Highlights
-
•
Viral membrane fusion proteins fall into three distinct structural classes (I–III).
-
•
In 2013, the E2 glycoproteins of pesti- and hepaciviruses were found to have novel folds.
-
•
E1 and E2 proteins of pesti- and hepaciviruses define a new class of fusion machinery.
-
•
Structural data suggest that fusion proteins evolved by host–virus or virus–virus transfer.
Keywords: bovine viral diarrhea virus, hepacivirus, host–virus coevolution, horizontal gene transfer, envelope glycoprotein, paleovirology
Abstract
Enveloped viruses must fuse their lipid membrane to a cellular membrane to deliver their genome into the cytoplasm for replication. Viral envelope proteins catalyze this critical membrane fusion event. They fall into three distinct structural classes. In 2013, envelope proteins from a pestivirus and hepatitis C virus were found to have two distinct novel folds. This was unexpected because these viruses are in the same family as flaviviruses, which have class II fusion proteins. We propose that the membrane fusion machinery of the closely related pestiviruses and hepatitis C virus defines a new structural class. This and other recently identified structural relationships between viral fusion proteins shift the paradigm for how these proteins evolved.
Virus cell entry by membrane fusion
In many viruses, the genome is enveloped in a lipid membrane. Viral envelope proteins anchored in the membrane fulfill indispensable functions throughout the life cycle of the virus. Envelope proteins drive virus assembly, form the protective outer shell of the virus, mediate cellular attachment and tropism, and catalyze the fusion of the viral and host cell membranes to deliver the viral genome into the cytoplasm for replication. Envelope proteins also provide a shield against the immune system of the host and bear most of the neutralizing antibody epitopes against any given virus. In viruses with more than one envelope protein, the proteins responsible for cellular attachment are as varied as the host cell receptors that they recognize. By contrast, the viral envelope proteins that catalyze the essential membrane fusion step in cell entry fall into three broad yet distinct structural classes (Figure 1 ). The influenza virus hemagglutinin (HA) is the prototype of class I fusion proteins [1], which encompass those of other orthomyxoviruses, and of paramyxoviruses [2], retroviruses 3, 4, filoviruses [5], and coronaviruses 6, 7. The unifying structural features of class I fusion proteins are a proteolytically generated N-terminal fusion peptide, and a core consisting of three bundled α-helices in the prefusion conformation, which refolds into a six-helix bundle in the postfusion conformation [8]. Class II fusion proteins are a structurally unrelated class found in flaviviruses [9], alphaviruses [10], and most recently in rubella virus (sole member of the rubivirus genus) [11] and Rift Valley fever virus (RVFV, from the phlebovirus genus) [12]. Class II proteins share a three-domain architecture consisting almost entirely of β-strands, with a tightly folded ‘fusion loop’ in the central domain serving as the anchor in the cellular membrane targeted for fusion (Figure 1) [13]. Class III fusion proteins, found in herpesviruses [14], rhabdoviruses [15], and baculoviruses [16], possess core helical bundles like class I proteins, and a central β-stranded domain bearing one or more fusion loops like class II proteins (Figure 1). However, similarities between class II and class III proteins are likely to have arisen from convergent evolution because the fold and connectivity of the fusion domains are different in the two classes. Notably, reoviruses encode a family of fusion-associated small transmembrane (FAST) proteins that function as dedicated cell–cell fusogens. The resulting multinuclear syncytia promote viral replication by obviating the need for cell-to-cell transmission (reviewed in [17]).
Figure 1.
