Abstract
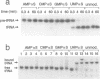
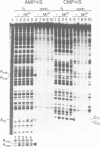
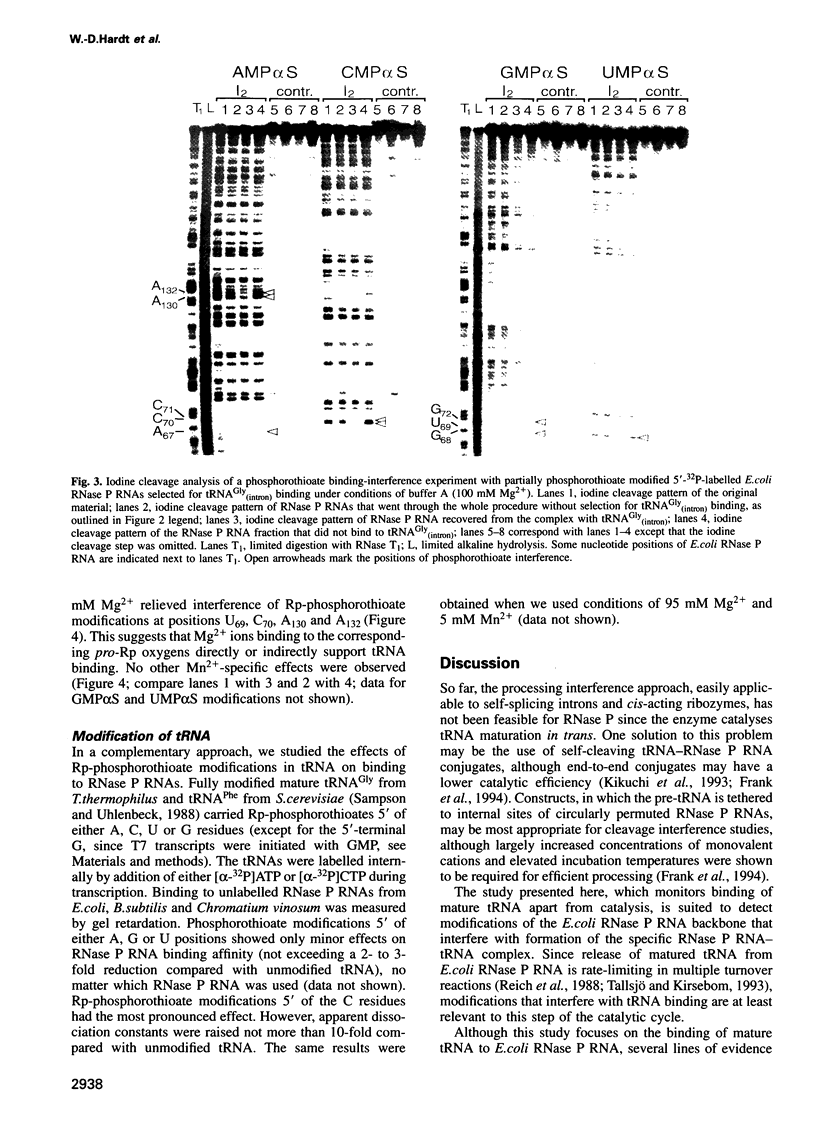
We have used Rp-phosphorothioate modifications and a binding interference assay to analyse the role of phosphate oxygens in tRNA recognition by Escherichia coli ribonuclease P (RNase P) RNA. Total (100%) Rp-phosphorothioate modification at A, C or G positions of RNase P RNA strongly impaired tRNA binding and pre-tRNA processing, while effects were less pronounced at U positions. Partially modified E. coli RNase P RNAs were separated into tRNA binding and non-binding fractions by gel retardation. Rp-phosphorothioate modifications that interfered with tRNA binding were found 5' of nucleotides A67, G68, U69, C70, C71, G72, A130, A132, A248, A249, G300, A317, A330, A352, C353 and C354. Manganese rescue at positions U69, C70, A130 and A132 identified, for the first time, sites of direct metal ion coordination in RNase P RNA. Most sites of interference are at strongly conserved nucleotides and nine reside within a long-range base-pairing interaction present in all known RNase P RNAs. In contrast to RNase P RNA, 100% Rp-phosphorothioate substitutions in tRNA showed only moderate effects on binding to RNase P RNAs from E. coli, Bacillus subtilis and Chromatium vinosum, suggesting that pro-Rp phosphate oxygens of mature tRNA contribute relatively little to the formation of the tRNA-RNase P RNA complex.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Kirsebom L., Talbot S. Recent studies of ribonuclease P. FASEB J. 1993 Jan;7(1):7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- Brown R. S., Dewan J. C., Klug A. Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry. 1985 Aug 27;24(18):4785–4801. doi: 10.1021/bi00339a012. [DOI] [PubMed] [Google Scholar]
- Burgin A. B., Pace N. R. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 1990 Dec;9(12):4111–4118. doi: 10.1002/j.1460-2075.1990.tb07633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian E. L., Yarus M. Analysis of the role of phosphate oxygens in the group I intron from Tetrahymena. J Mol Biol. 1992 Dec 5;228(3):743–758. doi: 10.1016/0022-2836(92)90861-d. [DOI] [PubMed] [Google Scholar]
- Christian E. L., Yarus M. Metal coordination sites that contribute to structure and catalysis in the group I intron from Tetrahymena. Biochemistry. 1993 May 4;32(17):4475–4480. doi: 10.1021/bi00068a001. [DOI] [PubMed] [Google Scholar]
- Ciesiolka J., Hardt W. D., Schlegl J., Erdmann V. A., Hartmann R. K. Lead-ion-induced cleavage of RNase P RNA. Eur J Biochem. 1994 Jan 15;219(1-2):49–56. doi: 10.1111/j.1432-1033.1994.tb19913.x. [DOI] [PubMed] [Google Scholar]
- Darr S. C., Zito K., Smith D., Pace N. R. Contributions of phylogenetically variable structural elements to the function of the ribozyme ribonuclease P. Biochemistry. 1992 Jan 21;31(2):328–333. doi: 10.1021/bi00117a003. [DOI] [PubMed] [Google Scholar]
