional zone (Fig. 4b). Increased sebocyte differentiation was accompanied by a diminished bulge that still expressed HFSC markers (Fig. 4c).
Following activation, the β-cat cKO niche never recovered. As differentiation progressed, the niche was eventually depleted of its stem cells (Fig. 4d). Together, these collective findings provide compelling evidence that the function of β-catenin in HFSCs resides not in homeostasis or proliferative potential per se but rather in hair follicle cell fate and/or fate-linked divisions.
β-catenin governs activation of a cohort of genes involved in hair follicle fate determination
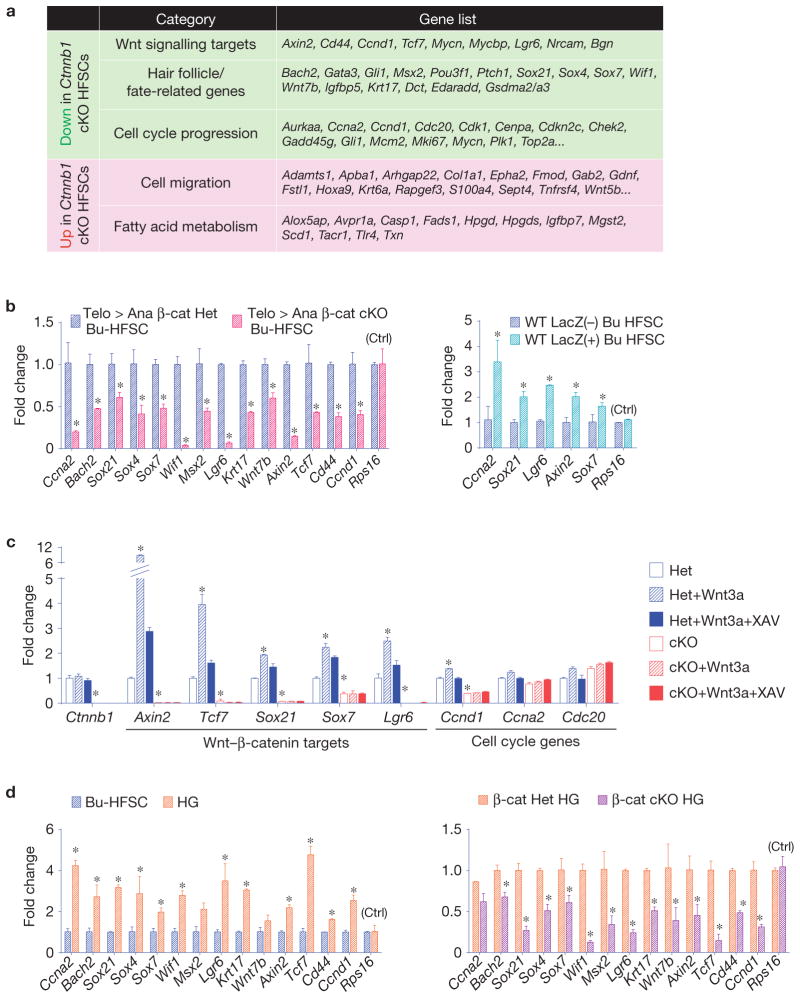
To ascertain how β-catenin functions to regulate hair follicle fate determination, we transcriptionally profiled CD34+α6+YFP+ Bu-HFSCs from control hair follicles and their matched β-cat cKO littermates at early anagen onset. Notably, a significant cohort of established Wnt–β-catenin target genes was all downregulated in β-cat cKO Bu-HFSCs (Fig. 5a). In addition, cell cycle and hair follicle/fate-related genes were poorly transcribed in these cells. Validation was carried out by qPCR on independent Bu-HFSC isolates (Fig. 5b, left). Interestingly, some of these genes were downregulated even earlier when β-cat cKO Bu-HFSCs were still in telogen (Supplementary Fig. 4a). Conversely, some of these genes were preferentially upregulated in LacZ(+) versus LacZ(−) Bu-HFSCs isolated from wild-type Axin2LacZ/+ hair follicles at the start of anagen (Fig. 5b, right).
Figure 5.
β-catenin influences expression of Wnt-responsive genes in bulge HFSCs that are first activated in the hair germ at anagen onset. (a) Comparisons of RNA-seq data sets reveal a cohort of genes whose expression is sensitive to the presence of β-catenin. Gene ontology categories for genes downregulated in β-cat cKO Bu-HFSCs are highlighted with a green background and upregulated genes with a red background. Representative genes for each category are shown. (b) qPCR validates the β-catenin responsiveness of a group of genes that are downregulated in β-cat cKO Bu-HFSCs (left); note that a number of these are conversely upregulated in Wnt-responsive Axin2–LacZ(+) Bu-HFSCs (right). Values are normalized to β-cat Het or LacZ(−) Bu-HFSC mRNAs. Rps16, mRNA control. (c) β-catenin target genes show Wnt-responsiveness in culture. Cultured control and β-cat cKO Bu-HFSCs were treated with recombinant Wnt3a protein and/or Wnt inhibitor, XAV939 (XAV), for 6 h, and their mRNAs were then collected for qPCR analysis. Note that although the levels of hair follicle fate genes in control (Het) Bu-HFSCs were elevated by Wnt3a, they show no Wnt-responsiveness in β-cat cKO (cKO) Bu-HFSCs. (d) A cohort of genes that are preferentially upregulated in hair germ at anagen onset (left) are downregulated in this compartment when β-catenin is lost (right). Note that many of these genes are also differentially sensitive to Wnt signalling in the bulge (b,c). Values are normalized to Bu-HFSC or β-cat Het hair germ mRNAs. Data in b–d are reported as average+ s.d. n = 3; *P < 0.05. Error bars for qPCR calculated from technical triplicates. Similar results were reproduced in 3 (b), 2 (c), and 4 (d) independent experiments.
To further test the Wnt-responsiveness of these genes, we treated cultured Bu-HFSCs with Wnt3a ± XAV939, a Wnt-signalling inhibitor (Fig. 5c). β-cat cKO Bu-HFSCs showed no Wnt-responsiveness with regards to the expression of these genes (shown). While hair follicle fate genes showed Wnt responsiveness in vitro, the known Wnt-responsive cell cycle gene, Ccnd1, was one of only a few cell cycle genes that exhibited appreciable sensitivity to Wnt–β-catenin in culture. This finding was in good agreement with our functional studies showing that multiple signalling pathways can influence HFSC proliferation status.
Consistent with the hair germ’s greater Wnt sensitivity of the over the bulge at anagen onset43 (Supplementary Fig. 2b), the Wnt-responsive β-catenin-dependent genes of activated Bu-HFSCs were indeed expressed at even higher levels in hair germ cells (Fig. 5d, left). In agreement with their dependency on β-catenin, expression of these genes was compromised in β-cat cKO hair germ (Fig. 5d, right). Thus, we identified a group of Wnt–β-cat-responsive genes that are upregulated in normal HFSCs as they transition to the activated state within the physiological context of their niche.