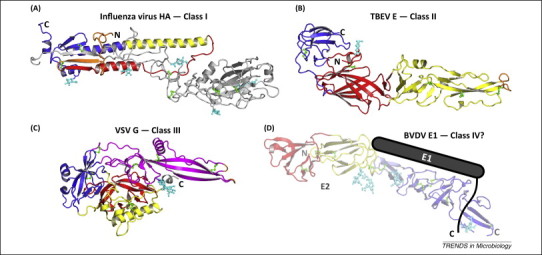
Representative structures of the ectodomains of viral membrane fusion proteins from different structural families in their prefusion conformations (drawn to scale).The N terminus of each ectodomain is labeled with an ‘N’, whereas the C terminus of each ectodomain (labeled with a ‘C’) connects to the transmembrane anchor, which is missing from the crystal structures. (A) Structure of the influenza A virus hemagglutinin (HA). HA forms trimers on the viral surface [Protein Data Bank (PDB) accession code 2HMG]. HA is the prototypic class I fusion protein. (B) Structure of envelope glycoprotein E from tick-borne encephalitis virus (TBEV E, PDB code 1SVB). This was the first class II protein structure to be determined. (C) Structure of vesicular stomatitis virus glycoprotein G (VSV G, PDB code 2J6J). This structure was the first class III protein structure to be determined in the prefusion conformation. (D) Structure of envelope glycoprotein E2 from bovine viral diarrhea virus (BVDV), a pestivirus related to hepatitis C viruses (hepaciviruses). E2 has a novel fold (PDB code 4JNT). This was unexpected because pesti- and hepaciviruses had been predicted to contain class II fusion proteins and belong to the same Flaviviridae family as flaviviruses, which have class II fusion proteins (such as TBEV E). Because the E2 structure lacks the hallmarks of a fusion protein, E1 is presumed to be the fusogen. Fusion motifs are colored orange. N-Linked glycans and disulfide bonds are shown in cyan and green, respectively.
Viruses from the Flaviviridae family, including flaviviruses, pestiviruses, and hepaciviruses (principally hepatitis C virus, HCV), share many key characteristics. Because flaviviruses contain prototypical class II membrane fusion proteins, pestiviruses and hepaciviruses had been expected to have similar class II fusion proteins 18, 19. However, in 2013 the larger envelope protein, E2, from the pestivirus BVDV (bovine viral diarrhea virus) was unexpectedly found to have a novel fold 20, 21. The structure of a core fragment of E2 from HCV was subsequently found to have a novel fold unrelated to that of BVDV E2 [22]. Moreover, BVDV E2 and HCV E2 both lack the structural hallmarks of fusion proteins. Together, these discoveries suggest that E1 is the fusogen and that pesti- and hepaciviruses contain a new class (or classes) of membrane fusion machinery. The evolutionary implications of this and other recently identified unexpected structural relationships between fusion proteins across virus families are discussed.
A conserved overall mechanism and topology of catalysis of viral membrane fusion
Structural studies of viral envelope proteins have revealed certain overarching commonalities in the membrane fusion mechanisms of viruses across different families. Crystal structures of fusion proteins from classes I, II, and III before and after the conformational change that catalyzes membrane fusion provide a molecular outline of their respective fusion mechanisms (reviewed in 1, 8, 23, 24). Complementing these pre- and postfusion structures, structures thought to represent fusion intermediates provide invaluable insights on the steps required for fusion 12, 24, 25, 26, 27, 28, 29, 30, 31. The paradigm that has emerged is that, despite the existence of three distinct fusion protein architectures and significant structure divergence within each class, all viral fusion proteins catalyze membrane fusion with a common overall mechanism and topology (Figure 2 ) 8, 16, 23, 24, 32, 33. Fusion proteins from all three classes respond to one or more environmental cues – such as low pH, coreceptor binding, or disulfide bond exchange – by exposing a hydrophobic fusion motif previously shielded from the solvent (Figure 2B). The fusion motif, an N-terminal ‘fusion peptide’ in class I proteins or internal fusion loops in class II and class III proteins, spontaneously inserts into the outer bilayer leaflet of the host cell membrane (Figure 2C). This extended conformation, postulated for all viral fusion proteins and recently observed in a bunyavirus protein [12], is called the prehairpin intermediate (Figure 2C). The fusion protein then folds back on itself, directing its C-terminal transmembrane anchor towards the fusion motif (Figure 2D). This fold-back forces the host cell membrane (held by the fusion motif) and the viral membrane (held by the transmembrane anchor) against each other, resulting in fusion of the outer leaflets of the two membranes to form a hemifusion intermediate (Figure 2E), followed by fusion of the distal leaflets to form a fusion pore and complete fusion [33] (Figure 2F). The oligomeric state of fusion proteins vary before fusion, but all fusion proteins undergo the fusogenic fold-back as trimers and are trimeric in the postfusion conformation (Figure 3 ). Moreover, postfusion trimers from all three classes have been reported to form interacting networks 34, 35, 36, 37, which have been proposed to be required for fusion pore expansion 38, 39. The conservation of trimeric postfusion states across structural classes may be coincidental, but it is possible that trimeric assemblies have been selected because they provide the optimal balance of stability and susceptibility to the first fold-back event. Indeed, because fusion requires multiple trimers to fold back cooperatively [38], prehairpins cannot be too short-lived but should fold back rapidly once fold back has been initiated. Like a three-legged stool, a trimeric prehairpin intermediate may have a favorable degree of stability (necessary for multiple prehairpins to accumulate on the viral surface), but is rapidly destabilized once the first subunit begins to fold back (allowing multiple trimers to refold cooperatively).
