Abstract
We here describe the first reported comprehensive analysis of Hsp90 paralogue affinity and selectivity in the clinical Hsp90 inhibitor chemotypes. This has been possible through the development of a versatile experimental assay based on a new FP-probe (16a) that we both describe here. The assay can test rapidly and accurately the binding affinity of all major Hsp90 chemotypes and has a testing range that spans low nanomolar to millimolar binding affinities. We couple this assay with a computational analysis that allows for rationalization of paralogue selectivity and defines not only the major binding modes that relay pan-paralogue binding or, conversely, paralogue selectivity, but also identifies molecular characteristics that impart such features. The methods developed here provide a blueprint for parsing out the contribution of the four Hsp90 paralogues to the perceived biological activity with the current Hsp90 chemotypes and set the ground for the development of paralogue selective inhibitors.
INTRODUCTION
The heat shock protein 90 (Hsp90) chaperones have been the subject of more than 30 years of intensive biological and translational research, the fruits of which are reflected in the more than 20 Hsp90 inhibitors that have been or are currently in clinical trials for the treatment of cancers.1,2 Despite such intense interest, however, it is surprising that while we know a lot about Hsp90, the cytoplasmic chaperone, we know actually very little about Hsp90, the four-paralogue family, and even less about how these paralogues influence the activity of the clinical Hsp90 inhibitors. Indeed, the biennial “Hsp90 Chaperone Machine” meeting held in Switzerland in September 2012 concluded with a specific challenge to understand the roles of Hsp90 paralogues in disease and their contribution to the observed activity of Hsp90 inhibitors.3
Hsp90 is a family of molecular chaperones that function to fold client proteins to their active conformation through their ATPase activity.4 There are four different paralogues of Hsp90; Hsp90α and Hsp90β in the cytoplasm, Grp94 in the endoplasmic reticulum, and Trap-1 in the mitochondria. It was generally believed that the cytoplasmic Hsp90α/β paralogues have the most relevance in cancer because both are overexpressed and because of the oncogenic nature of their client proteins.5 Many of these clients (i.e., HER2, EGFR, mutant ER, HIF1α, Raf-1, AKT, mutant p53) are involved in signal transduction pathways, cell-cycle regulation, and apoptosis, pathways commonly deregulated in cancer. As a result, inhibition of cytosolic Hsp90 has attracted much interest as an anticancer targeting modality, with numerous small-molecule inhibitors binding to the N-terminal nucleotide-binding domain (NBD) of Hsp90 being currently evaluated in clinical trials.1
However, it is becoming increasingly clear that the organelle-specific chaperones Grp94 and Trap-1 also play a role in cancer. An important function of Grp94 is to properly fold secreted and membrane proteins, and its client proteins include immunoglobulins, Toll-like receptors, and integrins.6 High Grp94 expression in tumors is also associated with increased proliferation, metastasis, and drug resistance.7,8 Trap-1 is another important cancer chaperone that is overexpressed in tumors9,10 and leads to multidrug resistance.10 In tumor cells, Trap-1, along with Hsp90α/β, act in maintaining mitochondrial integrity by protecting against oxidative stress and apoptosis,9 and these antiapoptotic functions are also exploited by cancer cells.
These advances in understanding paralogue biology underscore the importance of gaining parallel knowledge on how inhibition of each paralogue contributes to the biological activity observed with several Hsp90 chemotypes currently in clinical evaluation for cancers. The molecules that have thus far advanced into clinical trials are recognized as pan-Hsp90 inhibitors, but evidence that they may modulate paralogues with distinct affinity and that such selectivity, even if minor, may contribute to differences in phenotypes observed with such compounds, is scattered throughout the literature.11–14
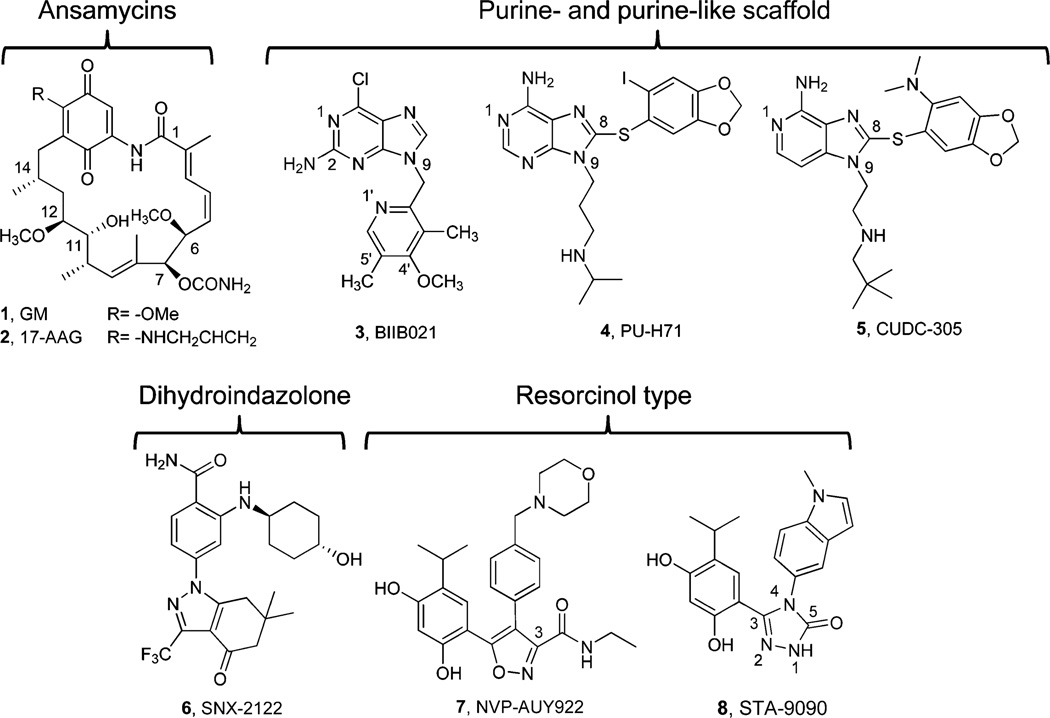
Geldanamycin (GM; 1) was the first Hsp90 inhibitor to be identified15 and is a naturally occurring benzoquinone ansamycin isolated from a fermentation broth of Streptomyces hygroscopicus.16 As a result of its overt toxicity in vivo as well as poor pharmaceutical properties, 1 never entered into clinical trials. A closely related analogue with improved toxicity properties, 17-allyl-17-demethoxygeldanamycin (17-AAG; 2),17 became the first Hsp90 inhibitor to advance into clinical trials. However, by this time, intense efforts by academia and industry were already underway to discover inhibitors of a more “drug-like” character. These efforts have resulted in the development of numerous diverse molecules with improved pharmaceutical properties to have entered into clinical trials for their evaluation in cancer. Some of these are shown in Figure 1 and include the purine-scaffold [3 (BIIB021),18 4 (PU-H71)19 and 5 (CUDC-305)20], dihydroindazolone derivatives [(1r,4r)-4-((2-carbamoyl-5-(6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)phenyl)amino)cyclohexyl 2-aminoacetate (SNX-5422), the glycine prodrug of 6 (SNX-2112)21], and resorcinol [7 (NVP-AUY922)22 and 8 (STA-9090)23].
Figure 1.
Structures of various Hsp90 inhibitors and the chemotypes they belong to. All but geldanamycin have been or currently are in clinical evaluation for cancers.
We here describe the development of a testing module that combines rapid experimental and computational tools and methods to analyze the paralogue specificity and paralogue-binding mode of Hsp90 inhibitors. We validate this module to provide the first reported analysis of the paralogue binding affinity, selectivity, and binding mode of the major Hsp90 chemotypes currently in clinical evaluation for cancers.