Even though most HFSC genes were downregulated in β-cat cKO Bu-HFSCs at the time when their counterparts show Wnt-responsiveness, some genes were upregulated (Fig. 5a and Supplementary Fig. 4b). Of these, fatty acid metabolism genes were notable, given that β-cat cKO Bu-HFSCs had shown signs of sebocyte transformation on wound-induced activation (Figs 5a and 4b–d). qPCR verified these changes in depilation-activated β-cat cKO Bu-HFSCs (Supplementary Fig. 4c).
TCF3 and TCF4 bind many of the same genes in HFSCs
To elucidate how canonical Wnt signalling directly affects gene expression in HFSCs, we first showed that of the four LEF1–TCFs, only Tcf7l1 and Tcf7l2 (encoding TCF4) are expressed and exhibit nuclear co-localization in Bu-HFSCs throughout the hair cycle (Fig. 6a). In contrast, TCF1 (encoded by Tcf7) and LEF1 are expressed and nuclear in hair germ (Fig. 6a) and in Wnt-activated transit-amplifying cells as they differentiate to make hair48 (Supplementary Fig. 5a). Interestingly, in the hair germ, TCF3/4 expression levels were reduced, and on Wnt activation, LEF1 and TCF1 were induced (Fig. 6a). This switch suggests differential roles for these member proteins in response to Wnt signalling.
Figure 6.
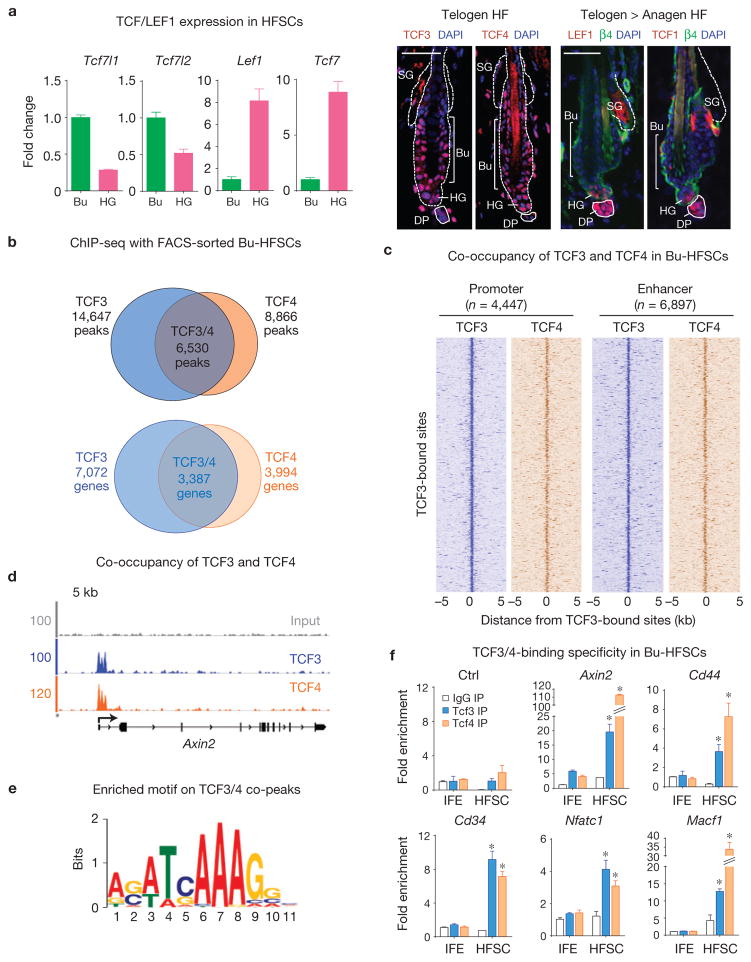
Co-occupancy of TCF3 and TCF4 on the bulge HFSC genome. (a) Of the four LEF1–TCF DNA-binding partners of β-catenin, TCF3 and TCF4 are expressed and nuclear in bulge HFSCs, while LEF1 and TCF1 are present in hair germ. mRNA expression (left) and protein patterns (right) of TCF3 (Tcf7l1), TCF4 (Tcf7l2), LEF1 (Lef1) and TCF1 (Tcf7) in bulge and hair germ compartments. Scale bar, 50 μm. (b) Venn diagrams showing overlap of TCF3 and TCF4 peaks (top) and genes (bottom) in Bu-HFSC chromatin. (c) TCF3 and TCF4 co-occupancy across the Bu-HFSC genome. Density maps of TCF3 (blue) and TCF4 (orange) ChIP-seq reads at TCF3-bound peaks. n, number of peaks within indicated region. For each peak (y axis), the normalized counts of TCF3 and TCF4 ChIP-seq reads are shown within the region centred on the TCF3 peak summit (x axis). (d) TCF3 and TCF4 occupy many common targets in Bu-HFSCs. Shown are representative examples (grey is input control). The y axis shows the number of mapped reads at each genomic position. (e) TCF3 and TCF4 overlapping peaks are most enriched for the classical LEF1–TCF binding motif. (f) ChIP experiments were performed with FACS-sorted IFE cells and Bu-HFSCs. Antibodies were against TCF3, TCF4 and IgG (negative control). Ctrl, region from ChIP-seq that lacks TCF3/4-binding peaks. Fold change corresponds to the immunoprecipitate enrichment over input. Data are reported as average+s.d. n = 3; *P < 0.01. Error bars for qPCR calculated from technical triplicates. Similar results were reproduced in 3 (a) and 2 (f) independent experiments.
Given the reduced sensitivity of Bu-HFSCs to Wnt–β-catenin-mediated gene activation, we searched for mechanisms that might extend beyond mere proximity to dermal papilla. We first conducted in vivo ChIP-seq analyses on FACS-purified Bu-HFSCs to address whether TCF3 and TCF4 bind to the same or different genes, and whether these genes overlap with those whose expression showed Wnt–β-catenin sensitivity. The selective specificities and quality of our antibodies were validated by their abilities to quantitatively immunoprecipitate TCF3 and TCF4 from wild-type Bu-HFSC lysates relative to Tcf7l1-null or Tcf7l2-null cells, and to IgG controls (Supplementary Fig. 5b). To control for all other technical aspects, our ChIP-seq data were analysed in comparison to input DNAs as established previously25,49 (see also ENCODE guideline50).
After aligning ChIP-seq reads to the mouse genome and removing redundant reads, we used the program MACS (ref. 51) for peak calling and identified 14,647 and 8,866 bound regions (that is, peaks) for TCF3 and TCF4, respectively. Among these, 6,530 peaks (3,387 genes) were co-occupied by both TCF3 and TCF4 (Fig. 6b,c). ~75% of peaks were located to promoters (±2 kilobases (kb) of transcription start sites) or ‘enhancers’ (−50 kb upstream to and +5 kb downstream of the body) of mouse RefSeq annotated genes (Supplementary Fig. 5c–d).
