Abstract
Single locus variants (SLVs) are bacterial sequence types that differ at only one of the seven canonical multilocus sequence typing (MLST) loci. Estimating the relative roles of recombination and point mutation in the generation of new alleles that lead to SLVs is helpful in understanding how organisms evolve. The relative rates of recombination and mutation for Campylobacter jejuni and Campylobacter coli were estimated at seven different housekeeping loci from publically available MLST data. The probability of recombination generating a new allele that leads to an SLV is estimated to be roughly seven times more than that of mutation for C. jejuni, but for C. coli recombination and mutation were estimated to have a similar contribution to the generation of SLVs. The majority of nucleotide differences (98 % for C. jejuni and 85 % for C. coli) between strains that make up an SLV are attributable to recombination. These estimates are much larger than estimates of the relative rate of recombination to mutation calculated from more distantly related isolates using MLST data. One explanation for this is that purifying selection plays an important role in the evolution of Campylobacter. A simulation study was performed to test the performance of our method under a range of biologically realistic parameters. We found that our method performed well when the recombination tract length was longer than 3 kb. For situations in which recombination may occur with shorter tract lengths, our estimates are likely to be an underestimate of the ratio of recombination to mutation, and of the importance of recombination for creating diversity in closely related isolates. A parametric bootstrap method was applied to calculate the uncertainty of these estimates.
Keywords: Single locus variant, MLST, Purifying, selection, Evolution of Campylobacter, Mutation, Recombination
Introduction
The genus Campylobacter is the major cause of gastroenteritis in many industrialized countries (Tauxe et al. 1992), with approximately 1 % of the population throughout the western world being affected by campylobacteriosis every year (The World Health Organization, cited in Humphrey et al. 2007). The species Campylobacter jejuni (C. jejuni) and Campylobacter coli (C. coli) are the main causes of bacterial food-borne disease in developed countries, compared to other members of the family Campylobacteriaceae (Konkel et al. 1999).
Substantial evidence for the presence of recombination at specific genes has been found in several studies (Suerbaum et al. 2001; Fearnhead et al. 2005). The relative contributions of recombination and point mutation to genetic diversity have also been investigated (Feil et al. 1999, 2000,2001; Sarkar and Guttman 2004). Although most research indicates that recombination contributes more to genetic diversity than mutation, there is considerable uncertainty about the relative number of events and the number of nucleotide differences that may be attributable to these two processes (Schouls et al. 2003; Richman et al. 2003; Fearnhead et al. 2005). This paper is focused on estimating the relative contributions of recombination and point mutation to the generation of new alleles that lead to single locus variants (SLVs), based on C. jejuni and C. coli from the seven gene multilocus sequence typing (MLST) scheme.
An SLV is a pair of sequence types (STs) that differ at exactly one of the seven alleles that make up the MLST profile (Feil et al. 2004). SLVs are pairs of STs that most likely share a very recent common ancestor and the analysis of SLVs can be helpful in understanding the evolution and molecular epidemiology of pathogens. The large collections of isolates that have been characterized by MLST provide a good opportunity to study SLVs in detail.
This research is based on distinct STs of C. jejuni and C. coli in the PubMLST database (http://pubmlst.org/campylobacter). In order to understand whether there are differences in the mechanisms that produce SLVs across the genome, SLVs were divided into groups depending on the locus at which the STs differ. The distribution of nucleotide differences within SLVs was explored. The nucleotide differences between two STs that form an SLV can be generated by two different kinds of events: recombination or mutation. Intuitively, SLVs that comprise two STs which differ at many nucleotide positions are more likely to be due to recombination, whereas those that differ at only a few nucleotide positions may be the result of point mutations. In this study, an EM algorithm was applied to allocate SLVs into either a point mutation only model or a recombination model. Two key parameters were estimated: the probability that an SLV arose due to point mutation(s) only, and the relative rate of recombination to mutation. In order to test the performance of our method, a simulation study was performed under a range of biologically realistic parameters. When the recombination tract length was longer than 3 kb our method performed well. Three kilobase pairs is the average of the estimated mean tract lengths suggested by previous research on Campylobacter (Schouls et al. 2003; Fearnhead et al. 2005; Wilson et al.2009; Biggs et al. 2011). When recombination occurs with shorter tract lengths, our estimates may underestimate the ratio of recombination to mutation.