RESULTS
Fluorescence Polarization As an Assay for Hsp90 Paralogues
Most assays published to date and used to assess binding of small molecules to the four Hsp90 paralogues, such as intrinsic tryptophan fluorescence, affinity-resin competitive binding, and isothermal titration calorimetry,24–28 are laborious and costly as they make use of significant amounts of proteins. For the cytosolic Hsp90, since we introduced the use of fluorescence polarization (FP),29,30 this assay has become one of the most extensively used to identify and test Hsp90 inhibitors.29–33 There are numerous reasons why FP is an ideal method for measuring protein–ligand interactions and why it has become a favorite tool for Hsp90. First, it is a quick, homogeneous, i.e., there is no necessity for separation of free and bound ligand, highly reproducible, and facile for automation assay. By simply mixing a protein with a fluorescently labeled ligand, FP is able to measure real-time protein–ligand interactions in solution where binding of the fluorescently labeled ligand, also referred to as an FP probe, to a protein results in increased polarization values and is directly proportional to the fraction of bound ligand.34 Its theory, first described in 1926 by Perrin, is based on the observation that fluorescent molecules in solution, excited with plane-polarized light, will emit light back into a fixed plane (i.e., the light remains polarized) if the molecules remain still during the excitation of the fluorophore. Molecules, however, rotate and tumble, and the planes into which light is emitted differ from the plane used for initial excitation. Nonetheless, upon binding of the small probe to a protein (i.e., a large, slowly rotating molecule), motility is reduced, leading to higher FP. Unlabeled ligands that bind to the protein will compete with the probe, leading to lower FP. FP therefore provides a direct readout of the extent of probe binding to a protein.35 Second, FP is also well suited for Hsp90 because it is an assay that requires no engineering of the protein. As reported, Hsp90 is a highly flexible molecular chaperone whose function is very sensitive to interference with its conformational modality,36 such as the attachment of labels may lead to, and thus FP is best suited for this class of proteins. Third, there are numerous Hsp90 inhibitor chemotypes for which extensive chemistry has been developed and binding to Hsp90 revealed by crystallography, and therefore the choice for the FP probe and knowledge over the site of its fluorescence labeling is available.21–23,37–44 To date, however, no FP assay that efficiently tests for affinity and selectivity of small molecules to all four Hsp90 paralogues has been reported.
Suitability of FP Probes for Pan-Hsp90 Paralogue Binding
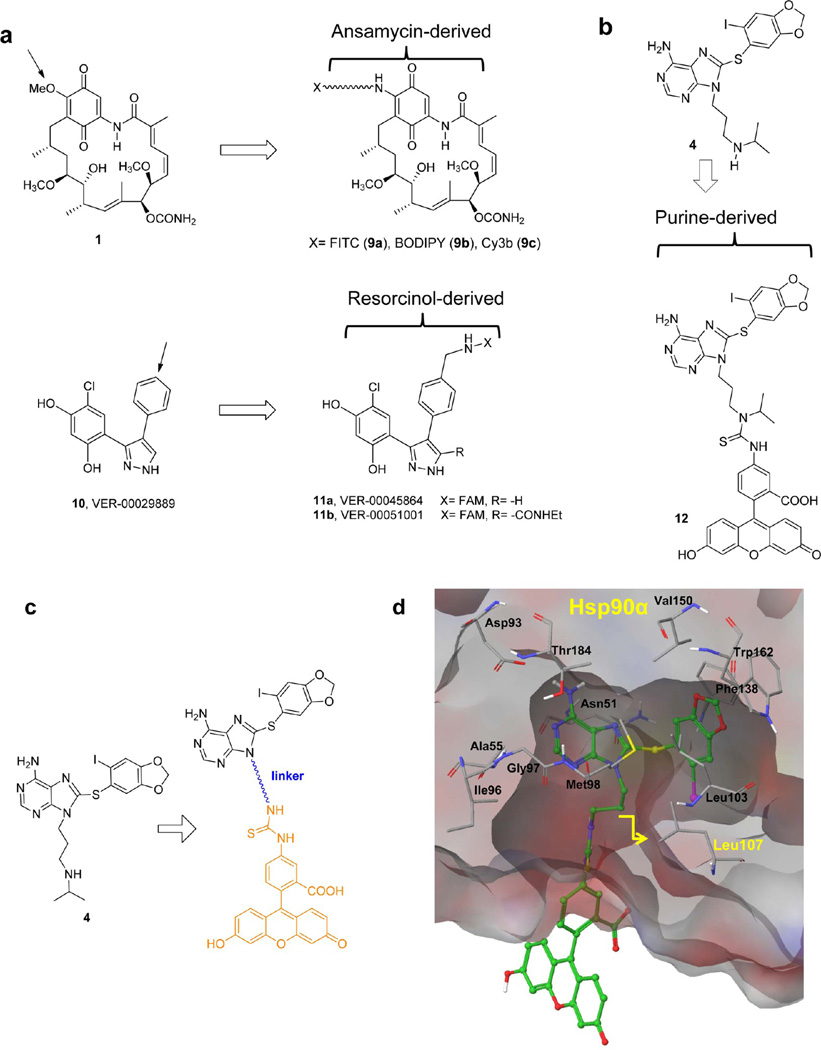
Several FP tracers for Hsp90 that bind to the NBD have been reported and include a number based on 1 [9a (GM-FITC),29 9b (GM-BODIPY),29 9c (GM-Cy3b),33 Figure 2a] and two carboxyfluorescein (FAM) probes based on the pyrazole scaffold [11a (VER-00045864)32 and 11b (VER-00051001),22 Figure 2a]. More recently, an FP probe of a derivative of Sansalvamide A-amide has been reported,45 which, in contrast to other probes, bindsHsp90 at the N-terminal/middle domains. It should also bementioned that a radicicol-based probe, RD-TAMRA, has been used in related confocal two-dimensional fluorescence intensity distribution analysis (2D-FIDA) assays in a high-throughput screening (HTS) format.46 However, radicicol based probes have the drawback of being unstable in assay buffer and requires preparation of fresh stock solution daily.46
Figure 2.
Selected reported fluorescently labeled Hsp90 inhibitors for use in FP (a) or flow cytometry and fluorescent microscopy (b). Arrow indicates point of attachment to ligand of linker-fluorophore. (c) Design of the FP probe based on the Hsp90 inhibitor 4. (d) Probe 16a docked into the Hsp90α ATP binding pocket (PDB ID: 2FWZ) as generated by Glide (version 5.0). Modeling shows potential steric clash between probe and Leu107 for linkers containing less than three carbons.
None of the aforementioned probes have been systematically assessed for their suitability as tracers in FP assays with each of the paralogues. While the geldanamycin and pyrazole probes have been extensively used to measure binding to Hsp90α, Hsp90β as well as to total Hsp90 in a cancer cell lysate, their use in measuring binding to Grp94 and Trap-1 is more limited.41,47,48 In our own hands, we have found 9c to be inadequate as a tracer for Trap-1 (see below). To get a suitable assay window, its use requires a considerable amount of protein and is therefore less suitable for large structure–activity relationship studies or HTS efforts.
We therefore proceeded here to design FP probes based on the Hsp90 inhibitor 4 labeled through different linkers with fluorescein (Figure 2b,c). We hypothesized that due to its known binding mode and well established chemistry, useful FP probes amenable for paralogue-selectivity testing may be created around this ligand. 12, a fluorescein isothiocyanate (FITC) derivative of 4 with optimal properties for flow cytometry and fluorescence microscopy, was also included in our analysis (Figure 2b).49
The linker and its attachment mode, both important in the synthesis of FP chemical probes because they can affect binding to the target protein, can be predicted for 4 from the available structural studies.39 For the preparation of a suitable FP probe, it is also important that the linker not be excessively long or flexible because of the propeller effect.50,51 Depolarization due to flexibility in the attachment of the dye, referred to as the “propeller effect”, distorts the relationship between FP and molecular weight. For this reason, it is generally preferable to use dyes without long aliphatic linkers between the fluorophore and the reactive group in the preparation of FP assay probes.
