Abstract
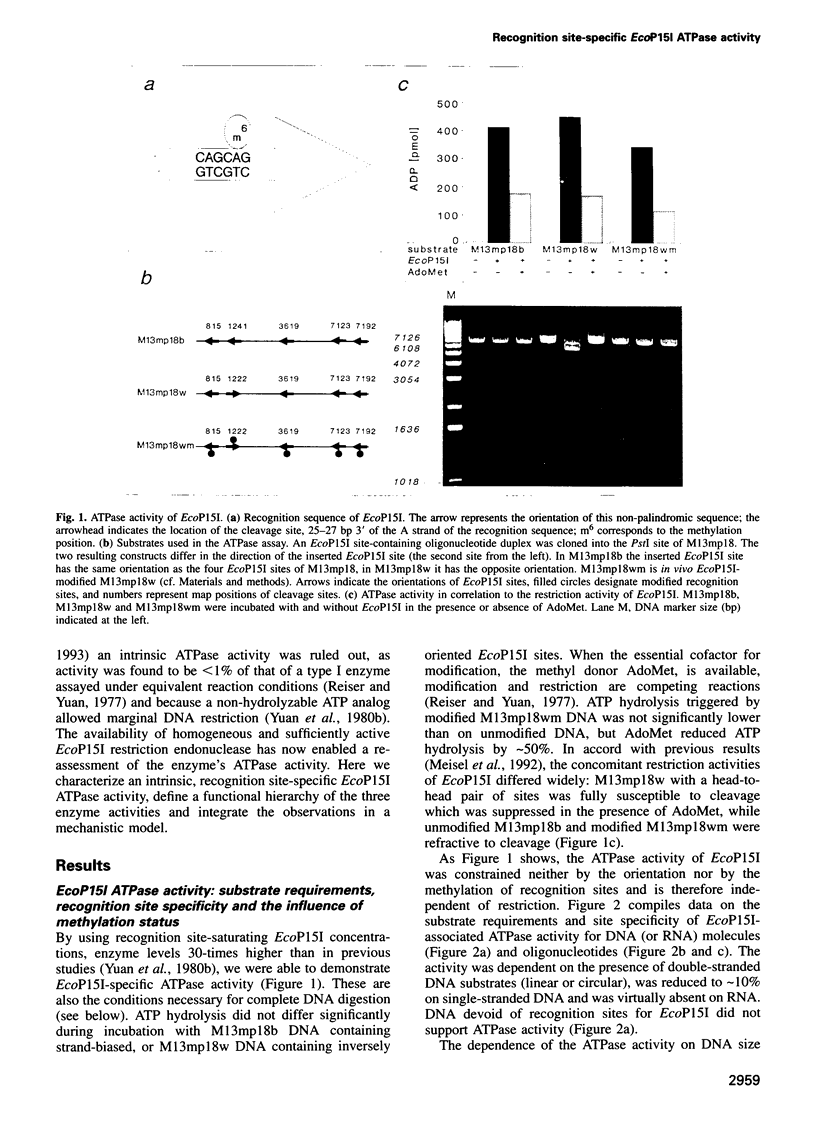
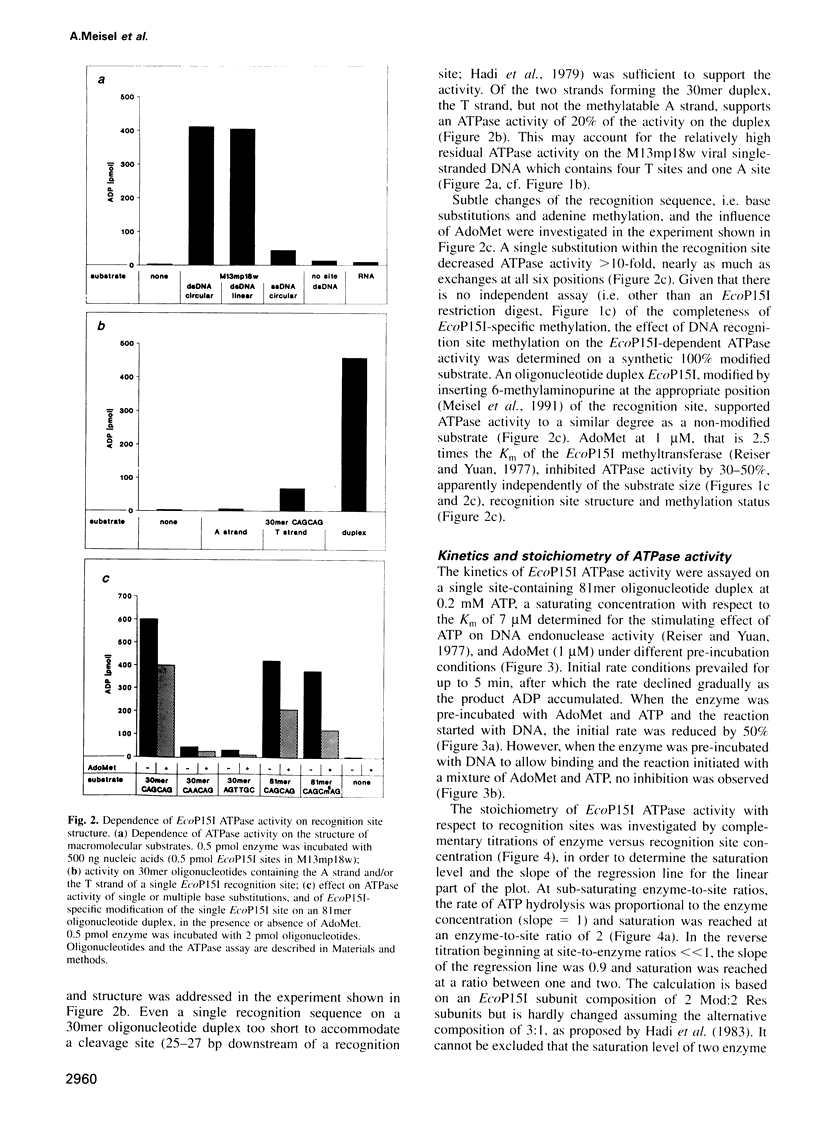
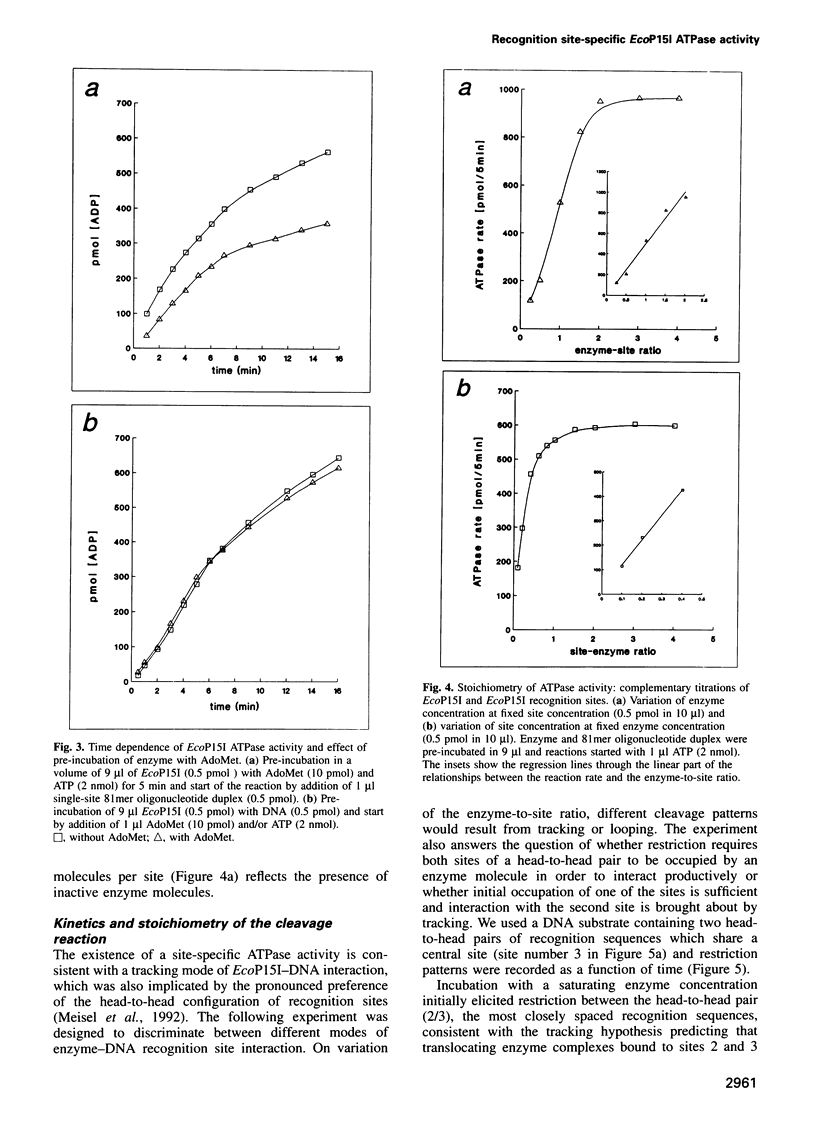
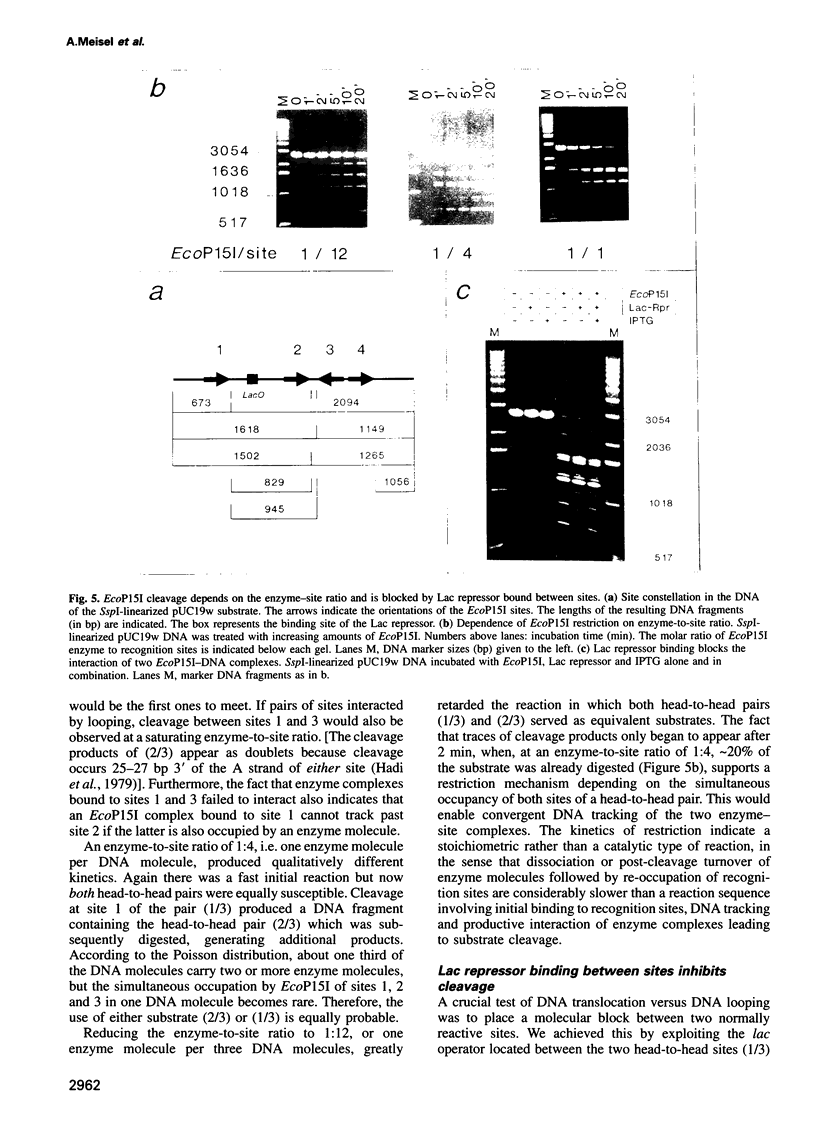
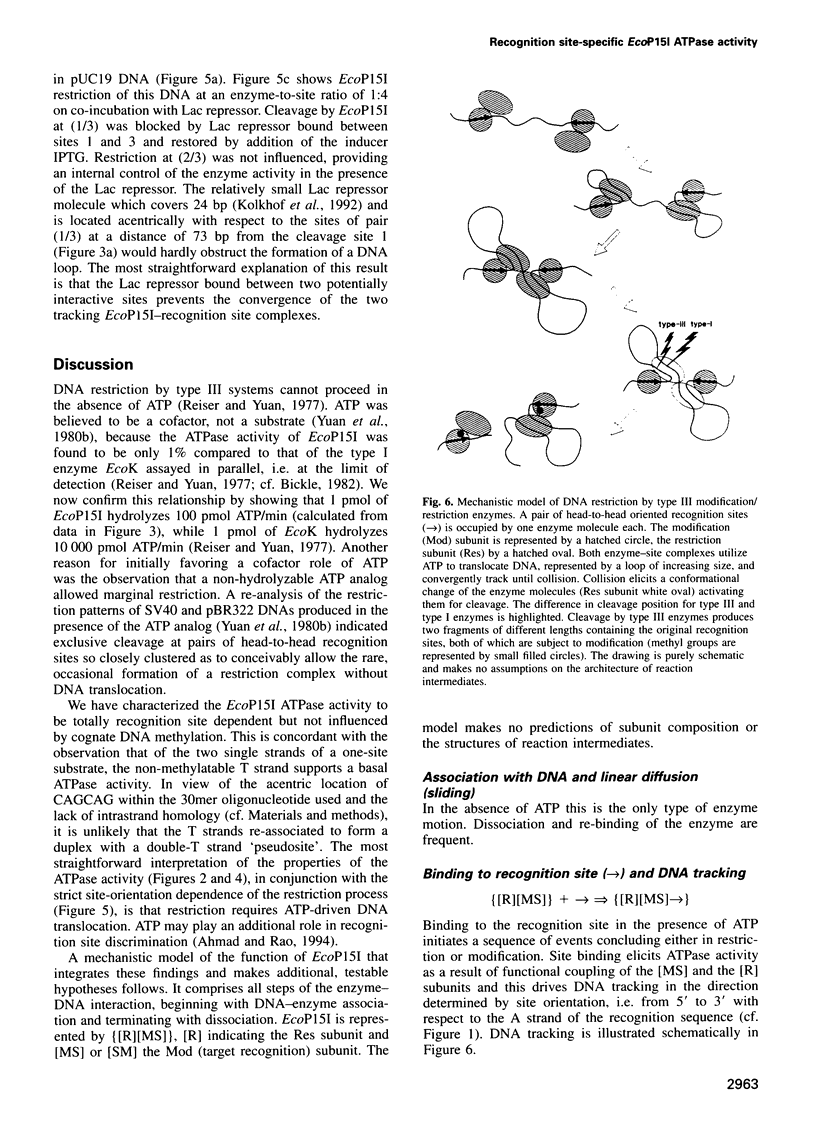
Type III restriction/modification systems recognize short non-palindromic sequences, only one strand of which can be methylated. Replication of type III-modified DNA produces completely unmethylated recognition sites which, according to classical mechanisms of restriction, should be signals for restriction. We have shown previously that suicidal restriction by the type III enzyme EcoP15I is prevented if all the unmodified sites are in the same orientation: restriction by EcoP15I requires a pair of unmethylated, inversely oriented recognition sites. We have now addressed the molecular mechanism of site orientation-specific DNA restriction. EcoP15I is demonstrated to possess an intrinsic ATPase activity, the potential driving force of DNA translocation. The ATPase activity is uniquely recognition site-specific, but EcoP15I-modified sites also support the reaction. EcoP15I DNA restriction patterns are shown to be predetermined by the enzyme-to-site ratio, in that site-saturating enzyme levels elicit cleavage exclusively between the closest pair of head-to-head oriented sites. DNA restriction is blocked by Lac repressor bound in the intervening sequence between the two EcoP15I