Figure 2.
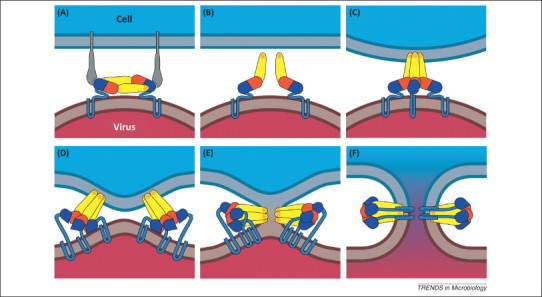
Membrane fusion mechanism of enveloped viruses. This figure was created with class II fusion proteins in mind but the overall mechanism and topology are conserved across all structural classes. (A) A viral envelope protein binds to a cell surface receptor. Most viruses are endocytosed. (B) Environmental cues such as low endosomal pH (shown here) or coreceptor binding (not shown) cause the ectodomain of the fusion protein to hinge away from the viral surface, exposing a hydrophobic fusion motif. (C) The fusion motif inserts into the cell membrane, promoting trimer formation if the prefusion conformation is not trimeric. (D) The fusion protein folds back on itself, directing the fusion loop towards the C-terminal transmembrane anchor. The refolding energy bends the apposed membranes. (E) Creation of new contacts during refolding of the fusion protein leads first to hemifusion and then, (F), to formation of a lipidic fusion pore.
Figure 3.
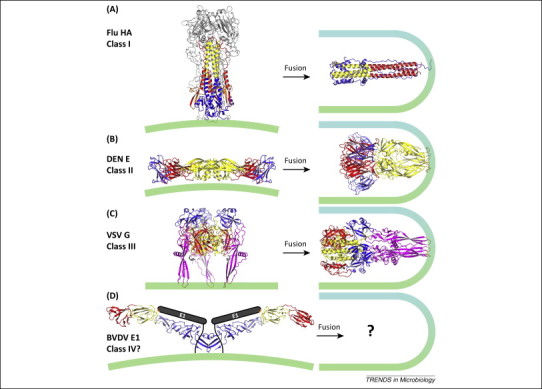
Conformational changes associated with membrane fusion in the different structural classes of membrane fusion proteins. In all classes, a fusion motif (orange) that is shielded from the solvent in the prefusion conformation (left column) becomes exposed in response to environmental cues (e.g., low pH or coreceptor binding). The fusion motif inserts into the cell membrane and the protein folds back on itself, forcing the fusion motif and the C-terminal transmembrane domain (not shown) anchored in the viral membrane towards each other. The proteins are trimeric in their postfusion conformations (right column). (A) In class I fusion proteins, such as influenza A virus hemagglutinin (Flu HA) shown here, membrane fusion is catalyzed by extensive refolding and secondary structure rearrangements of prefusion trimers to form a six-helix bundle [Protein Data Bank (PDB) codes 2HMG, 1HTM, 1QU1]. (B) Class II proteins usually form icosahedral shells in infectious virions. The envelope proteins respond to the reduced pH of an endosome with a repositioning of the three domains with only minor changes in secondary structure. The proteins form trimers during the fusion transition and the fusion loop in the central domain is directed towards the viral transmembrane anchor. The pre- and postfusion conformations of dengue type 2 virus E (DEN E) are shown here (PDB codes 1OKE, 1OK8). (C) Class III proteins are trimeric before and after fusion and undergo extensive refolding during the fusion transition like class I fusion proteins, but they contain internal fusion loops like class II proteins. The pre- and postfusion structures of vesicular stomatitis virus G (VSV G) are shown here (PDB codes 2J6J, 2CMZ). (D) The structure of envelope glycoprotein E2 from the pestivirus bovine viral diarrhea virus (BVDV) has been proposed to serve as a molecular scaffold for E1, which may define a new structural class of fusion machinery (PDB code 4JNT). The structure of envelope protein E1 (gray) and the nature of the fusogenic conformational change remain unknown. The outer leaflets of the viral and cellular membranes are represented in green and cyan, respectively.