To determine a linker length optimal for binding to all Hsp90 paralogues, we docked the linker-modified ligands of 4 into the respective paralogues of Hsp90, i.e., Hsp90α (PDB ID: 2FWZ39), Hsp90β (PDB ID: 3NMQ41), Grp94 (PDB ID: 3O2F,52 2EXL43), and Trap-1 (Homology Model52). On the basis of our molecular modeling analysis (Figure 2d), as well as our extensive experience with this class of molecule,39,49,53 we decided to attach FITC to the N9-position of the purine-scaffold via at least a three carbon linker as this would orient the probe toward solvent without affecting binding to the target protein (Figure 2c). A shorter linker would lead to clashes between the probe and a leucine residue positioned in all paralogues at the exit of the binding site (Leu107 in Hsp90α and Hsp90β, Leu163 in Grp94, and Leu172 in Trap-1) (Figure 2d for Hsp90α).
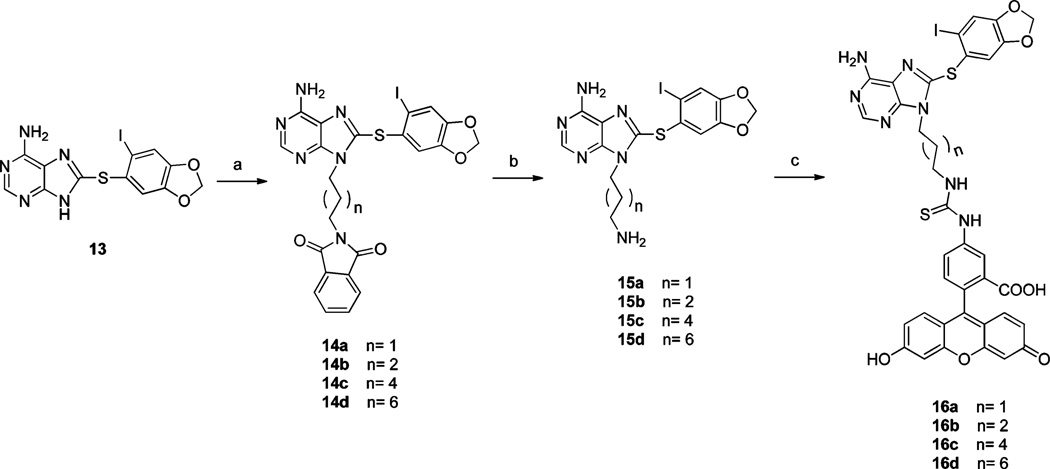
We also wanted to determine the optimal chain length for FP properties. To accomplish this, we synthesized a number of probes with linkers ranging from 3 to 8 carbons (Scheme 1). These were prepared by a three-step sequence from 13, commencing with N9-alkylation with ω-bromophthalimides to yield 14a–d (Scheme 1). Following unmasking of the amine with hydrazine and attachment of FITC, 16a–d were obtained after HPLC purification (Scheme 1).
Scheme 1a.
aReagents and conditions: (a) N-(ω-bromoalkyl)-phthalimide, Cs2CO3, DMF, rt; (b) hydrazine hydrate, MeOH, CH2Cl2, rt; (c) FITC, Et3N, DMF, rt.
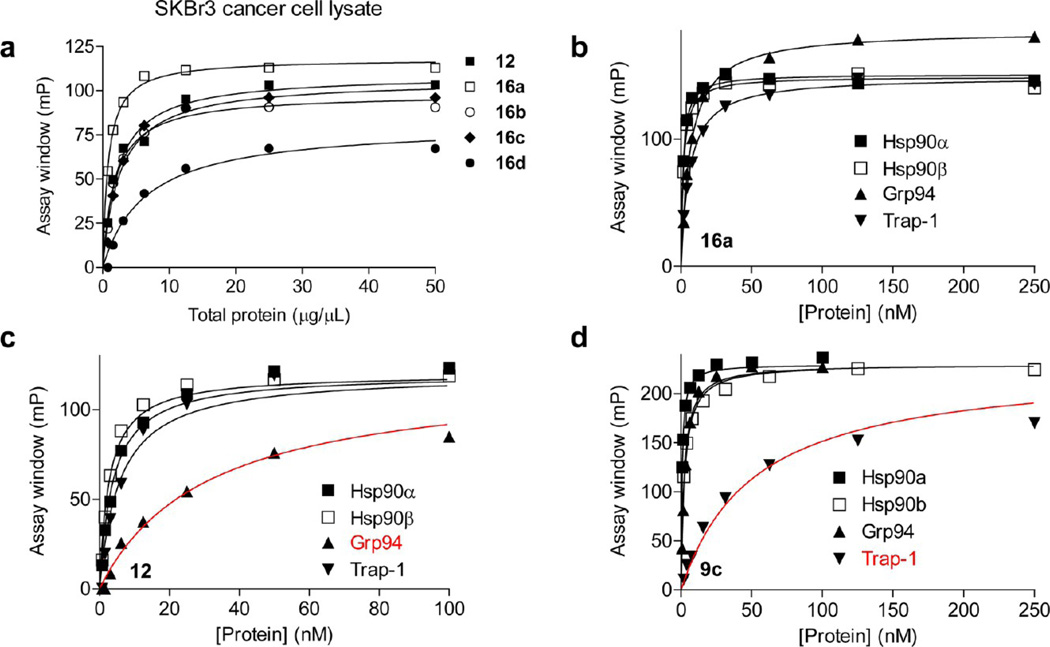
The synthesized FITC-derivatives were first evaluated for their potential as FP tracers for binding to Hsp90 in a cancer cell homogenate (Figure 3a). The decision to perform the primary screen with a cancer cell lysate was primarily an economic one because the cost to us of obtaining lysate was significantly less than using purified protein and partly out of our routine practice of using such lysates in FP assays to exploit the Hsp90 cancer complex. The potential tracers were initially evaluated by titration with increasing amounts of lysate up to 50 µg of total protein (Figure 3a). To be useful in FP, the binding affinity of the probe should be high and the binding range (i.e., assay window), defined as the millipolarization (mP) value at saturation minus the mP recorded for probe alone should be large. As observed in Figure 3a, as the amount of lysate, and thus of Hsp90, increased, so did the assay window. Good performance was observed for all probes, with an excellent assay window of >100mP for 16a. Similar to other Hsp90 FP assay probes, when measured at 4 °C to maintain proper folding of Hsp90, the binding assay between 16a and Hsp90 reached equilibrium by 8 h and remained stable for more than 24 h (not shown). While 16a, the analogue with a 3-carbon linker, was optimal, 12, the N-isopropyl analogue of 16a, and 16b and 16c, the 4- or 6-carbon linker compounds, respectively, performed acceptably well (Figure 3a). The significantly decreased assay window observed for 16d, the probe with an 8-carbon linker, can likely be attributed to the propeller effect.50,51
Figure 3.
Dose–response curve for the binding of indicated probes to the Hsp90 paralogues from a cancer cell extract (a) or to individual Hsp90 paralogues (b–d). Different amounts of protein were incubated with the ligand at 4 °C and the response measured at equilibrium (24 h). The assay window data were obtained by subtracting free probe values from values recorded in the presence of specified protein concentrations. Data were analyzed and plotted in Prism 4.0. Average values from duplicate experiments are presented.
We next determined, in a standard saturation binding experiment that measures ligand binding in the presence of varying concentration of protein, the ability of these ligands as probes for the Hsp90 paralogues (Figure 3b–d). For comparison, we also included 9c, a probe we have previously developed for FP assays of Hsp90.33 Taken as a whole, saturation binding experiments with 16a showed it to be an excellent tracer for each Hsp90 paralogue with an assay window of >150 mP and an apparent Kd = 1.4, 1.6, 6.6, and 5.9 nM for Hsp90α, Hsp90β, Grp94, and Trap-1, respectively (Figure 3b), and we proceed further here to use it as a probe in evaluating the paralogue-selective binding of Hsp90 inhibitors and Hsp90 endogenous ligands. Interestingly, 12 showed a 1-log preference for Hsp90α, Hsp90β, and Trap-1 over Grp94 (apparent Kd = 3.9, 2.8, 30.7, and 5.8 nM for Hsp90α, Hsp90β, Grp94, and Trap-1, respectively) with a poor assay window, i.e., less than 100 mP, for Grp94 (Figure 3c). On the other hand, the geldanamycin-derived probe 9c was a good tracer for the cytosolic Hsp90s andGrp94 but displayed a 1- to 2-log weaker binding and inferior assay window when evaluated for Trap-1 (9c apparent Kd = 0.7, 2.2, 2.5, and 51.8 nMforHsp90α, Hsp90β, Grp94, and Trap-1, respectively; Figure 3d).