TCF3/4 co-targets included a number of our identified β-catenin-dependent genes, including Axin2 (Fig. 6d). Importantly, the classic LEF1–TCF binding motif was the top enriched motif in TCF3/4 co-occupied peaks, revealing considerably greater enrichment over random genomic sequences (Fig. 6e and Supplementary Fig. 5e).
Approximately a third of TCF3/4 targets in Bu-HFSCs were also TCF3 targets in cultured mouse ESCs (ref. 28; Supplementary Fig. 5f). Although not necessarily correlating with transcriptional activity, established Wnt-signalling genes were among this group. Moreover, by ChIP–qPCR, clear specificity of TCF3/4 binding to target genes was seen with Bu-HFSC versus IFE chromatin (Fig. 6f). These data provide an example where two LEF1–TCF proteins share many of the same targets within a purified cell population.
Within the stem cell niche, elevated TCF3 suppresses HFSC activation, while loss of TCF3/4 lowers the activation threshold
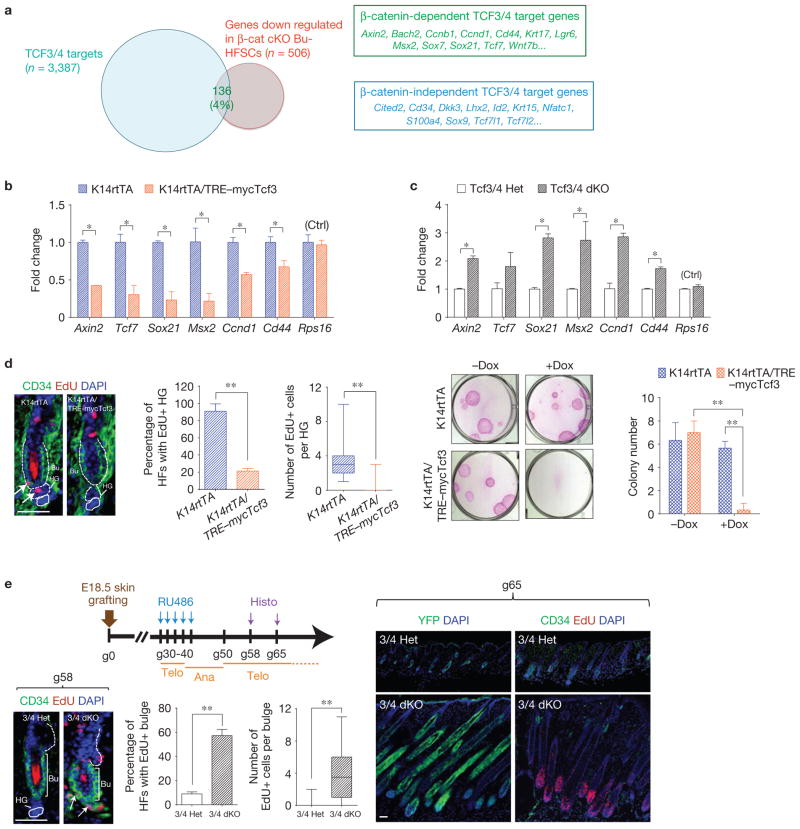
It was surprising that only 4% of TCF3/4-bound genes were downregulated in β-cat cKO Bu-HFSCs (Fig. 7a). That said, this cohort included established Wnt–β-catenin target genes (for example, Axin2, Ccnd1 and Cd44) and hair follicle/fate-related genes (for example, Msx2, Sox21 and Tcf7); conversely, many TCF3/4-bound genes that exhibited no sensitivity to β-catenin loss within the bulge niche were HFSC signature genes (for example, Lhx2, Sox9 and Tcf7/1; Fig. 7a).
Figure 7.
Within the stem cell niche, elevated TCF3 impairs bulge HFSC activation, whereas loss-of-TCF3/4 lowers the activation threshold. (a) Venn diagram showing that only a small number of genes are bound by TCF3/4 in Bu-HFSCs and downregulated on β-catenin loss (left). Examples of β-catenin-dependent and -independent TCF3/4 targets (right). Note that the list of 136 dependent genes includes those involved in fate determination. (b) TCF3 overexpression results in repression of Wnt–β-catenin-activated genes. qPCR of Wnt-activated target genes on FACS-isolated Bu-HFSCs from doxycycline-treated K14rtTA and K14rtTA/TRE–mycTcf3 skins (see Supplementary Fig. 6a). Values are normalized to K14rtTA Bu-HFSC mRNAs. (c) Depletion of TCF3 and TCF4 in HFSCs causes upregulation of Wnt–β-catenin-activated genes. qPCR on FACS-isolated Bu-HFSCs from RU486-treated K15CrePGR/Tcf7l1+/3;Tcf7l2+/−;Rosa26–YFP3/stop/3 (Tcf3/4 Het) and K15CrePGR;Tcf7l13/3;Tcf7l2−/−;Rosa26–YFP3/stop/3 (Tcf3/4 dKO) grafted skins (see Supplementary Fig. 6b). Values are normalized to TCF3/4 Het Bu-HFSC mRNAs. (d) Elevated TCF3 prohibits HFSC activation in vivo and proliferation in vitro. Left: doxycycline (Dox) was given throughout P19–22. EdU was then administered for 24 h and hair follicles were analysed. Immunofluorescence and quantifications show that increased TCF3 impairs HFSC proliferation at telogen-to-anagen. Right: TCF3 overexpression impedes Bu-HFSC proliferation in vitro. Two weeks after plating, colonies from telogen-phase HFSCs were fixed, Rhodamine B stained and quantified. (e) Loss of TCF3 and TCF4 in HFSCs causes precocious hair cycling. The schematic shows timelines for treatment, analyses and hair follicle cycle stage. Immunostained sections of hair follicle bulge/hair germ (bottom left) are from telogen skin 58d after grafting (g58) and 24 h after EdU injections. Sections taken at g65 show that Tcf7l1/Tcf7l2-null HFSCs (3/4 dKO) precociously initiated anagen relative to control HFSCs (3/4 Het), which remained quiescent. Lineage tracing shows YFP-marked HFSC progeny. Rps16, mRNA control. Scale bars, 50 μm (d,e). Data in b–e are reported as average + s.d. n = 3; *P < 0.05; **P < 0.01. d (middle), e (bottom right), box-and-whisker plots show distribution of EdU(+) cell numbers. Error bars for qPCR calculated from technical triplicates. Similar results were reproduced in 5 (b) and 3 (c) independent experiments.