- Drainas D., Zimmerly S., Willis I., Söll D. Substrate structural requirements of Schizosaccharomyces pombe RNase P. FEBS Lett. 1989 Jul 17;251(1-2):84–88. doi: 10.1016/0014-5793(89)81433-4. [DOI] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Altman S. External guide sequences for an RNA enzyme. Science. 1990 Aug 17;249(4970):783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990 Aug 10;62(3):407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- Frank D. N., Harris M. E., Pace N. R. Rational design of self-cleaving pre-tRNA-ribonuclease P RNA conjugates. Biochemistry. 1994 Sep 6;33(35):10800–10808. doi: 10.1021/bi00201a030. [DOI] [PubMed] [Google Scholar]
- Frey P. A., Sammons R. D. Bond order and charge localization in nucleoside phosphorothioates. Science. 1985 May 3;228(4699):541–545. doi: 10.1126/science.2984773. [DOI] [PubMed] [Google Scholar]
- Gaur R. K., Krupp G. Modification interference approach to detect ribose moieties important for the optimal activity of a ribozyme. Nucleic Acids Res. 1993 Jan 11;21(1):21–26. doi: 10.1093/nar/21.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. J., Vold B. S. Structural requirements for processing of synthetic tRNAHis precursors by the catalytic RNA component of RNase P. J Biol Chem. 1988 Jan 15;263(2):652–657. [PubMed] [Google Scholar]
- Griffiths A. D., Potter B. V., Eperon I. C. Stereospecificity of nucleases towards phosphorothioate-substituted RNA: stereochemistry of transcription by T7 RNA polymerase. Nucleic Acids Res. 1987 May 26;15(10):4145–4162. doi: 10.1093/nar/15.10.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Lumelsky N., Altman S. Specific interactions in RNA enzyme-substrate complexes. Science. 1989 Dec 22;246(4937):1578–1584. doi: 10.1126/science.2480641. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., McClain W. H., Altman S. Cleavage of tRNA precursors by the RNA subunit of E. coli ribonuclease P (M1 RNA) is influenced by 3'-proximal CCA in the substrates. Cell. 1984 Aug;38(1):219–224. doi: 10.1016/0092-8674(84)90543-9. [DOI] [PubMed] [Google Scholar]
- Haas E. S., Brown J. W., Pitulle C., Pace N. R. Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2527–2531. doi: 10.1073/pnas.91.7.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt W. D., Schlegl J., Erdmann V. A., Hartmann R. K. Gel retardation analysis of E. coli M1 RNA-tRNA complexes. Nucleic Acids Res. 1993 Jul 25;21(15):3521–3527. doi: 10.1093/nar/21.15.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt W. D., Schlegl J., Erdmann V. A., Hartmann R. K. Kinetics and thermodynamics of the RNase P RNA cleavage reaction: analysis of tRNA 3'-end variants. J Mol Biol. 1995 Mar 24;247(2):161–172. doi: 10.1006/jmbi.1994.0130. [DOI] [PubMed] [Google Scholar]
- Hardt W. D., Schlegl J., Erdmann V. A., Hartmann R. K. Role of the D arm and the anticodon arm in tRNA recognition by eubacterial and eukaryotic RNase P enzymes. Biochemistry. 1993 Dec 7;32(48):13046–13053. doi: 10.1021/bi00211a014. [DOI] [PubMed] [Google Scholar]