Materials and Methods
Campylobacter Data
The data were taken from the PubMLST database (September 27, 2010); at that time the PubMLST database contained 4676 distinct C. jejuni and C. coli STs. MLST is a way of typing strains that is based on nucleotide sequences (Maiden et al. 1998). Using the MLST technique (Dingle et al. 2001), these isolates are sequenced at seven housekeeping loci (aspA, glnA, gltA, glyA, pgm, tkt, and uncA). These seven loci are widely dispersed around the genome, which means there is a very low chance for one recombination to change two or more loci.
We separated ST datasets for C. jejuni and C. coli, and excluded the 22 STs found in both species. Furthermore, we separated C. coli by clades according to previous research (Sheppard et al. 2008, 2011), and we chose C. coli clade 1 to investigate in detail because C. coli clade 1 contains more STs, and is more diverse, compared to the other two clades (Sheppard et al. 2008). We selected clade 1 from C. coli by extracting all STs that are members of ST-828 clonal complex and ST-1150 clonal complex (Sheppard et al. 2010). There are 3654 STs for C. jejuni, and 606 distinct STs for C. coli clade1.
Methods Overview
Either mutation(s) or recombination(s) can generate SLVs. In this paper, mutation is defined as a single nucleotide change (a point mutation), whereas recombination represents the transfer of several adjacent nucleotides from one DNA source to another. An event is either a mutation or a recombination. An SLV can be generated by one or more events; however, recombination will tend to mask mutation. We model separately the mutation and recombination process to derive a probability model for the number of nucleotide differences between STs, under both the assumption that the SLV has been created solely by mutation, and that it has not. This then enables us to estimate the proportion of SLVs that have been caused solely by mutation, and also estimate the relative rate of recombination to mutation. More details of the analysis are given in the Supplementary Material.
Modeling SLV Evolution
The data consists of, for each SLV, the locus at which the pair of STs differ, and the number of nucleotide differences at that locus. From this, we aim to infer how likely it is that the differences observed at this locus arise from point mutation only, as opposed to being produced by recombination.
To do this, we first model the distribution of nucleotide differences we would expect at an SLV at a given locus if these differences are solely due to mutation. This can be done by first calculating the probability of an SLV given the number of point of mutations that have occurred in one locus as the likelihood function, introducing a prior distribution for the number of mutations to occur between two STs in that locus. The former probability is based on the need for all mutation events to occur at the same locus. Under the coalescent theory, a geometric distribution is chosen to use as the prior distribution (Hein et al. 2005). Under Bayesian theory, we can obtain the required conditional distribution (Equation 1 in Supplementary Material). The resulting conditional distribution of the number of nucleotide differences is concentrated on small numbers of nucleotide differences, and is robust to the choice of prior.
Second, the probability of observing h (h = 1, 2, 3…) nucleotide differences introduced by recombination was estimated using Bayesian methods. It was calculated by sampling the alleles based on their frequencies in the current database. Two (simplifying) assumptions for the recombination model were made: (1) if recombination occurs between two alleles it affects an entire locus rather than just part of a locus; and (2) we ignore the effect of any additional mutation events. Under these assumptions, our model suggests that in most cases recombination will introduce many more nucleotide differences than expected under the mutation only model. Note that our results are robust to the assumption in (1) unless recombination affects only small fragments of a locus, in these cases our assumption will tend to lead to overestimates of the proportion of SLVs due to mutation only. Hence, it will tend to underestimate the ratio of recombination to mutation.
Given these two models, we can then estimate the proportion of our SLVs at each locus that are due to mutation only. In practice, we use an expectation-maximization (EM) algorithm (Dempster et al. 1977) to infer this proportion. Finally, based on the estimated proportion of SLVs at a given locus that is due to mutation only we estimate the probability that the single event that led to the generation of a new allele was a mutation. The above analysis was carried out by an R script (available by request from the first author).