sites. These results rule out DNA looping and strongly suggest that cleavage is triggered by the close proximity of two convergently tracking EcoP15I-DNA complexes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad I., Rao D. N. Interaction of EcoP15I DNA methyltransferase with oligonucleotides containing the asymmetric sequence 5'-CAGCAG-3'. J Mol Biol. 1994 Sep 30;242(4):378–388. doi: 10.1006/jmbi.1994.1588. [DOI] [PubMed] [Google Scholar]
- BEAVEN G. H., HOLIDAY E. R. Ultraviolet absorption spectra of proteins and amino acids. Adv Protein Chem. 1952;7:319–386. doi: 10.1016/s0065-3233(08)60022-4. [DOI] [PubMed] [Google Scholar]
- Bickle T. A., Krüger D. H. Biology of DNA restriction. Microbiol Rev. 1993 Jun;57(2):434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dartois V., De Backer O., Colson C. Sequence of the Salmonella typhimurium StyLT1 restriction-modification genes: homologies with EcoP1 and EcoP15 type-III R-M systems and presence of helicase domains. Gene. 1993 May 15;127(1):105–110. doi: 10.1016/0378-1119(93)90623-b. [DOI] [PubMed] [Google Scholar]
- Dröge P. Protein tracking-induced supercoiling of DNA: a tool to regulate DNA transactions in vivo? Bioessays. 1994 Feb;16(2):91–99. doi: 10.1002/bies.950160205. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V. Endonuclease (R) subunits of type-I and type-III restriction-modification enzymes contain a helicase-like domain. FEBS Lett. 1991 Oct 21;291(2):277–281. doi: 10.1016/0014-5793(91)81301-n. [DOI] [PubMed] [Google Scholar]