- Harris M. E., Nolan J. M., Malhotra A., Brown J. W., Harvey S. C., Pace N. R. Use of photoaffinity crosslinking and molecular modeling to analyze the global architecture of ribonuclease P RNA. EMBO J. 1994 Sep 1;13(17):3953–3963. doi: 10.1002/j.1460-2075.1994.tb06711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Church G. M., Kim S. H. RNA-ligant interactions. (I) Magnesium binding sites in yeast tRNAPhe. Nucleic Acids Res. 1977 Aug;4(8):2811–2820. doi: 10.1093/nar/4.8.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Rhodes D., Brown R. S., Klug A. A crystallographic study of metal-binding to yeast phenylalanine transfer RNA. J Mol Biol. 1977 Apr 15;111(3):315–328. doi: 10.1016/s0022-2836(77)80054-5. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Michel F., Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J Mol Biol. 1994 Mar 11;236(5):1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Jaffe E. K., Cohn M. Diastereomers of the nucleoside phosphorothioates as probes of the structure of the metal nucleotide substrates and of the nucleotide binding site of yeast hexokinase. J Biol Chem. 1979 Nov 10;254(21):10839–10845. [PubMed] [Google Scholar]
- Jeoung Y. H., Kumar P. K., Suh Y. A., Taira K., Nishikawa S. Identification of phosphate oxygens that are important for self-cleavage activity of the HDV ribozyme by phosphorothioate substitution interference analysis. Nucleic Acids Res. 1994 Sep 11;22(18):3722–3727. doi: 10.1093/nar/22.18.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle D., Küst B., Krupp G. Phosphorothioates in pre-tRNAs can change the specificities of RNAses P or reduce the cleavage efficiencies. Biochimie. 1993;75(11):955–962. doi: 10.1016/0300-9084(93)90145-i. [DOI] [PubMed] [Google Scholar]
- Kahle D., Wehmeyer U., Krupp G. Substrate recognition by RNase P and by the catalytic M1 RNA: identification of possible contact points in pre-tRNAs. EMBO J. 1990 Jun;9(6):1929–1937. doi: 10.1002/j.1460-2075.1990.tb08320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakov S., Altman S. Site-specific cleavage by metal ion cofactors and inhibitors of M1 RNA, the catalytic subunit of RNase P from Escherichia coli. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9193–9197. doi: 10.1073/pnas.88.20.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Sasaki-Tozawa N., Suzuki K. Artificial self-cleaving molecules consisting of a tRNA precursor and the catalytic RNA of RNase P. Nucleic Acids Res. 1993 Oct 11;21(20):4685–4689. doi: 10.1093/nar/21.20.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom L. A., Svärd S. G. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994 Oct 17;13(20):4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom L. A., Svärd S. G. Identification of a region within M1 RNA of Escherichia coli RNase P important for the location of the cleavage site on a wild-type tRNA precursor. J Mol Biol. 1993 Jun 5;231(3):594–604. doi: 10.1006/jmbi.1993.1312. [DOI] [PubMed] [Google Scholar]
- Knap A. K., Wesolowski D., Altman S. Protection from chemical modification of nucleotides in complexes of M1 RNA, the catalytic subunit of RNase P from E coli, and tRNA precursors. Biochimie. 1990 Nov;72(11):779–790. doi: 10.1016/0300-9084(90)90187-l. [DOI] [PubMed] [Google Scholar]