To test the accuracy of our method for estimating the ratio of recombination to mutation, MLST data were simulated under different known ratios of recombination to mutation with different recombination tract lengths using SimMLST software (Didelot et al. 2009).
We used a parametric bootstrap to assess uncertainty in estimates. We simulated 100 datasets for both C. coli and C. jejuni. These datasets matched the true data in terms of number of STs, relative rate of mutation to recombination, and overall mutation rate across the seven gene loci. Within the simulations we assumed that the mutation rate and recombination rate were the same across loci. For our simulated data we estimated the probability of an event being a mutation, and calculated the variability of estimates of this quantity across the simulations: both for estimates for a single locus, and for the estimate obtained by averaging across loci. We consider estimates of this quantity as the variance of the estimates changed little when we varied the true value of the relative rate of recombination to mutation. Confidence intervals were then calculated using a normal approximation, and transformed to confidence intervals for the relative rate of recombination to mutation.
Results
SLV Analysis on the Campylobacter MLST Databases
From our downloaded dataset, there were 7417 SLVs (aspA: 992; glnA: 1045; gltA: 1250; glyA: 773; pgm: 1580; tkt: 1060; and uncA 717) for C. jejuni, and 1842 SLVs (aspA: 110; glnA: 179; gltA: 128; glyA: 292; pgm: 325; tkt: 647; and uncA: 161) for C. coli clade1. The difference in the number of SLVs at each locus suggests that it is worthwhile estimating the relative mutation and recombination rates separately for each locus.
The Distribution of Nucleotide Differences Between Each SLV for Each Locus
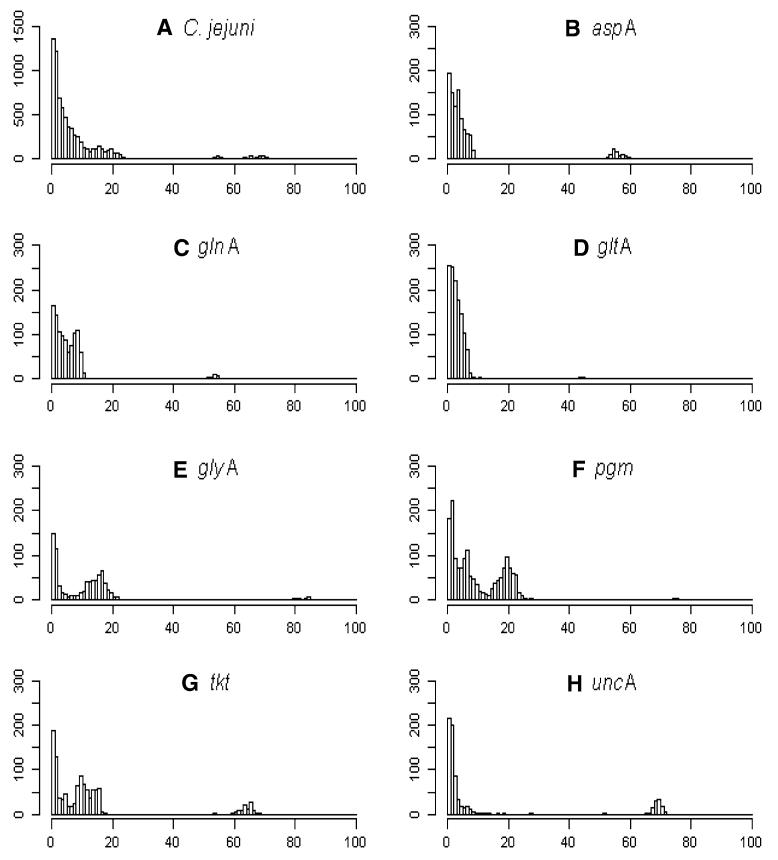
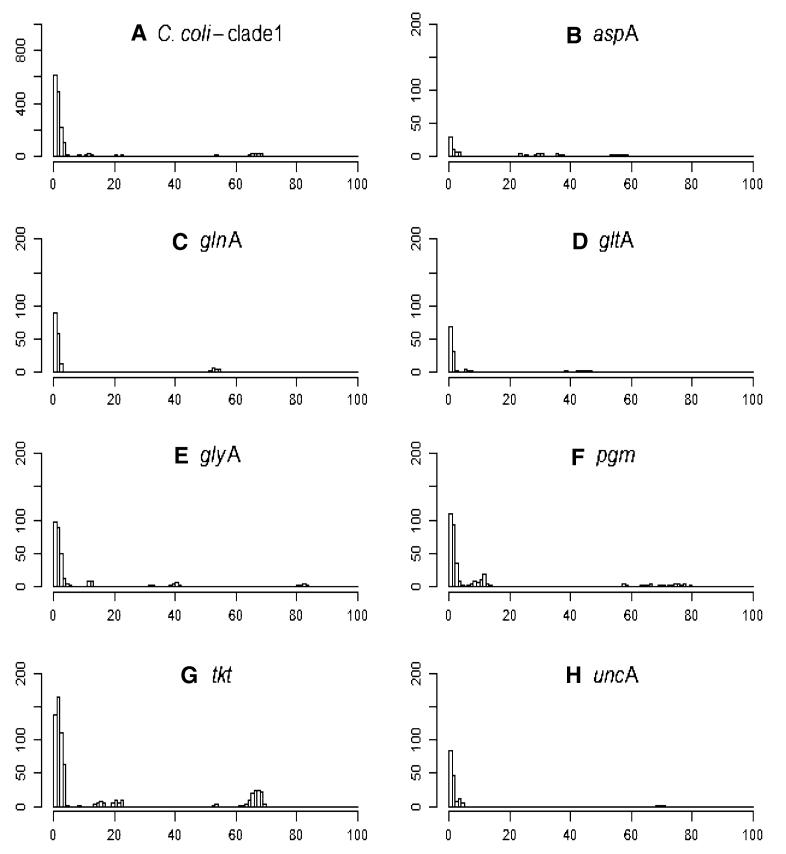
Each SLV relates to one pair of STs, and the plots (Figs. 1, 2) show the nucleotide differences that occurred within those pairs of STs at each MLST locus for C. jejuni and C. coli clade 1. These plots show that SLVs with a large number of nucleotide differences (>45) occurred in every locus. The pairs of STs with a large number of