Suitability of the FP Assay for Evaluating the Selectivity and Affinity of Small Molecules for the Hsp90 Paralogues
Having found a probe that binds effectively to all four Hsp90 paralogues, we next validated its ability to evaluate paralogue affinity and selectivity for small-molecule ligands. Specifically, we evaluated the binding of ATP and ADP, the two endogenous Hsp90 paralogue ligands, for which paralogue binding affinity has been extensively explored by means of intrinsic tryptophan fluorescence and isothermal titration calorimetry.24–28 We observe for these ligands relative affinities in line with what has been reported previously for each paralogue (Table 1). Specifically, the ADP interaction with Hsp90 was reported to be much tighter than that of ATP (41 versus 840 µM),24 which is very much in line with our findings (Table 1). ADP was reported to be a slightly weaker binder of Hsp90α than of Hsp90β (51 versus 34 µM),25 which is what we find (59 versus 42 µM, Table 1). Additionally, as previously reported, we show Grp94 and Trap-1 to show little discrimination between both nucleotides.26–28,54,55 Grp94 binds both nucleotides relatively well, with binding affinities reported ranging from 2.3 to 3.4 µM26 to 5 µM,56 which compares well with 3.2 and 11.4 µM we recorded for ATP and ADP, respectively (Table 1). As in our study, ATP was found to be a slightly tighter binder of Grp94 than ADP.27 Trap-1, which most closely resembles the bacterial Hsp90, HtpG, is reported to bind ATP with approximately 10-fold greater affinity than does Hsp90.28,55 The reported values for ATP (3355 and 34.3 µM28) are in close concordance with our recorded value of 31.3 µM (Table 1). These confirm that our assay records with high accuracy the binding affinity and selectivity profile of Hsp90 paralogue ligands, and moreover, does so by using substantially lower amounts of protein, i.e., low nanomols versus the micromols of protein needed in the classical isothermal titration calorimetry and intrinsic tryptophan fluorescence experiments, with less labor and time. When comparing it to the classical techniques, it is also important to note that our FP assay only probes for binding to the same pocket the labeled probe binds. This is both an advantage because it probes a single binding pocket but may be a disadvantage if one is concerned about secondary/nonspecific binding sites.
Table 1.
Hsp90 Paralogue Affinity Determined for Hsp90 Inhibitors in Clinical Development Using 16a as a FP Probea
| Hsp90α IC50 (nM) |
Hsp90β IC50 (nM) |
Grp94 IC50 (nM) |
Trap-1 IC50 (nM) |
|
|---|---|---|---|---|
| 1 | 28 | 22 | 10 | 661 |
| 2 | 46 | 45 | 31 | 1496 |
| 3 | 19 | 17 | 124 | 90 |
| 4 | 43 | 42 | 30 | 205 |
| 5 | 33 | 38 | 190 | 1586 |
| 6 | 29 | 25 | 578 | 726 |
| 7 | 20 | 16 | 12 | 38 |
| 8 | 5 | 5 | 10 | 51 |
| ADP | 59308 | 42159 | 11447 | 55594 |
| ATP | 861330 | 893677 | 3241 | 31303 |
The paralogue binding affinity of Hsp90-regulatory nucleotides is also presented.
We next used the newly developed FP assay to evaluate the paralogue affinity and selectivity of Hsp90 NBD inhibitors encompassing a variety of chemical classes (Figure 1). All but 1 have been or still are in clinical evaluation for cancers.1 The results for each paralogue of Hsp90 are summarized in Table 1 and show that all inhibitors effectively compete with 16a, demonstrating specificity of binding for the probe. The low nanomolar binding affinities for Hsp90α/β that we measured for these inhibitors correlate well with their biological activity determined in several cancer cells.20,23,48,57–60
Interestingly, while it is believed that the clinical Hsp90 inhibitors bind equally well to all paralogues, we determined a spectrum of paralogue binding preferences (Table 1). Of note, all these inhibitors bound approximately equally well and with low nanomolar affinity to the cytosolic Hsp90s, as indicated previously by the extensive interactions they form with the pocket.19,26–29
In contrast, we found a significant difference among the several agents with regards to their affinity for Grp94 and Trap-1. Most striking was an almost 2-log loss of affinity for Trap-1 recorded for 2 and 5 (Hsp90 vs Trap-1: 46 nM vs 1.5 µM for 2, 33 nM vs 1.5 µMfor 5). Lower binding efficacy for Trap-1 was also seen for the other agents, with a decrease ranging from 25-fold for 1 and 6, 10-fold for 8 to 5-fold for 3 and 4 and 2-fold for 7. The affinity of these agents for Grp94, while comparable to Hsp90 for most agents, was substantially lower for a few inhibitors. Specifically, an approximately 10-fold loss of affinity was noted for 3, 5, and 6 (Hsp90 vs Grp94: 19 vs 124 nM, 33 vs 190 nM, and 29 vs 578 nM, respectively).
Paralogue Affinity and Selectivity in the Hsp90 Inhibitor Chemotypes: A Computational Analysis
In an attempt to rationalize some of these differences and build a structural model that can be used to analyze and predict inhibitor binding to the Hsp90 paralogues, we overlaid the crystallographic and/or the docked pose of 4 (PDB: 2FWZ), 5 (docked in PDB: 2FWZ), 3 (PDB: 3QDD),44 6 (docked in PDB: 3D0B),42 7 (PDB: 2VCI),22 8 (PDB: 3TUH), and 1 (PDB: 1YET,37 2EXL43) into the binding site of each paralogue, i.e., Hsp90α (PDB ID: 2FWZ), Hsp90β (PDB ID: 3NMQ), Grp94 (PDB ID: 2EXL, 3O2F), and Trap-1 (Homology Model52).