The E2 envelope glycoproteins from BVDV and HCV adopt novel folds
The Flaviviridae family contains four genera: flavivirus, pestivirus, pegivirus (GB viruses), and hepacivirus (HCV). Among these genera, pestiviruses and pegiviruses are the most closely related to HCV, a serious and persistent global health threat [40]. Until 2013, envelope protein structures were available only from the flavivirus genus. Envelope proteins from pesti- and hepaciviruses had been predicted to have class II folds based on the disulfide-bonding pattern [18] and amino acid sequence analysis of the E1 and E2 envelope proteins [19]. It was therefore surprising when two groups discovered in 2013 that the larger envelope protein, E2, from the pestivirus BVDV is not a class II fusion protein 20, 21. Instead, the BVDV E2 ectodomain has a novel architecture, spanning a total of 140 Å and consisting of two immunoglobulin (Ig)-like domains followed by a unique elongated β-stranded domain (domain III) and a membrane anchor (Figure 1D). The overall fold and topology of BVDV E2 domain III bears no significant similarity to previously determined protein structures. The structure of a core fragment of HCV E2 was subsequently determined and found to have a novel globular architecture, distinct from that of BVDV E2, with an Ig-like β-sandwich at its core [22]. Notably, both BVDV E2 and HCV E2 lack an internal or terminal fusion motif with an obvious resemblance to those of other viral fusion proteins. Moreover, BVDV E2 forms tightly associated dimers but the dimerization interface, which contains a large cluster of conserved aromatic residues, is very different from that of flavivirus E proteins (Figure 3) [20]. The structures of BVDV E2 and HCV E2 provide striking examples of how structurally divergent viral envelope proteins can be within a single virus family.
E1 from pesti- and hepaciviruses may define a new class of membrane fusogen
The novel structures of BVDV E2 and HCV E2 lack the structural hallmarks of membrane fusion proteins such as a hydrophobic fusion motif (found in all classes of fusion protein), a helical core (as in classes I and III), or a flexible multidomain structure (as in classes II and III). This suggests that E1 is the fusion protein in pesti- and hepaciviruses. Indeed, E1 has been proposed to bear the fusion motif in both genera 20, 21, 41. As the fusion protein, E1 would have to at least transiently extend to span the distance between the cellular and viral membranes prior to membrane fusion, approximately 20 nm [28]. E1 would have to adopt a highly elongated fold in order to span 20 nm. With its ectodomain of less than 180 amino acids, E1 is too small to have a class II or class III fold that could span 20 nm. Additionally, secondary structure predictions suggest that approximately 30% of the E1 ectodomain is α-helical, which is inconsistent with a class II or class III architecture. Coiled-coils efficiently form rigid, highly elongated structures, thus it is possible that predicted α-helices in the central region of E1 form a helical bundle. However, E1 lacks a clear leucine zipper motif that would be indicative of a coiled coil. Moreover, E1 is unlikely to have a class I fold as it lacks key unifying features of class I proteins: E1 does not form trimers, is not subject to proteolytic activation, and does not appear to have an N-terminal fusion peptide. In fact, E1 cannot fold correctly or support cell entry without E2 20, 21, 42, 43. Thus, E2 appears to function as an essential molecular scaffold or chaperone to E1 as the fusogen 20, 21. Consistent with this view, fusogenic pesti- or hepacivirus particles contain E1–E2 disulfide links, and virus particles with mutations disrupting E1–E2 interactions (or lacking E2) are not fusogenic 42, 44, 45, 46, 47. Based on the structures of BVDV E2 and HCV E2, on available biochemical data, and on the amino acid sequences of E1 proteins, the possibility that pesti- and hepaciviruses utilize class I, class II, or class III architectures can now be ruled out. Hence, we propose that in pesti- and hepaciviruses, E1 defines a new class (or two distinct classes) of membrane fusion protein, and that E2 plays an accessory role as a molecular scaffold for E1.