nucleotide differences (50–80) are almost certainly due to recombination, as it is highly unlikely that more than 50 independent point mutations would occur at a single locus while the other six loci remained the same. These large differences are likely to be due to recombination between C. jejuni and C. coli (Sheppard et al. 2008; Wilson et al.2009). Species were designated according to the PubMLST data, and only those SLVs that comprised STs that were assigned 100 % C. jejuni or C. coli were plotted. Even with this strict species designation, there were still large nucleotide differences visible between SLVs within species. There were second peaks in the range of 15-20 differences at the loci glyA, pgm, and tkt for both C. jejuni and C. coli clade 1. These peaks are likely to be due to recombination as well. The first peak of most loci (except for pgm for C. jejuni and tkt for C. coli clade 1) represented approximately 100-200 SLVs for C. jejuni and around 100 SLVs for C. coli clade 1 with only one nucleotide difference; most of these are more likely to be due to mutation.
Fig. 1.
SLVs of PubMLST data. The x axes represent the number of nucleotide differences between STs that make up an SLV; y axes represent the number of recorded events. A represents the nucleotide differences for SLVs in the PubMLST database for C. jejuni; others are the nucleotide differences for SLVs by loci
Fig. 2.
SLVs of PubMLST data. The x axes represent the number of nucleotide differences between STs that make up an SLV; y axes represent the number of recorded events. A represents the nucleotide differences for SLVs in the PubMLST database for C. coli clade 1; others are the nucleotide differences for SLVs by loci
Relative Contributions of Recombination and Mutation Separately for C. jejuni and C. coli Clade 1
Tables 1 and 2 demonstrate that recombination contributed more to the generation of SLVs than did mutation for both the groups (C. jejuni and C. coli clade 1), but the range of estimates vary for the two groups. The average ratio of recombination events to mutation events from the seven loci is 6.96 (95 % CI 6.08, 8.09) for C. jejuni (Table 1), and 1.01 (95 % CI 0.78, 1.30) for C. coli clade 1 (Table 2).