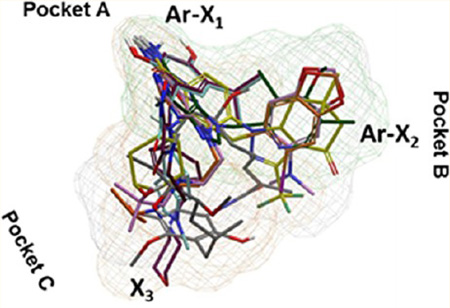
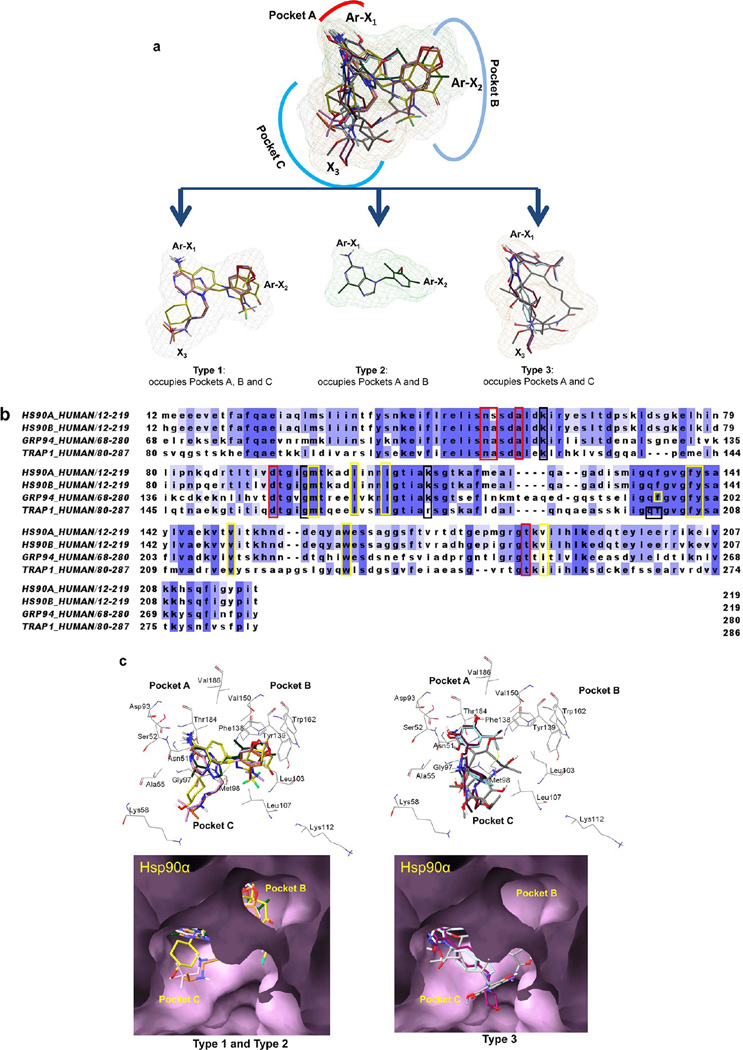
Common to all molecules are critical interactions with what we refer here as pocket A (Figure 4a). Specifically, a key interaction is formed with a conserved Asp93 of Hsp90α/β and a network of water molecules at the bottom of the ATP-binding pocket, pocket A. In the case of purine-scaffold compounds (3–5), this interaction is maintained by the exocyclic −NH2 and N1 of the adenine or imidazopyridine core or the resorcinol moiety for 7 and 8 or the −CONH2 of 6. These groups are referred to as X1 here and are essential elements for binding not only to Hsp90α/β but also to Grp94 (Asp149) and Trap-1 (Asp158). As can be seen in the overlaid structures (Figure 4a), the Ar-X1 group in each molecule remains static in the binding of each isoform. Naturally then, the origins for the differences in affinity observed to each isoform is as a result of suboptimal binding that occurs in other regions of the molecule.
Figure 4.
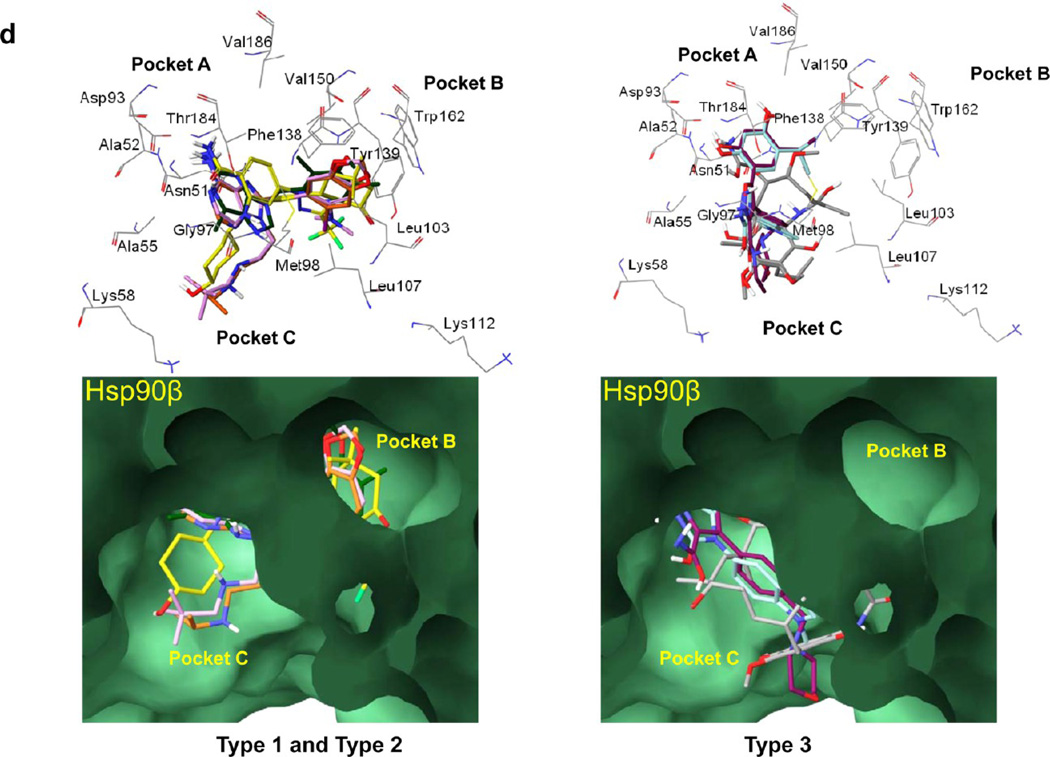
(a) Overlay of 1 (gray), 3 (dark-green), 4 (orange), 5 (plum), 6 (yellow), 7 (maroon), and 8 (turquoise) into the Hsp90 paralogue binding site. (b) Alignment of the N-terminal domains of Hsp90α (PDB ID: 2FWZ), Hsp90β (PDB ID: 3NMQ), Grp94 (accession no. P14625), and Trap-1 (accession no. Q12931). Dark-blue to light-blue: conserved to less conserved residues. Red, yellow, and black boxed letters: amino acids interacting with the ligands in pockets A, B, and C, respectively. (c) Interactions of type 1, 2, and 3 compounds with Hsp90α (PDB ID: 2FWZ). (d) Interactions of types 1, 2, and 3 compounds with Hsp90β (PDB ID: 3NMQ). Top, wire and bottom, surface representations of residues. 1 (gray), 3 (dark-green), 4 (orange), 5 (plum), 6 (yellow), 7 (maroon), and 8 (turquoise).
For the purposes of this discussion and in our attempt to explain these differences, we refer to these other groups as Ar-X2 and X3 (Figure 4a). The X2 group occupies a lipophilic pocket B, and the X3 group occupies the exit pocket C that is oriented toward solvent and observed differences in potencies are as a result of suboptimal binding in the B and C pockets. Upon closer inspection of the pockets they occupy, the Hsp90 inhibitor chemotypes can be further divided in type 1, type 2 and type 3. Type 1 occupies all three available pockets, whereas the binding of type 2 is restricted to pockets A and B and of type 3 to pockets A and C (Figure 4a).
We next investigated whether significant residue differences exist among these paralogues in pockets B and C that could account for such differences. Each of the paralogues contain three conserved domains: N-terminal nucleotide binding domain (NBD), a middle domain, and a C-terminal dimerization domain. The NBD is the most highly conserved domain and as mentioned previously is the binding site for each of the inhibitors described here. Alignment of the NBD for each of the isoforms shows the overall similarity of the N-terminal domain of these paralogues (Figure 4b). The amino acids depicted in boxes are those that directly interact with the ligands, and although the amino acids making up the individual binding pockets are highly similar, a number of differences do exist. In pocket A of Hsp90α, a serine residue (Ser52) replaces an alanine in each of the other isoforms (Ala52 in Hsp90β, Ala108 in Grp94, and Ala120 in Trap-1). In pocket B, valine in Hsp90 (Val186 in Hsp90α and Hsp90β) is replaced by isoleucine in Grp94 (Ile247) and Trap-1 (Ile253). In pocket C, lysine in Hsp90 (Lys112 in Hsp90α and Hsp90β) and Grp94 (Lys168) is replaced by arginine in Trap-1 (Arg177).
With this structural information combined with the findings of our experimental binding assays, we next proceed to analyze the affinity and selectivity of type 1, 2, and 3 compounds for the four Hsp90 paralogues.