Evolutionary implications of the structural relationships between fusion proteins from different virus families
The unexpected structural relationships between viral fusion proteins discovered in 2013 provide important insights on viral evolution. Viruses from the Flaviviridae family each have similar genetic organizations, coding strategies, morphologies, and cell entry pathways, suggesting that they might have evolved from a common virus ancestor (Box 1 ). However, the discovery of novel folds in BVDV E2 20, 21 and HCV E2 [22] implies that the envelope proteins of pestiviruses, hepaciviruses, and flaviviruses have different evolutionary origins. The discovery of a highly divergent class II fold in rubella virus glycoprotein E1 was also unexpected given that rubella virus belongs to the same Togaviridae family as alphaviruses, which have prototypical class II fusion proteins [11]. Rubella virus E1 is also the first class II protein to be identified that does not assemble into a rigid icosahedrally symmetric shell. Rubella virus only infects humans and is the first example of a virus with a class II fusion protein that does not alternate between arthropod and mammalian hosts. For these reasons, alphaviruses appear to be more closely related to flaviviruses than to rubella virus. Hence, an evolutionary model in which viruses within a given family evolved from a common ancestor virus seems overly simplistic.
Box 1. Outstanding questions.
-
•
Do the E1 proteins from pestiviruses and HCV have similar folds, despite the different folds of E2 proteins from these two genera?
-
•
What is the molecular mechanism of membrane fusion in pestiviruses and HCV?
-
∘What is the role of E2 in supporting the fusion activity of E1?
-
∘Is E1 a fusion protein with a novel architecture, or does it bear resemblance to one or more of the known structural classes?
-
∘Low pH is required but not sufficient for fusion in pesti- and hepaciviruses. What is the nature of the additional activation step required for fusion?
-
∘What is the role (if any) of disulfide bond exchange in pestivirus cell entry?
-
∘
-
•
Did pesti- and hepaciviruses evolve from a common ancestor virus genus?
-
•
What is the evolutionary origin of class I fusion proteins? Did they evolve from cellular SNARE proteins?
-
•
Did viral fusion proteins evolve by host-to-virus transfer of fusion protein genes?
-
∘Do eukaryotes contain as yet unidentified ancestral fusion proteins with class II, class III, or pestivirus-like folds? If so, what are their cellular functions?
-
∘
-
•
Can viral membrane fusion proteins be successfully targeted with small molecules or vaccines to treat and prevent infections?
Although fusion proteins within each class of fusion protein can have surprisingly divergent structures, proteins with a remarkable degree of structural conservation have also been identified in otherwise unrelated viruses. This was recently illustrated by the discovery of a class II fusion protein in RVFV, from the Bunyaviridae family, with striking structural similarity to flavivirus envelope proteins, which extends to the mode of protein dimerization in the outer icosahedral shell of the virus 12, 48. The presence of class II fusion proteins in these two unrelated virus families reveals that the class II fold is more prevalent and widely distributed across virus families than previously anticipated. Similarly, fusion proteins with conserved class I and class III folds have been identified in viruses from unrelated families (Table 1 ). This type of structural and functional conservation among fusion proteins, even in the absence of significant amino acid sequence similarity, is strongly suggestive of a common evolutionary origin. But what is the nature of this link? The structural conservation of certain fusion proteins across virus families and the structural divergence of other fusion proteins within the same virus family both point to a new evolutionary paradigm for how these proteins evolved. It is tempting to speculate that rather than diverging from a common ancestor virus, fusion proteins may instead have evolved independently from ancestral cellular membrane fusion proteins with the same folds. For example, although pesti- and flaviviruses may have evolved from a common Flaviviridae ancestor virus, they evidently borrowed their fusion machineries from different sources. These could presumably be different host fusion proteins, but alternatively different virus species could conceivably have borrowed fusion proteins from each other during coinfections with multiple viruses.
Table 1.