TCF3/4 were significantly downregulated in hair germ, and because Bu-HFSCs showed less sensitivity to Wnt signalling than hair germ (Fig. 6a), we wondered whether TCF3/4 might act as transcriptional repressors in Bu-HFSCs, thereby restricting the numbers of HFSCs that become activated at anagen onset. To test this possibility, we used doxycycline-regulatable TRE–mycTCF3 transgenic mice18 to manipulate TCF3/4 levels within the bulge (Supplementary Fig. 6a). Elevated TCF3 reduced transcriptional activities of Wnt target genes (Fig. 7b). Thus, Wnt is probably present, but TCF3/4 levels prevent it being sensed strongly by quiescent Bu-HFSCs. Furthermore, TCF3 overexpression represses rather than superactivates Wnt signalling.
To assess how loss of TCF3/4 function affects Wnt target genes in a native stem cell niche, we used the HFSC-specific K15CrePGR driver to conditionally inducibly ablate Tcf7l1 on a Tcf7l2-null; Rosa26 –YFPfl/stop/fl background. To circumvent lethality, embryonic day (E)18.5 Tcf7l2-null skins were grafted onto nude mice (referred to as grafted day 0; g0). Seven weeks later, when hair follicles lacking TCF4 were phenotypically normal and in an extended (second) telogen, RU486 was administered to specifically target Tcf7l1 alleles in quiescent HFSCs. qPCR and immunoblotting of FACS-isolated YFP(+) HFSCs showed that TCF3 was depleted by >90%, and as expected, TCF4 was absent (Supplementary Fig. 6b). Our cohort of canonical Wnt targets were all upregulated (Fig. 7c).
Given the inverse correlation between TCF3/4 and expression of Wnt-responsive genes, we wondered whether changing TCF3/4 levels within the stem cell niche might affect HFSC quiescence and/or fate determination, and if so how. TCF3 elevation in quiescent HFSCs blocked their telogen-to-anagen transition, which as discussed earlier affects both proliferation and fate determination (Fig. 7d, left). Depilation-induced proliferation and hair cycling were also blocked when TCF3 was elevated in HFSCs (Supplementary Fig. 6c). Interestingly, when HFSCs were cultured under likely low TCF3 conditions where proliferation is normally favoured, TCF3 overexpression significantly compromised colony-forming efficiency in vitro (Fig. 7d, right). Thus, elevated levels of TCF3 can override the proliferative effects generated by other signalling pathways. Moreover, the data suggest that the epidermal proliferation seen when TCF3 is elevated in young animals18 is probably due to secondary effects of compromising barrier function, rather than direct effects of TCF3.
Hair follicles do not form in engrafted embryonic TCF3/4-deficient skin17. A priori, this could be due to an inability of the HFSC niche to form, a wound-induced effect or an effect on HFSC maintenance and/or cell fate commitment. To distinguish between these possibilities, we induced Tcf7l1 ablation in telogen-phase Tcf7l2-null hair follicles after they had formed. TCF3/4 loss caused precocious proliferation and activation of HFSCs, followed by a seemingly normal hair cycle (Fig. 7e). Thus, TCF3/4 seem to function by maintaining HFSC quiescence and suppressing their transition from telogen-to-anagen.
TLE proteins act as co-repressors with TCF3/4 in quiescent HFSCs
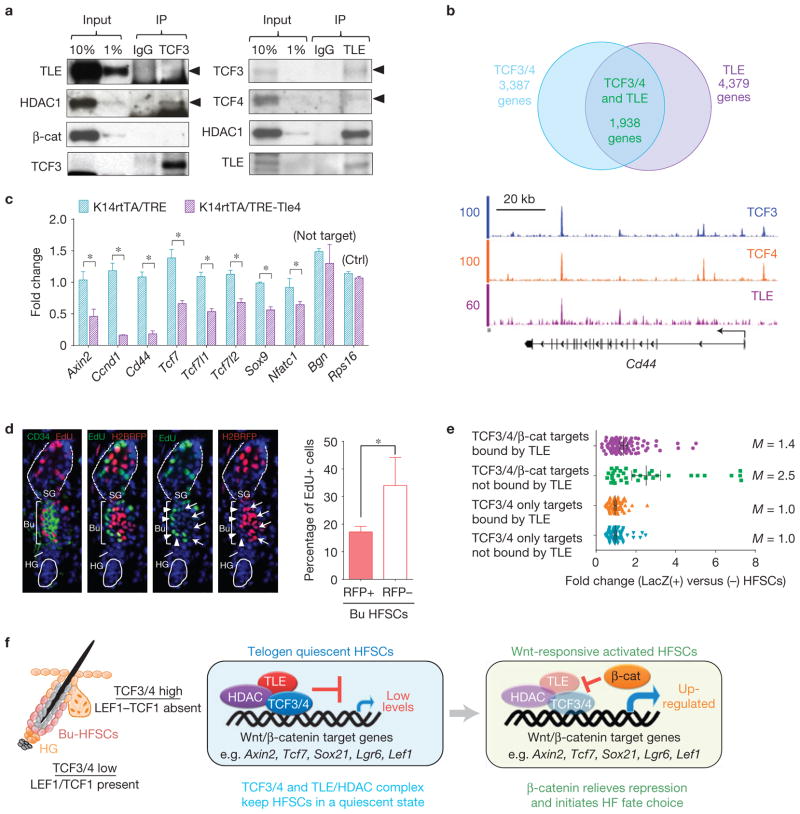
Interestingly, Groucho/TLE and histone deacetylase1 (HDAC1) physically associated with TCF3 in Bu-HFSCs in vivo (Fig. 8a). Notably, β-catenin was not a part of this complex, which contained TCF3, TLE and HDAC1. Similarly, TLE antibodies pulled down not only TLE but also TCF3, TCF4 and HDAC1.
Figure 8.