Type 1 Compounds
As previously reported for 8-benzyladenine series of inhibitor (purine-scaffold), a conformational change in the flexible lid of Hsp90α/β occurs upon ligand binding and creates a new binding channel (PDB: 1UYM).61 In this class, the 8-aryl moiety rearranges the flexible Hsp90α/β lid, exposing a lipophilic pocket (B) whereby the phenyl ring becomes stacked between Phe138 and Leu107 and forms further favorable interactions with Met98 and Leu103. The N9-chain is oriented toward an exit pocket (C) that is blocked somewhat by Leu107 (Figure 4c,d; type 1). Although the first and second atoms of the alkyl chain form hydrophobic interactions with Leu107 and Met98 and are important for binding, the remainder of the side chain is solvent exposed. Each of the purine and purine-like scaffold compounds (4, 5) bind in a more or less similar way, however, there are certain structural differences that can explain preferences in paralogue binding. 6, a compound of a rather unique scaffold, also inserts into pockets A, B, and C (Figure 4c,d; type 1). The gem-dimethyl groups of the indazolone ring are buried in the lipophilic pocket (B) formed by Phe138, Leu107, and Trp162, while the carbonyl group may form a favorable H-bonding interaction with the side-chain of Tyr139. The trans-4-hydroxycyclohexylamino group occupies the exit pocket (C) and makes favorable hydrophobic interactions with Met98 and Ala55, whereas the 4-hydroxy group may form an H-bond with side chain of Lys58.21
Type 2 Compounds
3, while also based on the purine-scaffold, incorporates a concomitant shift of the exocyclic amine and of the aryl group to the 2- and 9-position, respectively, of the purine core. This shift results in orientation of these groups similar to those of the amino and 8-aryl groups of 4 and 5. Because of this shift in position of the above functionalities 3 lacks the chain that extends, as seen in 4 and 5, inside the exit pocket C. Thus, binding of 3 is restricted to pockets A and B (Figure 4a,c,d; type 2).
Type 3 Compounds
We classify ansamycins (i.e., 1 and 2) as well as the resorcinols, 7 and 8, as type 3 compounds as they extend primarily into pockets A and C, and although each make some interactions with amino acids lining the entrance of pocket B, do not extend into it like type 1 and 2 compounds (Figure 4a,c,d). For resorcinols, it is known that the potency of these molecules is largely derived from the phenolic hydroxyls (Ar-X1) that form a very extensive network of H-bonds, and compounds of this class are recognized as some of the tightest known inhibitors. When bound to Hsp90α/β, the resorcinol moiety of 7 and 8 form hydrogen bonding interactions with Asp93 and Asn51 in pocket A (Figure 4c,d; type 3). Additionally, the oxygen atom of isoxazole ring of 7 is making a H-bond with the sidechain hydroxyl of Thr184, while the carbonyl oxygen and amido group on the 3-position of the isoxazole ring is making a H-bond with the side-chain of Lys58 and the carbonyl backbone of Gly97, respectively, in exit pocket C. Similar to 7, the N2 of the 1,2,4-triazol-5(4H)-one ring of 8 may make H-bonding interaction with Thr184 while 1-NH and the 5-carbonyl oxygen form H-bonding interaction with the backbone carbonyl of Gly97 and the side-chain amine of Lys58, respectively, in pocket C. Although the isopropyl group on the resorcinol moiety of both 7 and 8 does not extend into pocket B, it does however make hydrophobic interactions with amino acids lining the entrance, e.g., Phe138, Leu107, Val150, Val186.
The amido group of C7-carbamate moiety of 1 is making H-bonding interactions with Asp93 in pocket A (Figure 4c,d; type 3). The hydroxyl group at C11 is making H-bond with the side chain amino group of Lys58 and the benzoquinone carbonyl group may make H-bond with side chain amino group of Lys112 in pocket C. The C1-carbonyl oxygen of 1 forms H-bonding interaction with the backbone amino of Phe138 in close proximity to pocket C. Although 1 does not extend into pocket B, the C6-methoxy group does make some hydrophobic interactions with amino acids lining the entrance of the pocket as mentioned earlier, i.e., Phe138, Leu107, Val150, and Val186.
According to our classification, ATP/ADP would be a type 3 ligand because it binds only to pockets A and C. The adenine ring occupies pocket A (similar to the adenine ring of the purine-scaffold class) and the ribose occupies pocket C (similar to the N9-alkyl chain of 4). ATP/ADP is not able to bind within pocket B and therefore cannot make interactions with it. In fact, this pocket is closed (i.e., helical closed conformation) in crystal structures for ATP or ADP and was only first revealed when the structure of 9-butyl-8-(3,4,5-trimethoxybenzyl)-9H-purin-6-amine (PU3), a purine scaffold inhibitor, was obtained which showed what was at the time an unexpected conformational shift of residues 104–111 which was required in order to accommodate the 8-aryl substituent into pocket B.61
To summarize our analyses, type 3 compounds (as well as ADP/ATP) do not bind into pocket B. A conformational change in the B pocket region occurs when type 1 or type 2 compounds bind and the change in conformation observed for these compounds is identical, which causes the opening of the B pocket.
All of the molecules described above bind exceptionally well to Hsp90, which should not be of any surprise, because aside from the ansamycins, each were optimized to target Hsp90. We now describe below how differences in affinity for Grp94 and Trap-1 are a result of suboptimal binding that occurs in pockets B and C of these paralogues.
Analysis of Grp94 Binding
Grp94 is distinct from the other Hsp90 paralogues in that binding of ATP/ADP induces an opening of the lid region covering the nucleotide-binding pocket rather than closing it.54,62 The conformational changes leading to this opening are required in order to avoid a steric clash of the phosphate groups with Gly196 and result in a twisted “V” conformation whereby the N-terminal domains are oriented in opposite directions.54 Therefore, whereas the ligand free form of Grp94 adopts a closed conformation, binding of ATP results in an open conformation. Binding of geldanamycin into the ATP pocket also requires a conformational shift in order to avoid a steric clash with Gly196, however, such shifts are much less dramatic and the GM-bound structure more closely resembles the closed structure rather than the open structure.43 This is also the case for radicicol (PDB: 1U0Z), and it is likely that most of the pan-Hsp90 inhibitors described here induce conformational changes to various degrees in order to enable binding within the ATP-pocket. Such conformational changes in Grp94 are possible due to an insertion of amino acids in Grp94 (a 5-amino acid insertion, 182QEDGQ186; Figure 4b) not observed for the other three paralogues. Thus, the Gly196 induced conformational change, which results in reorientation of Phe199 in Grp94, is not possible in Hsp90α/β and Trap-1.