Comprehensive list of viral membrane fusion proteins for which atomic structures are available, and their structural and phylogenetic classifications
| Coding strategy | Familya | Genusa | Virus | Fusion protein (PDB code)b | Receptorc, d | Structural class |
|---|---|---|---|---|---|---|
| Positive single-stranded(ss)RNA | Toga- | Alpha- | Semliki forest | E1 (1I9W, 1RER) | Heparan sulfate | Class II |
| Sindbis | E1–E2 (3MUU) | Heparan sulfate | Class II | |||
| Chikungunya | E1–E2–E3 (3N41) | Prohibitin | Class II | |||
| Rubi- | Rubella | E1 (4ADI) | MOG | Class II | ||
| Flavi- | Flavi- | Tick-borne encephalitis | E (1SVB, 1URZ) | Heparan sulfate | Class II | |
| Dengue | E (1OAN, 1OK8) | Heparan sulfate | Class II | |||
| West Nile | E (2I69) | Heparan sulfate | Class II | |||
| Japanese encephalitis | E (3P54) | Hsp70 | Class II | |||
| Pesti- | Bovine viral diarrhea | E1 (structure unknown) | CD46 | New class | ||
| Hepaci- | Hepatitis C virus | E1 (structure unknown) | CD81 | New class | ||
| Corona- | Betacorona- | Murine hepatitis | S (1WDF) | Ceacam1 | Class I | |
| Negative ssRNA | Orthomyxo- | Influenza- | Influenza A | HA (1HTM) | Sialic acid | Class I |
| Rhabdo- | Vesiculo- | Vesicular stomatitis | G (2CMZ, 2J6J) | LDLR | Class III | |
| Bunya- | Phlebo- | Rift Valley fever | Gc (4HJ1) | DC-SIGN | Class II | |
| Filo- | Ebola- | Ebola | GP, GP2 (1EBO, 3CSY) | TIM-1, NPC1 | Class I | |
| Paramyxo- | Rubula- | Parainfluenza 5 | F (1SVF, 2B9B) | Sialic acid | Class I | |
| Mumps | F (2FYZ) | Sialic acid | Class I | |||
| Pneumo- | Respiratory syncytial | F (1G2C) | Nucleolin | Class I | ||
| Avula- | Newcastle disease | F (1G5G, 1USR) | Sialic acid | Class I | ||
| Arena- | Arena- | Guanarito | GP2 (4C53) | Transferrin receptor 1 | Class I | |
| Retroviruses | Retro- | Lenti- | HIV-1 | gp120, gp41 (1GC1, 1AIK) | CD4, CCR5 | Class I |
| Double-stranded DNA | Herpes- | Lymphocrypto- | Epstein–Barr | B (3FVC) | CD21, integrins, MHC-II | Class III |
| Simplex- | Herpes simplex | B (2GUM) | Heparan sulfate, Nectin-1, integrins, HVEM | Class III | ||
| Baculo- | Betabaculo- | Autographa californica nucleopolyhedro- | Gp64 (3DUZ) | Unknown | Class III |
Virus families end in ‘-viridae’ and genera end in ‘-virus’.
Only the first available or most representative PDB codes are listed.
In many viruses a viral protein other than the fusion protein binds to the cellular receptor(s). For example, in pesti- and hepaciviruses, E2 binds to CD46 and CD81, respectively; in paramyxoviruses, the hemagglutinin-neuraminidase (HN) binds to the cellular receptor (most commonly sialic acid); and in herpesviruses, cellular attachment can be mediated by several glycoproteins including gC, gB, gD, and gH/gL.
Abbreviations: MOG, myelin oligodendrocyte glycoprotein; Hsp70, heat shock protein 70; LDLR, low density lipoprotein receptor; DC-SIGN, dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin; TIM-1, T cell immunoglobulin and mucin domain 1; NPC1, Niemann–Pick C1; CCR5, C–C chemokine receptor type 5; CD, cluster of differentiation; MHC-II, major histocompatibility complex class II; HVEM, herpesvirus entry mediator.
The conservation of an α-helical coiled coil architecture in class I viral proteins and in the SNARE family of intracellular vesicle fusion proteins provides a possible precedent for the evolutionary transfer of a structural membrane fusion fold between host and virus during evolution. Although similarities between class I fusion proteins and SNAREs have long been recognized [49], the link was strengthened by a recent study demonstrating that a paramyxovirus class I fusion protein resembles SNAREs in that it has α-helical transmembrane anchors in both membranes prior to fusion, with subsequent zippering of the coiled coils during fusion resulting in a bundle of helical hairpins that extends across the fused membrane 50, 51.