TLE proteins act as co-repressors of TCF3/4 in quiescent bulge HFSCs. (a) TCF3 and TCF4 interact with TLE proteins and HDAC1 in Bu-HFSCs. Co-immunoprecipitation of endogenous TCF3 (left) and TLE (right) in FACS-isolated Bu-HFSCs. Complexes (IP) and whole-cell extracts (Input) were analysed by immunoblotting. Note that TLE and HDAC1, but not β-catenin, were pulled down in TCF3-immunoprecipitation. (b) Co-occupancy of TCF3, TCF4 and TLE. Top: Venn diagrams showing overlap of TCF3/4 and TLE genes in Bu-HFSCs. Note that >50% of TCF3/4 targets are also bound by TLE. Bottom: ChIP-seq signals represent binding of TCF3 (blue), TCF4 (orange) and TLE (purple) at Cd44 locus. (c) TLE4 overexpression represses expression of TCF3/4-bound genes. K14rtTA/TRE–Tle4 mice were generated using in utero infection (H2BRFP+; ref. 52; see Supplementary Fig. 7b). Shown are qPCR analysis of mRNAs from FACS-isolated RFP(+) HFSCs of doxycycline-induced K14rtTA/TRE (control) and K14rtTA/TRE–Tle4 mice. Control Bgn is not a TLE target. Values are normalized to K14rtTA Bu-HFSC mRNAs for each set. Data are reported as average + s.d. n = 3; *P < 0.05. Error bars calculated from technical triplicates. Similar results were reproduced in 3 independent experiments. (d) TLE4 overexpression compromises depilation-induced Bu-HFSC proliferation. Doxycycline-induced K14rtTA/TRE–Tle4 skins were waxed and EdU-labelled. RFP(+) Tle4-overexpressing Bu-HFSCs exhibit less proliferation than RFP(−) non-expressing Bu-HFSCs. Data are reported as average+ s.d. n = 5; *P < 0.05. (e) TLE-binding increases the threshold for Wnt-signalling activation of β-catenin-activated TCF3/4-bound targets. Dot plot showing fold changes of indicated target genes responding to Wnt activity in anagen-HFSCs (Axin2–LacZ(+) versus (−) Bu-HFSCs; see Supplementary Table 1). Mean (M) values of fold changes for each group are shown at the right. β-catenin-activated TCF3/4 targets showed greater induction to Wnt activity in anagen Bu-HFSCs if they were not bound by TLE while in telogen (not bound, n = 35; bound by TLE, n = 100). Random selected TCF-only targets are controls (n = 100). (f) Model summarizing results. In quiescent Bu-HFSCs, where TCF3/TCF4 are highly expressed, TLE repressors and HDAC1 are recruited to TCF3/4-bound genes to keep expression of Wnt–β-catenin targets low. At telogen-to-anagen when Wnt activity rises, nuclear β-catenin relieves this repression and activates its targets to initiate hair follicle fate choice. Uncropped images of blots are shown in Supplementary Fig. 8.
To determine whether this interacting complex was recruited to TCF3/4 target genes in Bu-HFSCs, we conducted in vivo ChIP-seq analysis for TLE using a pan-TLE antibody. Intriguingly, >50% of TCF3/4 target genes also showed occupancy of TLE proteins (Fig. 8b). Moreover, many of these genes showed close or overlapping TCF3, TCF4 and TLE binding activities within a promoter or enhancer, as revealed by the distribution of TLE ChIP-seq signals across TCF3-binding sites (Supplementary Fig. 7a). Notably, Wnt-activated TCF3/4 targets, such as Cd44, were bound by TLE in quiescent HFSCs (Fig. 8b).
To determine whether TLE levels influence the expression of Wnt-activated TCF3/4-bound genes, we generated a mouse model in which we could efficiently induce TLE4 overexpression in HFSCs when hair follicles were in their second telogen (Supplementary Fig. 7b). Notably, TCF3/4 and TLE co-targets, including Wnt-activated and HFSC signature genes, were downregulated on TLE4 overexpression, compared with the control (transduced with TRE alone; Fig. 8c). Although the targets of TLE4 included HFSC stemness genes, its repressive effects were greatest on Wnt-activated genes. Moreover, genes bound by TCF3/4 but not TLE (for example, Bgn) remained unchanged, suggesting that the effects of elevated TLE were specific to the natural TLE/TCF3/4-targeted genes (Fig. 8c).
To test whether TLE4 overexpression might block HFSC proliferation by virtue of its ability to downregulate Wnt targets, we performed hair plucking on TLE4-transduced skins. Depilation-induced proliferation in Bu-HFSCs was blocked by TLE4 overexpression when compared with those neighbouring not-transduced control Bu-HFSCs (Fig. 8d). Finally, β-catenin-activated targets bound by only TCF3/4 and not TLE showed greater expression in Wnt-responsive [Axin2–LacZ(+)] HFSCs than those bound by TCF3/4/TLE (Fig. 8e, M = 2.5 versus 1.4; Supplementary Table 1).
DISCUSSION
Despite the commonly held view that cells thrive in a Wnt-rich environment, increasing evidence suggests that this does not hold for quiescent niches, such as the bulge. Moreover, had a low level of endogenous Wnt activity been critical for sustaining stemness, we should have seen, but did not observe, an effect on stemness genes and/or long-term maintenance of quiescent HFSCs lacking β-catenin. The ability of β-cat cKO HFSCs to survive within their quiescent niche rendered the issue irrelevant physiologically as to whether there is low versus no Wnt signalling in the niche.
Our in vivo ChIP-seq analyses revealed co-occupancy of TCF3 and TCF4 across much of the HFSC genome. Thus, although TCF4 acts as a transcriptional activator in intestinal stem cells3, in Bu-HFSCs, both TCF3 and TCF4 are co-expressed, bind many common targets, and show opposing effects to those of Wnt–β-catenin signalling. Notably, many of the TCF3/4-bound genes were also bound by TLE/Groucho, whose action in quiescent HFSCs seemed to be as a global dampener of TCF3/4-bound genes.
Overall, our findings expose TCF3/4 as a repressive rheostat that governs bulge stem cell quiescence and hair follicle fate determination. When binding with TLE/HDAC in quiescent HFSCs, TCF3/4-bound fate determinants are protected from activation. When responding to Wnt activity at anagen onset, these targets are then activated by β-catenin and relieved of their repression, subsequently initiate fate choice first in hair germ where TCF3/TCF4 are low and LEF1–TCF1 are present. Given that both Lef1 and Tcf7 are TCF3/4 direct targets and also induced on Wnt–β-catenin activation, the expression switch from TCF3/TCF4 in Bu-HFSCs to LEF1–TCF1 in hair germ suggests that as TCF3/TCF4 levels drop and LEF1–TCF1 levels rise, cells become increasingly more sensitive to Wnt-mediated gene activation. These mechanisms are intriguing in light of our findings that although β-catenin is required for fate determination, it is not essential for HFSC maintenance or proliferation. This seemed to be the case both in the native niche and in culture, where other signalling pathways were sufficient to stimulate β-cat cKO Bu-HFSCs to proliferate. That said, high levels of TCF3/4/TLE overrode these other pathways and kept HFSC quiescent. In this way, Wnt–β-catenin and TCF3/4 essentially create proliferative-permissive versus non-permissive states, thereby collaborating to control stem cell quiescence. Summarized in Fig. 8f, this model places TCF3/4 levels at the crossroads of whether stem cells will remain in a quiescent, undifferentiated state or whether they will commit to actively generate tissue, a process likely to involve asymmetric but not symmetric cell divisions.
METHODS
Mice, doxycycline induction, and in utero injections
Mice were housed in the AAALAC-accredited Comparative Bioscience Center (CBC) at The Rockefeller University in accordance with NIH guidelines. Axin2LacZ/+ mice (C57BL6) were obtained from the Jackson Laboratory42; P62 (telogen) and P83 (telogen-to-anagen) are shown in Fig. 1a; P24 was used for FACS and RNA-seq analysis. K14rtTA/TRE–mycTcf3 mice (FVB/N) were generated as described previously18; P50–56 (second telogen) animals were used for analyses. Inducible knockout mouse lines were generated by crossing K15CrePGR (ref. 53; C57BL6), Tcf7l1fl/fl (ref. 17; C57BL6/129Sv), Tcf7l2−/− (C57BL6; kind gift from H. Clevers, Hubrecht Institute, Netherlands), Rosa26–YFPfl/stop/fl (ref. 54; C57BL6), or Ctnnb1fl/fl (C57BL6/129Sv; kind gift from R. Kempler, University of Freiburg, Germany). Ages of inducible mouse lines for each experiment are indicated in individual schemes shown in the figures.