Table 1.
Allele lengths for each locus; estimates for C. jejuni for each housekeeping locus of the probability of an SLV being caused by mutation only (p); the expected number of mutations for an SLV; the relative rate of recombination to mutation; 95 % CI for the estimated relative rate of recombination to mutation; and the % of nucleotide differences of an SLV that were introduced by recombination
| Locus | Allele lengths (bp) | p | Expected number mut. | Relative rate of rec | 95 % CI | % Diff due to rec |
|---|---|---|---|---|---|---|
| aspA | 477 | 0.09 | 0.11 | 8.91 | (5.98, 16.06) | 99 |
| glnA | 477 | 0.09 | 0.11 | 8.86 | (5.96, 15.91) | 98 |
| gltA | 402 | 0.10 | 0.12 | 7.84 | (5.43, 13.12) | 97 |
| glyA | 507 | 0.21 | 0.32 | 2.97 | (2.40, 3.77) | 97 |
| pgm | 498 | 0.10 | 0.15 | 7.14 | (5.06, 11.41) | 98 |
| tkt | 459 | 0.11 | 0.15 | 6.81 | (4.87, 10.66) | 99 |
| uncA | 489 | 0.12 | 0.16 | 6.21 | (4.53, 9.37) | 99 |
| Average | 472.71 | 0.12 | 0.16 | 6.96 | (6.08, 8.09) | 98 |
Table 2.
Allele lengths for each locus; estimates for C. coli clade 1 for each housekeeping locus of the probability of an SLV being caused by mutation only (p); the expected number of mutations for an SLV; the relative rate of recombination to mutation; 95 % CI for the estimated relative rate of recombination to mutation; and the % of nucleotide differences of an SLV that were introduced by recombination
| Locus | Allele lengths (bp) | p | Expected number mut. | Relative rate of rec | 95 % CI | % Diff due to rec |
|---|---|---|---|---|---|---|
| aspA | 477 | 0.18 | 0.65 | 2.23 | (1.13, 5.69) | 98 |
| glnA | 477 | 0.80 | 0.84 | 0.23 | (0.03, 0.53) | 60 |
| gltA | 402 | 0.56 | 0.61 | 0.72 | (0.35, 1.37) | 80 |
| glyA | 507 | 0.63 | 0.72 | 0.54 | (0.24, 1.04) | 86 |
| pgm | 498 | 0.47 | 0.54 | 1.04 | (0.54, 2.03) | 90 |
| tkt | 459 | 0.51 | 0.62 | 0.85 | (0.43, 1.63) | 91 |
| uncA | 489 | 0.39 | 0.44 | 1.44 | (0.75, 3.01) | 89 |
| Average | 472.71 | 0.51 | 0.63 | 1.01 | (0.78, 1.30) | 85 |
For each locus, we also estimated the proportion of nucleotide differences introduced by recombination as opposed to mutation, and this ranged from 97 % (gltA and glyA) to 99 % (aspA, tkt, and uncA) for C. jejuni, and from 60 % (glnA) to 98 % (aspA) for C. coli clade 1.
We also investigated the robustness of the mutation model to different prior distributions of the probability of events caused by mutations only. These suggest that the results in Supplementary Table 1 and 2 are conservative regarding the importance of recombination in producing new variation for C. jejuni and C. coli clade 1.
We see evidence for differences in the relative role of recombination to mutation across the genes (Tables 1, 2). In particular, the parametric bootstrap results show that there is evidence for a lower rate of recombination in glnA for C. coli and for glyA in C. jejuni, and for a higher rate in aspA in C. coli. To assess the strength of this evidence, we looked at the lowest (and the highest) estimated value of the relative rate of recombination to mutation across the seven genes in our simulated data divided by the average of estimated rate across the seven genes. For both C. coli and C. jejuni, we never observed an estimate as low as that for glnA and glyA, respectively, across the 100 simulations in each case (the lowest estimates were 0.36 and 0.62 for C. coli and C. jejuni, respectively, compared to observed values of 0.23 and 0.43) or as high as aspA for C. coli (highest estimate was 2.04, compared to an observed value of 2.21).