- LaGrandeur T. E., Hüttenhofer A., Noller H. F., Pace N. R. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J. 1994 Sep 1;13(17):3945–3952. doi: 10.1002/j.1460-2075.1994.tb06710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N., DaLio A., Strobel M., Engelke D. Effects of tRNA-intron structure on cleavage of precursor tRNAs by RNase P from Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Mar 25;16(6):2537–2552. doi: 10.1093/nar/16.6.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J. G., Svärd S. G., Kirsebom L. A. Characterization of the Borrelia burgdorferi RNase P RNA gene reveals a novel tertiary interaction. J Mol Biol. 1994 Aug 5;241(1):1–6. doi: 10.1006/jmbi.1994.1467. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Murphy F. L., Cech T. R. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J Mol Biol. 1994 Feb 11;236(1):49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- Nichols M., Söll D., Willis I. Yeast RNase P: catalytic activity and substrate binding are separate functions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1379–1383. doi: 10.1073/pnas.85.5.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan J. M., Burke D. H., Pace N. R. Circularly permuted tRNAs as specific photoaffinity probes of ribonuclease P RNA structure. Science. 1993 Aug 6;261(5122):762–765. doi: 10.1126/science.7688143. [DOI] [PubMed] [Google Scholar]
- Oh B. K., Pace N. R. Interaction of the 3'-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 1994 Oct 11;22(20):4087–4094. doi: 10.1093/nar/22.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecoraro V. L., Hermes J. D., Cleland W. W. Stability constants of Mg2+ and Cd2+ complexes of adenine nucleotides and thionucleotides and rate constants for formation and dissociation of MgATP and MgADP. Biochemistry. 1984 Oct 23;23(22):5262–5271. doi: 10.1021/bi00317a026. [DOI] [PubMed] [Google Scholar]
- Pley H. W., Flaherty K. M., McKay D. B. Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature. 1994 Nov 3;372(6501):111–113. doi: 10.1038/372111a0. [DOI] [PubMed] [Google Scholar]
- Reich C., Olsen G. J., Pace B., Pace N. R. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science. 1988 Jan 8;239(4836):178–181. doi: 10.1126/science.3122322. [DOI] [PubMed] [Google Scholar]
- Rubin J. R., Sundaralingam M. Lead ion binding and RNA chain hydrolysis in phenylalanine tRNA. J Biomol Struct Dyn. 1983 Dec;1(3):639–646. doi: 10.1080/07391102.1983.10507471. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegl J., Fürste J. P., Bald R., Erdmann V. A., Hartmann R. K. Cleavage efficiencies of model substrates for ribonuclease P from Escherichia coli and Thermus thermophilus. Nucleic Acids Res. 1992 Nov 25;20(22):5963–5970. doi: 10.1093/nar/20.22.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegl J., Hardt W. D., Erdmann V. A., Hartmann R. K. Contribution of structural elements to Thermus thermophilus ribonuclease P RNA function. EMBO J. 1994 Oct 17;13(20):4863–4869. doi: 10.1002/j.1460-2075.1994.tb06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. E., Bennett J. L., Dairaghi D. J., Clayton D. A. Secondary structure of RNase MRP RNA as predicted by phylogenetic comparison. FASEB J. 1993 Jan;7(1):208–213. doi: 10.1096/fasebj.7.1.7678563. [DOI] [PubMed] [Google Scholar]