No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. Mouse experiments performed were on sex-matched, age-matched and strain-matched pairs (usually littermates). The differences shown in figures were reproducible in both male and female pairs. Experiments were repeated on ≥3 pairs of sample sets (for example, Het versus cKO).
To make inducible TLE mice, we took E9.5 K14rtTA embryos (CD1/FVB/N), expressing a tetracycline-inducible transcriptional activator, and introduced a lentivirus harbouring a doxycycline-inducible Tle4 gene (TRE–Tle4) and a reporter (H2BRFP; see Supplementary Fig. 7b). Transduced embryos were born and grew to adults. When hair follicles were in second telogen (P50–56), doxycycline was administered. Real-time PCR on FACS-isolated RFP(+) HFSCs showed that doxycycline treatment efficiently elevated Tle4 messenger RNA levels. rtTA was activated by continuously feeding mice with doxycycline diet and intraperitoneal injection of doxycycline (0.5 mg per mouse). EdU (50 μg g−1) was injected intraperitoneally 24 h before lethal administration of CO2. Ultrasound-guided lentiviral injection and related procedures were performed as described previously52.
Chamber grafting and skin grafting
Chamber grafts were performed as described previously38. Cultured HFSCs (5 × 106) and freshly purified dermal fibroblasts (107) from newborn pups were injected into a silicon chamber implanted onto the back of nude mouse (Nu/Nu; from Charles River). Skin grafts were performed as described previously55. Skins from E18.5 K15CrePGR/Tcf7l1fl/fl;Tcf7l2−/−;Rosa–YFPfl/stop/fl embryos were grafted onto nude mice. Cre-recombinase was induced by intraperitoneal injection of 1% of RU486 in sesame oil and topical treatment of 4% RU486 in ethanol.
Antibody information
The following antibodies were used for immunostaining: TCF3 (1:100; Fuchs laboratory18), TCF4 (1:200; Cell Signaling Technology, 2569), LEF1 (1:1,000; Cell Signaling Technology, 2230), TCF1 (1:200; Cell Signaling Technology, 2203), CD34 (1:100; eBiosciences, 13-0341), SOX9 (1:1,000; Fuchs laboratory56), β-catenin (1:500; BD, clone 14), GFP (1:5,000; Abcam, ab13970), Keratin 5 (1:200; Covance, PRB-160P), Keratin 10 (1:200; Covance, PRB-159P), Integrin β4 (1:100; BD, 553745), PPARγ (1:100; Santa Cruz Biotechnology, H-100, sc-7196). The following antibodies were used for immunoprecipitation and immunoblotting: TCF3 (for immunoprecipitation, Fuchs laboratory18; for immunoblotting, 1:1,000; Santa Cruz Biotechnology, V-17, sc-12491), TCF4 (1:1,000; Santa Cruz Biotechnology, N-20, sc-8631), TLE (1:1,000; Santa Cruz Biotechnology, C-19, sc-13373), HDAC1 (1:1,000; Abcam, ab7028), β-catenin (1:2,000; Sigma, C7207), α-tubulin (1:2,000; Millipore, 05829). The following antibodies were used for FACS: integrin α6 (1:500; eBiosciences, PE-conjugated, 12-0495; PE-Cy7-conjugated, 25-0495), CD34 (1:100; eBiosciences, Alexa Fluor 660-conjugated, 50-0341), Sca1 (1:1,000; eBiosciences, PerCP-Cy5.5-conjugated, 45-5981).
ChIP-seq and data analyses
All materials, methods and sequencing for ChIP-seq have been described previously49. Independent immunoprecipitations were performed on FACS-sorted populations. For each ChIP-seq run, 107 cells were used. Antibodies used for ChIP-seq were TCF3 (Fuchs laboratory18); TCF4 (Santa Cruz Biotechnology, N-20); pan-TLE (Santa Cruz Biotechnology, C-19). ChIP-seq reads were aligned to the mouse genome (mm9, build 37) using the Bowtie aligner57. Peak overlap is defined as two peaks sharing at least one base pair along the entire spans of the two peaks. The MACS software51 was used for calling peaks with data from ChIP input as controls, while peak heights were computed with in-house software. Annotated mouse RefSeq genes with a peak at their promoter proximal (±2 kb of the transcription start site, TSS), promoter distal (−50 kb to −2 kb of the TSS), or gene body (+2 kb of the TSS to ±2 kb of transcription end site), were considered as targets. The MEME software suite58 was applied to 150-base-pair (bp) sequences around the ChIP-seq peak summits for enriched motifs, with the program MEME for motif discovery and MAST for motif scanning (P value <0.0005). ChIP-seq signal tracks were presented by IGB (Integrated Genome Browser) software. Gene ontology was analysed using Ingenuity IPA software. ChIP-seq raw data have been submitted to http://www.ncbi.nlm.nih.gov/geo. More details are provided in the Supplementary Methods.
To determine sequence motifs enriched in the peaks identified from TCF3/4 and TLE ChIP-seq analysis, we carried out a two-stage data analysis using the on-line motif analysis software MEME-ChIP (ref. 59). First, we applied the program MEME (ref. 58) to analyse 150-bp sequences around the summits of the top 600 TCF3/4 peaks for de novo motif discovery. Figure 5e and Supplementary Fig. 5d show the top three or two motifs from each of our data sets, indicating a significant enrichment of known LEF1–TCF- and TLE-binding sites in our called peaks. Next, we applied the position-specific scoring matrix for these motifs and scanned all of our called peaks (150-bp) to determine the percentages of peaks with a motif, using the program MAST within the MEME package (P < 0.0005). We used a stricter P value than the default setting (P < 0.001) because of the shorter length of TCF motifs. We also generated 50,000 random genomic regions as controls. Overall, the de novo motif discovery program MEME indicates that the enrichment is statistically significant as shown.
We obtained a total of 29,246,663 reads from TCF3 ChIP-seq, resulting in a total of 14,647 peaks. To assess whether we have identified most TCF3-bound sites, we randomly selected 5–28 million reads (1 million increment) from our total reads to call peaks at different sequencing depth; the result showed that we would have called ~90 and ~98% of the final peaks at the depth of 5–6 million and 10 million, indicating that we have reached sufficient sequencing coverage relative to the amount of immunoprecipitated DNAs. We also computed the fraction of reads in peaks (PRiP) as recommended by the ENCODE consortium for evaluating the success of an immunoprecipitation50, and obtained 4–10% PRiP for our TCF3/4 and TLE ChIP-seq data, supporting good quality of our ChIP experiments (>1% is considered good).