Fusion proteins from the other structural classes have not yet been identified in eukaryotes. However, if fusion proteins do in fact evolve as independent modules, they may have been hijacked from host cells by different viruses at different times throughout evolution. This would suggest that there are as yet unidentified membrane fusion proteins in eukaryotes with these folds.
If the virus-to-virus and host-to-virus horizontal transfer mechanisms described in this section are valid, the question of whether membrane fusion proteins can be transferred from viruses to their host arises. Indeed, there is evidence that this type of transfer has occurred. When retroviruses infect germline cells, copies of the viral genome integrated into the host genome often become fixed as inheritable endogenous retroviral sequences. Although most of these sequences are rapidly disrupted during evolution, a few endogenized retroviral genes have been conserved and still encode functional proteins. Over the past few years it has become clear that envelope glycoproteins from endogenous retroviruses have been domesticated by mammals to catalyze an essential membrane fusion reaction during placental development. These proteins, called syncytins, contribute to the formation of the placental fused cell layer called the syncytiotrophoblast, at the maternal–fetal interface (reviewed in [52]). Remarkably, the capture of syncytins has occurred independently from different endogenous retroviruses in various mammalian species including humans more than 10 million years ago [52]. Moreover, it is thought that the immunosuppressive domain embedded within retroviral envelope glycoproteins and conserved in syncytins may contribute to tolerance of the fetus by the maternal immune system [52]. There has also been speculation that the capture of a founding syncytin-like gene could have been instrumental in the dramatic transition from egg-laying to placental mammals [52].
Concluding remarks
In conclusion, the structural relationships that have recently emerged between envelope proteins across different virus families shift the paradigm for how these proteins evolved away from a divergent model with a common virus ancestor for each virus family. In the new evolutionary model, fusion proteins evolve as independent evolutionary units, transferring from host to virus, or from virus to virus in coinfected hosts. Hence, fusion proteins, and possibly other individual components in viruses, compete as ‘selfish genes’ [53], often crossing the boundaries between virus species or between virus and host.
Acknowledgments
Work on this article was supported by a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease Award and grant R01 GM102869 from the National Institutes of Health (NIH).
References
- 1.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 2.Lamb R.A., Jardetzky T.S. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan D.C. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 4.Weissenhorn W. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 5.Weissenhorn W. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 6.Xu Y. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y. Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core. J. Biol. Chem. 2004;279:30514–30522. doi: 10.1074/jbc.M403760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schibli D.J., Weissenhorn W. Class I and class II viral fusion protein structures reveal similar principles in membrane fusion. Mol. Membr. Biol. 2004;21:361–371. doi: 10.1080/09687860400017784. [DOI] [PubMed] [Google Scholar]
- 9.Rey F.A. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 10.Lescar J. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 11.DuBois R.M. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature. 2013;493:552–556. doi: 10.1038/nature11741. [DOI] [PubMed] [Google Scholar]
- 12.Dessau M., Modis Y. Crystal structure of glycoprotein C from Rift Valley fever virus. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1696–1701. doi: 10.1073/pnas.1217780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modis Y. Relating structure to evolution in class II viral membrane fusion proteins. Curr. Opin. Virol. 2014;5:34–41. doi: 10.1016/j.coviro.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heldwein E.E. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 15.Roche S. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 16.Kadlec J. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- 17.Boutilier J., Duncan R. The reovirus fusion-associated small transmembrane (FAST) proteins: virus-encoded cellular fusogens. Curr. Top. Membr. 2011;68:107–140. doi: 10.1016/B978-0-12-385891-7.00005-2. [DOI] [PubMed] [Google Scholar]
- 18.Krey T. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garry R.F., Dash S. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology. 2003;307:255–265. doi: 10.1016/s0042-6822(02)00065-x. [DOI] [PubMed] [Google Scholar]