- Haberman A. The bacteriophage P1 restriction endonuclease. J Mol Biol. 1974 Nov 15;89(4):545–563. doi: 10.1016/0022-2836(74)90035-7. [DOI] [PubMed] [Google Scholar]
- Hadi S. M., Bächi B., Iida S., Bickle T. A. DNA restriction--modification enzymes of phage P1 and plasmid p15B. Subunit functions and structural homologies. J Mol Biol. 1983 Mar 25;165(1):19–34. doi: 10.1016/s0022-2836(83)80240-x. [DOI] [PubMed] [Google Scholar]
- Hadi S. M., Bächi B., Shepherd J. C., Yuan R., Ineichen K., Bickle T. A. DNA recognition and cleavage by the EcoP15 restriction endonuclease. J Mol Biol. 1979 Nov 5;134(3):655–666. doi: 10.1016/0022-2836(79)90372-3. [DOI] [PubMed] [Google Scholar]
- Heitman J. On the origins, structures and functions of restriction-modification enzymes. Genet Eng (N Y) 1993;15:57–108. doi: 10.1007/978-1-4899-1666-2_4. [DOI] [PubMed] [Google Scholar]
- Kolkhof P., Teichmann D., Kisters-Woike B., von Wilcken-Bergmann B., Müller-Hill B. Lac repressor with the helix-turn-helix motif of lambda cro binds to lac operator. EMBO J. 1992 Aug;11(8):3031–3038. doi: 10.1002/j.1460-2075.1992.tb05373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel A., Bickle T. A., Krüger D. H., Schroeder C. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature. 1992 Jan 30;355(6359):467–469. doi: 10.1038/355467a0. [DOI] [PubMed] [Google Scholar]