RNA-seq assay and analysis
RNAs from FACS-sorted populations were purified using a Directzol RNA MiniPrep kit (Zymo Research) per the manufacturer’s instructions. The quality of the RNA for sequencing was determined using an Agilent 2100 Bioanalyzer; all samples used had RNA integrity numbers (RIN) > 8. Library preparation using the Illumina TrueSeq mRNA sample preparation kit was performed at the Weill Cornell Medical College Genomic Core facility, and RNAs were sequenced on Illumina HiSeq 2000 machines. RNA-seq reads (51 bp, single-ended) were aligned to the mouse genome (NCBI37/mm9) using the software TopHat (version 2.0.4; ref. 60). The resulting alignment data from TopHat were then fed to an assembler, Cufflinks (version 2.1.1; ref. 61), to assemble aligned RNA-seq reads into transcripts. These transcripts were compared to the gene model for annotated transcripts obtained from the Ensembl database (72/GRCm38). The category of transcripts is described at http://vega.sanger.ac.uk/info/about/gene_and_transcript_types.html. Transcript abundances were measured in FPKM (fragments per kilobase of exon per million fragments mapped). Expression profiles were quantile-normalized across samples for comparison. Significant differences in gene expression were determined according to the following criteria: fold change ≥2 for Axin2–LacZ(+) versus (−) samples; fold change ≥1.75 for Ctnnb1 cKO versus Het samples, and FPKM in upregulated samples ≥1.
Florescence-activated cell sorting
FACS purification procedures were as previously described49. Bu-HFSCs were sorted on the basis of surface expression of integrin α6 and CD34 and/or cytosolic YFP or RFP. Cells expressing bacterial β-galactosidase were purified using the FluoReporter lacZ Flow Cytometry kit (Molecular Probes). Hair germ cells were purified on the basis of the expression of YFP and integrin α6, but negative for CD34 and Sca1 markers. Cells were collected into either Trizol LS reagent (Invitrogen) for RNA purification, or culture media for colony formation, or pre-coated 15-ml Flacon tubes for ChIP-seq and protein purification.
Cell culture and growth factor treatment
HFSCs were FACS-purified from the skins of mice; unless otherwise specified, isolations were performed 10 d after targeting ablation of the desired gene(s). Isolated HFSCs were cultured and expanded on mitomycin C-treated J2 3T3 mouse fibroblast feeders in medium containing 0.3 mM calcium as described previously38. For growth factor treatment, high passaged (~10 passages) HFSCs were taken off from feeders and starved with medium containing 0.1% of FBS for 16 h and then treated with desired growth factors for 6 h before collection for mRNA purification. Treatment conditions were: vehicle (PBS or dimethylsuphoxide), Wnt3a (R&D Systems, 100 ng ml−1), XAV939 (Selleckchem, 5 μM).
Colony-formation assay and statistical analysis
HFSCs were FACS-isolated and cultured with mitomycin C-treated J2 3T3 mouse fibroblast feeders in 6-well plate, and 100,000 sorted quiescent HFSCs were divided equally into three wells. After ~14 days, colonies were then fixed with 4% paraformaldehyde for 10 min and stained with 1% rhodamine B for 30 minutes. Colony numbers and sizes in 3 independent wells (n = 3) from one sample set were counted and measured for analysis and presented using GraphPad Prism 5. Similar results were reproduced in 2 independent sample sets. For long-term passaged colonies, 3 independent groups (5–8 colonies for each group) were monitored. Error bars were calculated from these 3 independent groups. Statistical analyses were determined by the unpaired two-tailed Student’s t-test. The 0.05 level of confidence (P < 0.05) was accepted for statistical significance.
Whole-mount preparation of adult skins, immunostaining and statistical analysis
To prepare whole-mount samples, we incubated mouse back skins in dispase (Gibco, 2 mg ml−1) with 20 mM EDTA at room temperature for 1–2 h and then at 4 °C for 2 days. Skin epidermis with attached hair follicles was then peeled off from dermis and fixed in 4% paraformaldehyde/PBS at room temperature for 30 min. Washed whole-mount skins could be kept at 4 °C or directly processed for immunostaining. Each representative image shown in figures was selected from ≥5 individual hair follicles in ≥2 independent experiments.
Y chromosome in situ hybridization was detected using a Cy3 Star*FISH detection kit (Cambio). The proliferation in hair follicles was determined by EdU incorporation assay. Incorporated EdU was detected using a Click-iT Imaging Kit (Invitrogen). For statistical analyses, percentages of hair follicles containing EdU+ bulges (Figs 4a and 7e and Supplementary Fig. 6c) or EdU+ hair germ cells (Fig. 7d and Supplementary Fig. 3a) or with bulge (Fig. 4d) were calculated among 5–12 individual hair follicles for each set of samples. Error bars were calculated from 3 individual sets, and similar results were reproduced in 2 independent experiments. For box and whisker plots, numbers of EdU+ Bu-HFSCs (Figs 4a and 7e and Supplementary Fig. 6c) or hair germ cells (Fig. 7d and Supplementary Fig. 3a) were counted from 15–38 hair follicles (combined from 3 individual sets described above). For transduced hair follicles (Fig. 8d), percentages of EdU+/RFP+ or EdU+/RFP-Bu-HFSCs were calculated among 5–6 transduced hair follicles for each set of samples. Error bars were calculated from 5 individual sets. Statistical analyses were determined by the unpaired two-tailed Student’s t-test. The 0.05 level of confidence (P < 0.05) was accepted for statistical significance.
Reverse transcription and real-time PCR confirmation
Total RNAs from FACS-sorted cells were purified with the RNeasy Micro Kit (Qiagen) or Direct-zol RNA MiniPrep kit (Zymo Research), and then reverse transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen). cDNAs were amplified with primers in SYBR Green qPCR assay on the Applied Biosystems 7900HT Fast Real-Time PCR system. Expression levels were normalized to PCR amplification with primers for Ppib2. Primers were designed using Primer 3 online software to amplify regions spanning ~100–120 nucleotides and encompassing exon/intron boundaries. Information on the primers used for mRNA expression and ChIP-seq validation is provided in individual Excel files, Supplementary Tables 2 and 3, respectively. Error bars for qPCR analyses were calculated from technical triplicates. Similar results for each experiment were reproduced independently ≥2 times in each indicated figure legend. Exclusion was applied only on the rare occasion that one of the three wells of the triplicate analyses was a significant outlier; however, for each gene, triplicate qPCRs were themselves performed at least ≥2 times in biological independent experiments, which validated the exclusion.