Discussion
We have analyzed SLVs to infer the relative importance of recombination and mutation to generate differences between closely related C. jejuni and C. coli clade 1 isolates. The higher average estimates for C. jejuni compared to C. coli demonstrates higher recombination in C. jejuni, compared to C. coli. This is consistent with the existing population structure (three clades) of C. coli, but not with apparent subclade structure in C. jejuni (Sheppard et al.2008). We estimate that recombination contributes between 2.97 and 8.91 times more than mutation to events that generate new alleles for C. jejuni, depending on the MLST locus, and between 0.23 and 2.23 for C. coli clade 1. The variations between housekeeping genes within species also show the different evolution pressure on different genes. For C. jejuni, glyA has less recombination contribution, compared to the other six genes. For C. coli, glnA has less recombination contribution, compared to the other six genes.
Our analysis has similarities to that of Schouls et al. (2003), who used the approach described by Feil et al. (2000) to estimate the relative rate of recombination and mutation for C. jejuni. The original idea of Feil et al.’s method (2000) is put forward by Guttman and Dykhuizen (1994). However, their method overestimates the ratio of recombination to mutation, compared to ours. They also analyzed SLVs, though restricted to pairs of SLVs within the same clonal complex. Furthermore, rather than the model-based approach we consider, they used a simple rule to classify which SLVs had been caused by mutation as opposed to recombination. The rule was that if a pair of SLVs varies by a single nucleotide difference and one of the MLST alleles at the locus was unique, it is due to a mutation, whereas all other pairs of SLVs are caused by recombination. This means that, under this algorithm, SLVs that differ by two nucleotide differences could not have arisen by two independent mutation events, and recombination events that mask mutation events are not considered. Both assumptions may lead to an underestimate of mutation. The analysis of Schouls et al. (2003) estimates that recombination is approximately eight times more likely to change an allele than mutation. This is larger than our estimate, which is likely to be due to these biases in the method used by Schouls et al. (2003). According to Feil et al.’s method (2000), Schouls et al. (2003) estimated a recombination size of about 3.3 kb. We implemented a simplified version of Feil et al.’s method (2000) (details in the Supplementary Material), and the results show that under the 3 kb recombination size, the ratio is overestimated.
Our estimates suggest a more important role for recombination in producing new diversity into C. jejuni than more recent studies which have analyzed samples of C. jejuni isolates from different source populations. Fearnhead et al. (2005) estimate that recombination rates are, if anything, less than the mutation rates. While Vos and Didelot (2008), and Wilson et al. (2009) give estimates of the proportion of nucleotide differences introduced by recombination as opposed to mutation which are much smaller than the ones we obtained. Both the studies concluded that the number of nucleotide differences introduced by recombination are only approximately twice as many as those introduced by point mutation: 2.2 for Vos and Didelot (2008) and 2.67 (95 % CI 1.39, 4.95) for Wilson et al. (2009).
The difference between our study and these is that we analyze only SLVs, which means we are looking at closely related STs for which there has been less time for selection to act. Intuitively, selection is likely to be the strongest against recombination events that introduce large differences, although it is possible that some recombination events may introduce a section of DNA from an organism that is highly adapted and “successful” in the given environment. Therefore, although we estimate that recombination is introducing more differences than previously thought in our closely related and recently evolved STs, many of these differences may be subsequently purged from the population due to weak purifying selection. This is consistent with the effects of purifying selection described in Wilson et al.’s paper (2009).
Whole genome analysis may provide a greater insight into the genome-wide evolution of Campylobacter and provide further explanations for the apparent differences between previous estimates of recombination and mutation. Recently, Biggs et al. (2011) analyzed the genomes of two closely related Campylobacter ST-474 isolates that also had identical flaA SVR regions and compared them to available C. jejuni reference strains. They estimated that around 97 % of the nucleotide differences between these two closely related isolates were caused by recombination. This estimate is similar to ours, and suggests that the importance of recombination for driving changes in C. jejuni is not just confined to the MLST housekeeping genes we have studied.