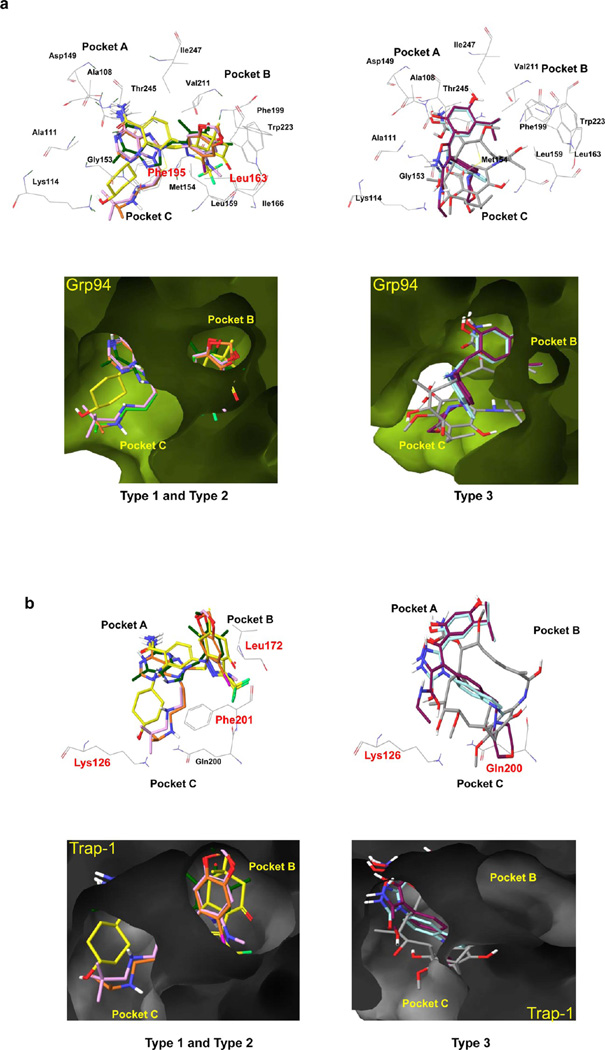
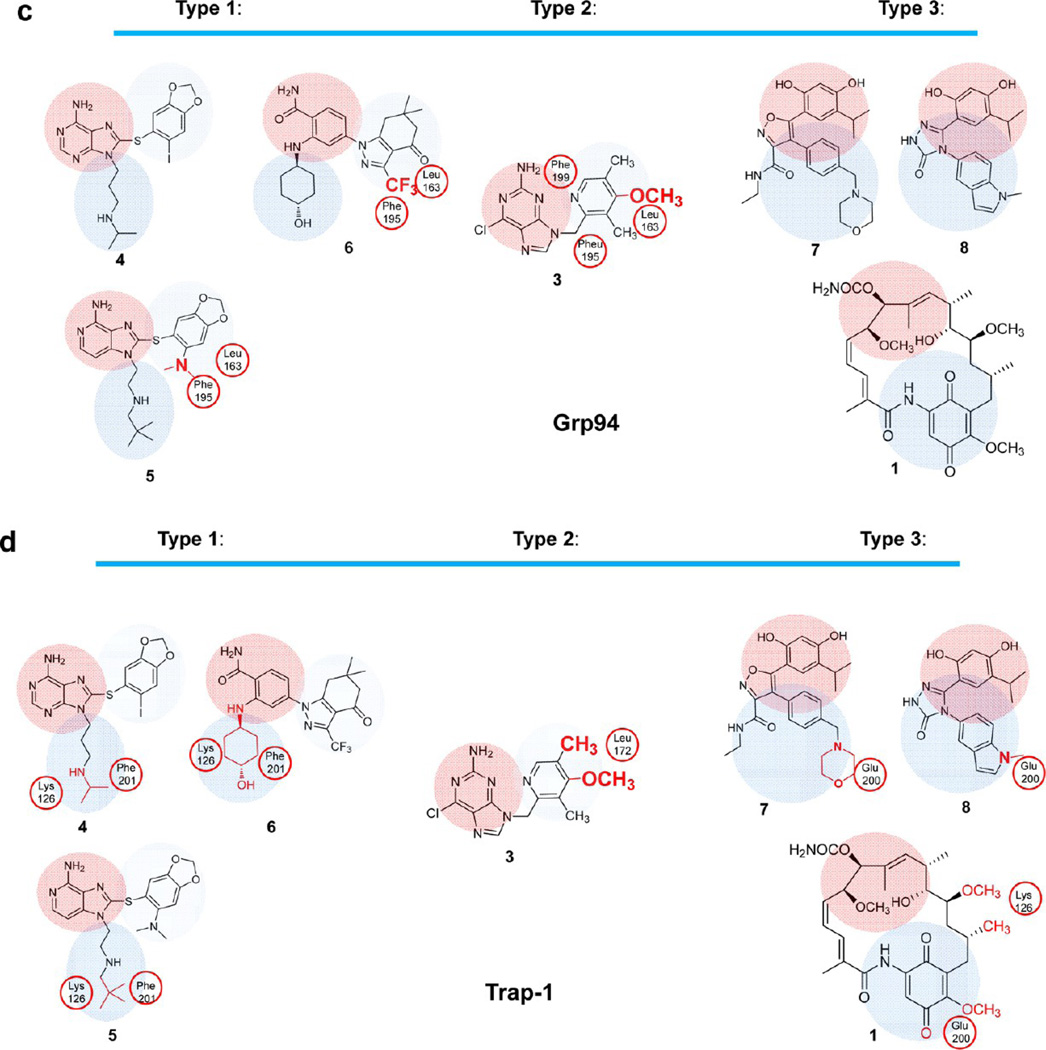
Our data shows that whereas 4 binds equally well to Hsp90α/β and Grp94, 5 and 3 each show approximately 6-fold decreased affinity for Grp94, while 6 has a 20-fold loss in affinity (Table 1). A crystal structure for any of the type 1 and 2 compounds in complex with Grp94 has yet to be reported, and modeling of these compounds into the ATP binding pockets of existing structures of nucleotide or GM-bound Grp94 did not reveal differences that could account for the observed binding affinity. We next analyzed a recently reported crystal structure of a derivative of 4, 8-((2,4-dimethylphenyl)thio)-3-(pent-4-yn-1-yl)-3H-purin-6-amine (PU-H54).52 In this structure, we observed that the helix 1,4,5 “lid” region in Grp94 adopted a novel “partially closed” conformation, whereby strand 1 and helix 1 were pulled away from the core of the N domain and helices 4 and 5 shifted upward to straddle the top of helix 1. These rearrangements also repositioned helix 3 of Grp94, resulting in a somewhat larger ATP binding pocket. We therefore used this pocket to understand the binding differences observed in type 1 and 2 compounds. While other lid conformations are possible for Grp94 and have been reported for this protein,43,54,62 we reasoned that the high affinity exhibited by these ligands, which are also in line with their affinity for Hsp90, can more likely be explained by insertion of these molecules in a pocket with an architecture similar to that seen in Hsp90. Thus, to investigate the binding mode of type 1 and 2 compounds into Grp94, we performed an induced fit docking of 4 into this pocket then overlaid other ligands onto the structure. The resulting analysis showed that the nature and size of groups on Ar-X2 are the likely cause of diminished binding as a result of suboptimal interaction with two residues located in pocket B (i.e., Leu163 and Phe195) of Grp94 (Figure 5a,c; type 1 and 2). 4 has an iodine atom, which is potentially of ideal size and lipophilicity to make strong hydrophobic contacts with the lipophilic residues in pocket B (Figure 5a,c). The relatively more polar −N(CH3)2, −OMe, and −CF3 groups in 5, 3, and 6, respectively, occupy the same space as the iodine in 4, and it is likely that suboptimal fit and interaction with the lipophilic residues in pocket B are the cause for the observed loss in activity of these compounds (Figure 5a,c; type 1 and 2).
Figure 5.
(a) Interactions of type 1, 2, and 3 compounds with Grp94 (PDB ID: 3O2F). (b) Interactions of type 1, 2, and 3 compounds with Trap-1 (homology model); 1 (gray), 3 (dark-green), 4 (orange), 5 (plum), 6 (yellow), 7 (maroon), and 8 (turquoise). (c,d) Schematic representation of Hsp90 inhibitors and the residues proposed to interfere with binding of these ligands into Grp94 (c) and Trap-1 (d).
The ansamycins and resorcinols we find to have good affinity for Grp94, and in concordance with our analysis thus far, can be explained by their inability to extend into pocket B (Figure 5a,c; type 3). The compounds 1 (and 2), 7, and 8 are type 3 compounds and are not dependent upon binding interactions within this pocket for affinity. As a result, they are unaffected by potential losses in affinity that might occur as a result of steric clashes and/or suboptimal interactions within pocket B, as observed for most of the type 1 and 2 compounds described above.