- Meisel A., Krüger D. H., Bickle T. A. M.EcoP15 methylates the second adenine in its recognition sequence. Nucleic Acids Res. 1991 Jul 25;19(14):3997–3997. doi: 10.1093/nar/19.14.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Daniel A. S., Cowan G. M., Sharp P. M. Conservation of motifs within the unusually variable polypeptide sequences of type I restriction and modification enzymes. Mol Microbiol. 1993 Jul;9(1):133–143. doi: 10.1111/j.1365-2958.1993.tb01675.x. [DOI] [PubMed] [Google Scholar]
- Rao D. N., Page M. G., Bickle T. A. Cloning, over-expression and the catalytic properties of the EcoP15 modification methylase from Escherichia coli. J Mol Biol. 1989 Oct 20;209(4):599–606. doi: 10.1016/0022-2836(89)90597-4. [DOI] [PubMed] [Google Scholar]
- Reiser J., Yuan R. Purification and properties of the P15 specific restriction endonuclease from Escherichia coli. J Biol Chem. 1977 Jan 25;252(2):451–456. [PubMed] [Google Scholar]
- Rosamond J., Endlich B., Linn S. Electron microscopic studies of the mechanism of action of the restriction endonuclease of Escherichia coli B. J Mol Biol. 1979 Apr 25;129(4):619–635. doi: 10.1016/0022-2836(79)90472-8. [DOI] [PubMed] [Google Scholar]