The aim of this study was to increase our understanding of the evolution of C. jejuni and C. coli by investigating the generation of SLVs. The availability of the large database of C. jejuni and C. coli isolates provides a good opportunity to investigate the evolution of C. jejuni and C. coli using SLVs. Using seven independent housekeeping loci we used the method proposed in this paper to estimate that recombination contributes roughly seven times as much as mutation to the generation of SLVs for C. jejuni, and equally for C. coli, which provides further evidence that recombination plays a more important role in the evolution of C. jejuni and C. coli than mutation.
Our results also point to important differences in terms of the forces driving evolution for C. jejuni and C. coli; and suggest that the relative role of recombination to mutation may differ between genes, and these differences themselves may be different for C. jejuni and C. coli. Understanding what is causing these differences will be important for fully understanding how these bacteria may evolve in the future. However, the fact that we observed differences in recombination between C. jejuni and C. coli is consistent with the introgression hypothesis of Sheppard et al.’s paper (2011), which implies that patterns of genetic exchange have changed over time. The research on SLVs described in this paper could be extended either by considering more genes, such as flagellin genes (flaA and flaB) (Meinersmann and Hiett 2000), and porA, the gene encoding the major outer membrane proteins (MOMPs) (Zhang et al. 2000; Clark et al. 2005), or by considering other species of Campylobacter.
Supplementary Material
Acknowledgments
The authors acknowledge the Marsden Fund project 08-MAU-099 (Cows, starlings and Campylobacter in New Zealand: unifying phylogeny, genealogy, and epidemiology to gain insight into pathogen evolution) for funding this project. This publication made use of the Campylobacter Multi Locus Sequence Typing website (http://pubmlst.org/campylobacter/) developed by Keith Jolley and sited at the University of Oxford (Jolley and Maiden 2010, BMC Bioinformatics, 11:595). The development of this site has been funded by the Wellcome Trust. BRH acknowledges the Australian Research Council (Grant FT100100031).
Contributor Information
Shoukai Yu, Infectious Disease Research Centre, Institute of Veterinary, Animal, and Biomedical Sciences, Massey University, Palmerston North, New Zealand; S.Yu1@massey.ac.nz.
Paul Fearnhead, Department of Mathematics and Statistics, Lancaster University, Lancaster, UK.
Barbara R. Holland, School of Mathematics and Physics, University of Tasmania, Hobart, Australia
Patrick Biggs, Infectious Disease Research Centre, Institute of Veterinary, Animal, and Biomedical Sciences, Massey University, Palmerston North, New Zealand.
Martin Maiden, Department of Zoology, University of Oxford, Oxford, UK.
Nigel French, Infectious Disease Research Centre, Institute of Veterinary, Animal, and Biomedical Sciences, Massey University, Palmerston North, New Zealand; Allan Wilson Centre for Molecular Ecology and Evolution, Massey University, Palmerston North, New Zealand; n.p.french@massey.ac.nz.