- 20.Li Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc. Natl. Acad. Sci. U.S.A. 2013;110:6805–6810. doi: 10.1073/pnas.1300524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Omari K. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013;3:30–35. doi: 10.1016/j.celrep.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong L. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kielian M., Rey F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baquero E. Intermediate conformations during viral fusion glycoprotein structural transition. Curr. Opin. Virol. 2013;3:143–150. doi: 10.1016/j.coviro.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-San Martin C. A stable prefusion intermediate of the alphavirus fusion protein reveals critical features of class II membrane fusion. Cell Host Microbe. 2008;4:600–608. doi: 10.1016/j.chom.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu R., Wilson I.A. Structural characterization of an early fusion intermediate of influenza virus hemagglutinin. J. Virol. 2011;85:5172–5182. doi: 10.1128/JVI.02430-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y.H. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20992–20997. doi: 10.1073/pnas.1116034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardone G. Visualization of the two-step fusion process of the retrovirus avian sarcoma/leukosis virus by cryo-electron tomography. J. Virol. 2012;86:12129–12137. doi: 10.1128/JVI.01880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albertini A.A. Characterization of monomeric intermediates during VSV glycoprotein structural transition. PLoS Pathog. 2012;8:e1002556. doi: 10.1371/journal.ppat.1002556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao S., Zhang W. Characterization of an early-stage fusion intermediate of Sindbis virus using cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13362–13367. doi: 10.1073/pnas.1301911110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modis Y. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 33.Chernomordik L.V., Kozlov M.M. Membrane hemifusion: crossing a chasm in two leaps. Cell. 2005;123:375–382. doi: 10.1016/j.cell.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 34.Gibbons D.L. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell. 2003;114:573–583. doi: 10.1016/s0092-8674(03)00683-4. [DOI] [PubMed] [Google Scholar]
- 35.Stiasny K. Characterization of a membrane-associated trimeric low-pH-induced form of the class II viral fusion protein E from tick-borne encephalitis virus and its crystallization. J. Virol. 2004;78:3178–3183. doi: 10.1128/JVI.78.6.3178-3183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Libersou S. Distinct structural rearrangements of the VSV glycoprotein drive membrane fusion. J. Cell Biol. 2010;191:199–210. doi: 10.1083/jcb.201006116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maurer U.E. The structure of herpesvirus fusion glycoprotein B-bilayer complex reveals the protein–membrane and lateral protein–protein interaction. Structure. 2013;21:1396–1405. doi: 10.1016/j.str.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanovic T. Influenza-virus membrane fusion by cooperative fold-back of stochastically induced hemagglutinin intermediates. eLIFE. 2013;2:e00333. doi: 10.7554/eLife.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leikina E. Influenza hemagglutinins outside of the contact zone are necessary for fusion pore expansion. J. Biol. Chem. 2004;279:26526–26532. doi: 10.1074/jbc.M401883200. [DOI] [PubMed] [Google Scholar]
- 40.Shepard C.W. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 41.Drummer H.E. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J. Gen. Virol. 2007;88:1144–1148. doi: 10.1099/vir.0.82567-0. [DOI] [PubMed] [Google Scholar]
- 42.Ronecker S. Formation of bovine viral diarrhea virus E1-E2 heterodimers is essential for virus entry and depends on charged residues in the transmembrane domains. J. Gen. Virol. 2008;89:2114–2121. doi: 10.1099/vir.0.2008/001792-0. [DOI] [PubMed] [Google Scholar]
- 43.Patel J. The transmembrane domain of the hepatitis C virus E2 glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology. 2001;279:58–68. doi: 10.1006/viro.2000.0693. [DOI] [PubMed] [Google Scholar]
- 44.Durantel D. Study of the mechanism of antiviral action of iminosugar derivatives against bovine viral diarrhea virus. J. Virol. 2001;75:8987–8998. doi: 10.1128/JVI.75.19.8987-8998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiland E. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J. Virol. 1990;64:3563–3569. doi: 10.1128/jvi.64.8.3563-3569.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Branza-Nichita N. Antiviral effect of N-butyldeoxynojirimycin against bovine viral diarrhea virus correlates with misfolding of E2 envelope proteins and impairment of their association into E1-E2 heterodimers. J. Virol. 2001;75:3527–3536. doi: 10.1128/JVI.75.8.3527-3536.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vieyres G. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J. Virol. 2010;84:10159–10168. doi: 10.1128/JVI.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Modis Y. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skehel J.J., Wiley D.C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 50.Donald J.E. Transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3958–3963. doi: 10.1073/pnas.1019668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stein A. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dupressoir A. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. 2012;33:663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 53.Gardner A., Welch J.J. A formal theory of the selfish gene. J. Evol. Biol. 2011;24:1801–1813. doi: 10.1111/j.1420-9101.2011.02310.x. [DOI] [PubMed] [Google Scholar]