- Smith D., Burgin A. B., Haas E. S., Pace N. R. Influence of metal ions on the ribonuclease P reaction. Distinguishing substrate binding from catalysis. J Biol Chem. 1992 Feb 5;267(4):2429–2436. [PubMed] [Google Scholar]
- Svärd S. G., Kirsebom L. A. Determinants of Escherichia coli RNase P cleavage site selection: a detailed in vitro and in vivo analysis. Nucleic Acids Res. 1993 Feb 11;21(3):427–434. doi: 10.1093/nar/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svärd S. G., Kirsebom L. A. Several regions of a tRNA precursor determine the Escherichia coli RNase P cleavage site. J Mol Biol. 1992 Oct 20;227(4):1019–1031. doi: 10.1016/0022-2836(92)90518-o. [DOI] [PubMed] [Google Scholar]
- Tallsjö A., Kirsebom L. A. Product release is a rate-limiting step during cleavage by the catalytic RNA subunit of Escherichia coli RNase P. Nucleic Acids Res. 1993 Jan 11;21(1):51–57. doi: 10.1093/nar/21.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N. K., Schaff S., Thill G., Petit-Koskas E., Crain-Denoyelle A. M., Westhof E. A three-dimensional model of hepatitis delta virus ribozyme based on biochemical and mutational analyses. Curr Biol. 1994 Jun 1;4(6):488–498. doi: 10.1016/s0960-9822(00)00109-3. [DOI] [PubMed] [Google Scholar]
- Thurlow D. L., Shilowski D., Marsh T. L. Nucleotides in precursor tRNAs that are required intact for catalysis by RNase P RNAs. Nucleic Acids Res. 1991 Feb 25;19(4):885–891. doi: 10.1093/nar/19.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranguch A. J., Engelke D. R. Comparative structural analysis of nuclear RNase P RNAs from yeast. J Biol Chem. 1993 Jul 5;268(19):14045–14055. [PubMed] [Google Scholar]
- Wang M. J., Davis N. W., Gegenheimer P. Novel mechanisms for maturation of chloroplast transfer RNA precursors. EMBO J. 1988 Jun;7(6):1567–1574. doi: 10.1002/j.1460-2075.1988.tb02981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Altman S. Three-dimensional working model of M1 RNA, the catalytic RNA subunit of ribonuclease P from Escherichia coli. Proc Natl Acad Sci U S A. 1994 May 24;91(11):5133–5137. doi: 10.1073/pnas.91.11.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Winker S., Gutell R. R. Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc Natl Acad Sci U S A. 1990 Nov;87(21):8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Altman S. Substrate recognition by human RNase P: identification of small, model substrates for the enzyme. EMBO J. 1995 Jan 3;14(1):159–168. doi: 10.1002/j.1460-2075.1995.tb06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Hwang E. S., Altman S. Targeted cleavage of mRNA by human RNase P. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8006–8010. doi: 10.1073/pnas.89.17.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinkar P. P., Williamson J. R. Kinetic intermediates in RNA folding. Science. 1994 Aug 12;265(5174):918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- Zito K., Hüttenhofer A., Pace N. R. Lead-catalyzed cleavage of ribonuclease P RNA as a probe for integrity of tertiary structure. Nucleic Acids Res. 1993 Dec 25;21(25):5916–5920. doi: 10.1093/nar/21.25.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]