References
- Biggs PJ, Fearnhead P, Hotter G, Mohan V, Collins-Emerson J, Kwan E, Besser TE, Cookson A, Carter PE, French NP. Whole-genome comparison of two Campylobacter jejuni isolates of the same sequence type reveals multiple loci of different ancestral lineage. PLoS One. 2011;6(11):e27121. doi: 10.1371/journal.pone.0027121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CG, Bryden L, Cuff WR, Johnson PL, Jamieson F, Ciebin B, Wang G. Use ofthe Oxford multilocus sequence typing protocol and sequencing of the flagellin short variable region to characterize isolates from a large outbreak of waterborne Campylobacter sp. strains in Walkerton, Ontario, Canada. J Clin Microbiol. 2005;43:2080. doi: 10.1128/JCM.43.5.2080-2091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J Roy Statist Soc Ser B. 1977;39(1):1–38. [Google Scholar]
- Didelot X, Lawson D, Falush D. Simmlst: simulation of multilocus sequence typing data under a neutral model. Bioinformatics. 2009;25:1442. doi: 10.1093/bioinformatics/btp145. [DOI] [PubMed] [Google Scholar]
- Dingle KE, Colles FM, Wareing DRA, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJL, Urwin R, Maiden MCJ. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead P, Smith NGC, Barrigas M, Fox A, French N. Analysis of recombination in Campylobacter jejuni from MLST population data. J Mol Evol. 2005;61:333–340. doi: 10.1007/s00239-004-0316-0. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Maiden MC, Achtman M, Spratt BG. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol Biol Evol. 1999;16:1496. doi: 10.1093/oxfordjournals.molbev.a026061. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Smith JM, Enright MC, Spratt BG. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics. 2000;154:1439. doi: 10.1093/genetics/154.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Holmes EC, Bessen DE, Chan MS, Day NPJ, Enright MC, Goldstein R, Hood DW, Kalia A, Moore CE, et al. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci. 2001;98:182. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman DS, Dykhuizen DE. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- Hein J, Schierup MH, Wiuf C. Gene genealogies, variation and evolution: a primer in coalescent theory. Oxford University Press; Oxford: 2005. [Google Scholar]
- Humphrey T, O’Brien S, Madsen M. Campylobacters as zoonotic pathogens: a food production perspective. Int J Food Microbiol. 2007;117:237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Jolley KA, Maiden MCJ. BIGSdb: scalable analysis of bacterial genome variationat thepopulation level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel ME, Gray SA, Kim BJ, Garvis SG, Yoon J. Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J Clin Microbiol. 1999;37:510. doi: 10.1128/jcm.37.3.510-517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden MCJ, Bygraves JA, Feil EJ, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Nat Acad Sci USA. 1998;95:3140. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinersmann RJ, Hiett KL. Concerted evolution of duplicate fla genes in Campylobacter. Microbiology. 2000;146:2283. doi: 10.1099/00221287-146-9-2283. [DOI] [PubMed] [Google Scholar]
- Richman AD, Herrera LG, Nash D, Schierup MH. Relative roles of mutation and recombination in generating allelic polymorphism at an MHC class II locus in Peromyscus maniculatus. Genet Res. 2003;82:89–99. doi: 10.1017/s0016672303006347. [DOI] [PubMed] [Google Scholar]
- Sarkar SF, Guttman DS. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl Environ Microbiol. 2004;70:1999. doi: 10.1128/AEM.70.4.1999-2012.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouls LM, Reulen S, Duim B, Wagenaar JA, Willems RJL, Dingle KE, Colles FM, Van Embden JDA. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J Clin Microbiol. 2003;41:15. doi: 10.1128/JCM.41.1.15-26.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, McCarthy ND, Falush D, Maiden MCJ. Convergence of Campylobacter species: implications for bacterial evolution. Science. 2008;320:237–239. doi: 10.1126/science.1155532. [DOI] [PubMed] [Google Scholar]
- Sheppard SK, Dallas JF, Wilson DJ, Strachan NJC, McCarthy ND, et al. Evolution of an agriculture-associated disease causing Campylobacter coli clade: evidence from National Surveillance Data in Scotland. PLoS ONE. 2010;5(12) doi: 10.1371/journal.pone.0015708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, McCarthy ND, Jolley KA, Maiden MCJ. Introgression in the genus Campylobacter: generation and spread of mosaic alleles. Microbiology. 2011;157:1066–1074. doi: 10.1099/mic.0.045153-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S, Lohrengel M, Sonnevend A, Ruberg F, Kist M. Allelic diversity and recombination in Campylobacter jejuni. J Bacteriol. 2001;183:2553. doi: 10.1128/JB.183.8.2553-2559.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauxe RV, Nachamkin I, Blaser MJ, Tompkins LS. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. MBio; Washington, DC: 1992. [Google Scholar]
- Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2008;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- Wilson DJ, Gabriel E, Leatherbarrow AJH, Cheesbrough J, Gee S, Bolton E, Fox A, Hart CA, Diggle PJ, Fearnhead P. Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol Biol Evol. 2009;26:385. doi: 10.1093/molbev/msn264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Meitzler JC, Huang S, Morishita T. Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein. Infect Immun. 2000;68:5679. doi: 10.1128/iai.68.10.5679-5689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.