- Schroeder C., Jurkschat H., Meisel A., Reich J. G., Krüger D. Unusual occurrence of EcoP1 and EcoP15 recognition sites and counterselection of type II methylation and restriction sequences in bacteriophage T7 DNA. Gene. 1986;45(1):77–86. doi: 10.1016/0378-1119(86)90134-4. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Bandyopadhyay P. K. Model for how type I restriction enzymes select cleavage sites in DNA. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Giaever G. N. Action at a distance along a DNA. Science. 1988 Apr 15;240(4850):300–304. doi: 10.1126/science.3281259. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Young M. C., Kuhl S. B., von Hippel P. H. Kinetic theory of ATP-driven translocases on one-dimensional polymer lattices. J Mol Biol. 1994 Feb 4;235(5):1436–1446. doi: 10.1006/jmbi.1994.1099. [DOI] [PubMed] [Google Scholar]
- Yuan R., Hamilton D. L., Burckhardt J. DNA translocation by the restriction enzyme from E. coli K. Cell. 1980 May;20(1):237–244. doi: 10.1016/0092-8674(80)90251-2. [DOI] [PubMed] [Google Scholar]
- Yuan R., Hamilton D. L., Hadi S. M., Bickle T. A. Role of ATP in the cleavage mechanism of the EcoP15 restriction endonuclease. J Mol Biol. 1980 Dec 25;144(4):501–519. doi: 10.1016/0022-2836(80)90334-4. [DOI] [PubMed] [Google Scholar]
- Yuan R., Reiser J. Steps in the reaction mechanism of the Escherichia coli plasmid P15-specific restriction endonuclease. J Mol Biol. 1978 Jul 15;122(4):433–445. doi: 10.1016/0022-2836(78)90420-5. [DOI] [PubMed] [Google Scholar]