Immunoprecipitation assay
FACS-isolated HFSCs were pelleted and resuspended (3 × 106 cells 100 μl−1) in Pierce IP Lysis Buffer (Thermo Scientific) with proteinase inhibitors (Roche) and phosphatase inhibitors (Roche). Protein extracts were then incubated with Dynabeads Protein G (Invitrogen) and indicated antibodies at 4 °C overnight. Dynabeads with pulled-down protein complexes were washed with IP Lysis Buffer three times and proteins then released in 20 μl of 1× LDS sample buffer (Invitrogen) at 100 °C for 10 min. Input (whole protein extract) and immunoprecipitated samples were subjected for immunoblotting assay. The immunoprecipitation experiment with FACS-sorted HFSCs was repeated independently twice. All of immunoblots were fully scanned and the whole images are shown in Supplementary Fig. 8.
Accession numbers
The complete ChIP-seq and RNA-seq data were submitted to the NCBI GEO repository and are available as GSE48878.
Supplementary Material
Acknowledgments
We thank S. Dewell for assistance in high-throughput sequencing (RU Genomics Resource Center), X. Guo for help in ChIP-seq analysis (Zheng laboratory), and S. Mazel, L. Li, S. Semova and S. Tadesse for FACS sorting (RU FACS facility). We also thank Fuchs’ laboratory members N. Stokes, D. Oristian and A. Aldeguer for assistance in mouse research (Fuchs laboratory); A. Rodriguez-Folgueras for assistance in confocal microscopy; and T. Chen, A. Rodriguez-Folgueras, S. Luo and B. Keyes for discussions and comments. W-H.L. was supported by a Harvey L. Karp Postdoctoral Fellowship and a Jane Coffin Child Fellowship. E.F. is an HHMI Investigator. This work was supported by a grant (to E.F.) from the NIH/NIAMS (R01AR31737) and partially by NIH/NIMH (R21MH099452, R01MH073164, D.Z.).
Footnotes
Note Supplementary Information is available in the online version of the paper
AUTHOR CONTRIBUTIONS
W-H.L. and E.F. designed experiments, analysed data and wrote the paper. W-H.L. performed all experiments, collected all data and prepared the figures. L.P. carried out the grafting and lentiviral injection procedure. M.L. analysed RNA-seq data. D.Z. performed ChIP-seq and other bioinformatics analyses. K.L. assisted in performing some experiments during the revision. E.F. supervised the study.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at www.nature.com/reprints
References
- 1.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Willert K, Nusse R. β-catenin: a key mediator of Wnt signaling. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 3.Korinek V, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 4.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 5.Willert K, et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 6.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 7.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 8.Merrill BJ, Gat U, DasGupta R, Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001;15:1688–1705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wray J, et al. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyashenko N, et al. Differential requirement for the dual functions of β-catenin in embryonic stem cell self-renewal and germ layer formation. Nat Cell Biol. 2011;13:753–761. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi F, Pereira L, Merrill BJ. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells. 2008;26:1951–1960. doi: 10.1634/stemcells.2008-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ten Berge D, et al. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shy BR, et al. Regulation of Tcf7l1 DNA binding and protein stability as principal mechanisms of Wnt/β-catenin signaling. Cell Rep. 2013;4:1–9. doi: 10.1016/j.celrep.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi F, et al. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat Cell Biol. 2011;13:762–70. doi: 10.1038/ncb2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merrill BJ, et al. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- 16.Gat U, DasGupta R, Degenstein L, Fuchs E. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated β-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen H, et al. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat Genet. 2009;41:1068–1075. doi: 10.1038/ng.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 19.Wu CI, et al. Function of Wnt/β-catenin in counteracting Tcf3 repression through the Tcf3-β-catenin interaction. Development. 2012;139:2118–2129. doi: 10.1242/dev.076067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van de Wetering M, et al. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 21.Korinek V, et al. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 22.Angus-Hill ML, Elbert KM, Hidalgo J, Capecchi MR. T-cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc Natl Acad Sci USA. 2011;108:4914–4919. doi: 10.1073/pnas.1102300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang W, et al. A genome-wide RNAi screen for Wnt/β-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci USA. 2008;105:9697–9702. doi: 10.1073/pnas.0804709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman JA, Wu CI, Merrill BJ. Tcf7l1 prepares epiblast cells in the gastrulating mouse embryo for lineage specification. Development. 2013;140:1665–1675. doi: 10.1242/dev.087387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trompouki E, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verzi MP, et al. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc Natl Acad Sci USA. 2010;107:15157–15162. doi: 10.1073/pnas.1003822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boj SF, et al. Diabetes risk gene and Wnt effector Tcf7l2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell. 2012;151:1595–1607. doi: 10.1016/j.cell.2012.10.053. [DOI] [PubMed] [Google Scholar]
- 28.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavallo RA, et al. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 30.Brantjes H, Roose J, van De Wetering M, Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurlstone A, Clevers H. T-cell factors: turn-ons and turn-offs. EMBO J. 2002;21:2303–2311. doi: 10.1093/emboj/21.10.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 33.Liu C, et al. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 34.Lo MC, Gay F, Odom R, Shi Y, Lin R. Phosphorylation by the β-catenin/MAPK complex promotes 14-3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell. 2004;117:95–106. doi: 10.1016/s0092-8674(04)00203-x. [DOI] [PubMed] [Google Scholar]
- 35.Hikasa H, et al. Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Dev Cell. 2010;19:521–532. doi: 10.1016/j.devcel.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park MH, et al. Phosphorylation of β-catenin at serine 663 regulates its transcriptional activity. Biochem Biophys Res Commun. 2012;419:543–549. doi: 10.1016/j.bbrc.2012.02.056. [DOI] [PubMed] [Google Scholar]
- 37.Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Millar SE. Molecular mechanisms regulating hair follicle development. J Invest Dermatol. 2002;118:216–225. doi: 10.1046/j.0022-202x.2001.01670.x. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt-Ullrich R, Paus R. Molecular principles of hair follicle induction and morphogenesis. Bioessays. 2005;27:247–261. doi: 10.1002/bies.20184. [DOI] [PubMed] [Google Scholar]
- 41.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 42.Lustig B, et al. Negative feedback loop of Wnt signaling through upregulation of con-ductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greco V, et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowry WE, et al. Defining the impact of β-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19:1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Posthaus H, et al. β-Catenin is not required for proliferation and differentiation of epidermal mouse keratinocytes. J Cell Sci. 2002;115:4587–4595. doi: 10.1242/jcs.00141. [DOI] [PubMed] [Google Scholar]
- 46.Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen T, et al. An RNA interference screen uncovers a new molecule in stem cell self-renewal and long-term regeneration. Nature. 2012;485:104–108. doi: 10.1038/nature10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]
- 49.Lien WH, et al. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beronja S, Livshits G, Williams S, Fuchs E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat Med. 2010;16:821–827. doi: 10.1038/nm.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito M, et al. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 54.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaufman CK, et al. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–22. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–555. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.