Analysis of Trap-1 Binding
No crystal structure of Trap-1 has been reported so far. Intrinsic fluorescence experiments suggest that binding of ATP to Trap-1 induces conformational changes that result in a predominantly closed conformation, similar to that observed in Hsp90.28 However, in contrast to Hsp90, this is insufficient to commit Trap-1 to hydrolysis because reopening occurs significantly faster. This suggests considerable conformational flexibility, and coupled with the current lack of available crystal structures of Trap-1, either apo or ligand bound, makes analysis of the observed binding affinities difficult. To overcome this problem and to better understand the requirements for Trap-1 binding, we created a homology model that presents Trap-1 in a conformation also seen for Hsp90α and Hsp90β. When creating this model, we hypothesized that the 2-amino acid insertion noted in Trap-1 (190AE192) would be insufficient to allow for a conformational change such as observed in Grp94. This homology model incorporated a number of elements we believed to be essential. First, we felt it essential that the B pocket be accessible to binding in order to allow the type 1 and 2 compounds to bind. Second, we required that the relative lipophilic nature of the Trap-1 binding pocket be adequately represented. To meet these two objectives, we used Hsp90α (PDB ID: 2FWZ) and Grp94 (PDB ID: 3O2F) crystal structures as a template for the amino acids. While similar in several respects to the Hsp90 structure, the model revealed several important differences not evident by simply aligning the two proteins (Figure 4b). Specifically, differences in the reorientation of several pocket amino acids such as Phe201, Lys126, and Asn171 were seen, which in this model are positioned toward the exit of the pocket. Other differences were noted in the orientation of analogous amino acids such as Leu107 and Ala111 (Hsp90α/β) and Leu172 and Ala176 (Trap-1).
With this model in hand, we next proceeded to overlay the crystallographic structures of the clinical candidates into our homology model.52 As mentioned above, in the case of Trap-1, the binding affinity for 4, 3, and 5 diminished 5-, 5-, and 50-fold, respectively, when compared to Hsp90 (Table 1). For 6, this drop was 25-fold. The homology model predicted numerous causes for such substantial loss of affinity when compared to Hsp90. One such cause is the nature of the exit channel, pocket C. Whereas for Hsp90α/β there are no amino acids interfering with the chains/functionalities inserted into pocket C by these inhibitors, in Trap-1, both Lys126 and Phe201 partially occlude the path of the chain to solvent (Figure 5b,d; type 1 and 2). Specifically important is the clash of the bulkier tert-butyl group in 5 with these residues. In 4, the smaller isopropyl group leads to less interference and thus a smaller drop in affinity. The cyclohexyl moiety in 6 also interferes with Lys126 and Phe201, leading to the 25-fold drop in affinity.
Another potential for reduced affinity with Trap-1 may come from unfavorable interactions that occur between Ar-X2 functionalities and Leu172. Such is the case for 3, where the lack of an X3 moiety precludes clashes within the exit pocket C, and its 5-fold decrease in affinity could be attributed to unfavorable steric interactions between its 4′-OMe and 5′-Me groups with Leu172 (Figure 5b,d; type 1 and 2). This bulky residue may also sterically interfere with the iodine of 4 and −N(CH3)2 of 5 and hence be partly responsible for the observed loss in affinity.
As mentioned above, the potency of the type 3 compounds 7 and 8 is largely derived from the resorcinol moiety (Ar-X1), which forms a very extensive network of H-bonds in pocket A. Although these remain potent for Trap-1, some loss in potency for both these compounds could be due to unfavorable interactions within the exit channel C, as the morpholine group of 7 and the indole N-methyl of 8 orient toward Glu200 (Figure 5b,d; type 3). Similarly, a loss of affinity for Trap-1 was also observed with 1 and 2, whereby the quinone ring (Figure 5b,d; type 3) unfavorably neighbors Glu200 while the 12-methoxy and 14-methyl substituents are determined to have unfavorable steric interactions with Lys126.
CONCLUSION
In conclusion, we here describe the first reported comprehensive analysis of Hsp90 paralogue affinity and selectivity in the clinical Hsp90 inhibitor chemotypes. This has been possible through the development of a versatile experimental assay based on a new FP-probe (16a) that we both describe here. The assay, we show, can test rapidly and accurately the binding affinity of all major Hsp90 chemotypes and has a testing range that spans low nanomolar to millimolar binding affinities. We couple this assay with a computational analysis that allows for rationalization of paralogue selectivity and defines not only the major binding modes that relay pan-paralogue binding or conversely, paralogue selectivity, but also identifies molecular characteristics that impart such features.
Our analysis has enabled us to classify each of the inhibitors into one of three distinct types based upon their binding interactions within three specified pockets (A, B, C). Type 1 inhibitors bind to all three pockets A, B, and C and include 4–6. The only type 2 inhibitor described here, 3, binds to pockets A and B. Type 3 inhibitors bind to pockets A and C and include the ansamycins 1 and 2 as well as resorcinols 7 and 8. As previously described, each of the inhibitors has high affinity for cytoplasmic Hsp90. However, when tested for binding to the organellar isoforms, Grp94 and Trap-1, we observe a spectrum of binding affinities. Our analysis shows that affinity to Grp94 is largely affected by interactions within pocket B and compounds which extend into this pocket (i.e., type 1 and 2) generally exhibit decreased binding relative to Hsp90. Of the type 1 and 2 compounds described here, only 4 retained affinity comparable with Hsp90, which we ascribe to the ideal size and lipophilicity of the iodine atom to interact with the lipophilic residues in pocketB. It is likely that diminished affinity for 5, 3, and 6 is as a result of suboptimal fit and interaction with lipophilic residues, such as Phe195 and Leu163 within pocket B. Conversely, type 3 inhibitors, which do not extend into pocket B and are not dependent upon interactions with it nor are affected by suboptimal binding within it, remain highly potent inhibitors of Grp94.
Analysis of the results for Trap-1 shows that without exception each of the inhibitors exhibit diminished binding. Our computational analysis has identified the exit channel C as being the main determinant for this, where both Lys126 and Phe201 partially occlude the path to solvent. This satisfactorily explains results with type 1 and 3 compounds, which each exhibit binding into pocket C and suffer from varying degrees of interference with both Lys126 and Phe201 but does not explain results with 3, which does not extend into this pocket. Our analysis shows that binding of 3 to Trap-1 may suffer from unfavorable steric interactions between its 4′-OMe and 5′-Me groups with Leu172 in the B pocket.
Because the residues that make up the adenine-binding cavities of Hsp90s are highly conserved, it was generally believed that the discovery of paralogue selective inhibitors was a challenging task. From the results described here, we hypothesized that screening of a focused library of compounds (i.e., purine-scaffold) would provide a good means for potentially identifying selective inhibitors and overcome the limitations of structure-based drug design approaches imposed upon by the high sequence similarity between each isoform. Although not described here, we show in a parallel paper the utility of such an approach in the discovery of a highly selective Grp94 inhibitor.52
There are potentially significant therapeutic implications in understanding the paralogue selectivity of Hsp90 inhibitors. It is at the moment unknown how much of the activity/toxicity of these perceived pan-Hsp90 inhibitors is as a result of inhibition of Hsp90α/β, Grp94, or Trap-1. As it is becoming increasingly apparent that individual Hsp90 paralogues have distinct biological roles, it may very well be the case that pan-Hsp90 inhibition may result in undesirable side effects resulting in enhanced toxicity. Alternatively, it may be that maximal anticancer activity is observed with pan-Hsp90 inhibition. These may only be known when the profile of the current Hsp90 inhibitors is well understood, as we attempt to do here, or when paralogue selective molecules become available.
Paralogue selective drugs may represent a good strategy to develop better, safer agents with the prospect for enhanced personalized medicine. It may very well be that particular tumors are dependent on a specific paralogue and therefore would be more responsive to one class of inhibitor over another. It may be possible that particular cancers are more suited to therapy by a particular isoform inhibitor and when additional therapeutic factors are taken into consideration (i.e., toxicity) should potentially enhance the prospects for personalized medicine. This should further drive the discovery of potent selective inhibitors for each isoform.
In conclusion, our work provides a testing paradigm to investigate Hsp90 paralogue selectivity and affinity of small molecules. We believe these tools will allow for the analysis of paralogue contribution to the observed biological activity of the current Hsp90 inhibitor classes, as exemplified here by the analysis of the clinical Hsp90 inhibitor chemotypes but also the identification of paralogue-selective small molecules both as potential therapeutics in tumors dependent on a particular Hsp90 paralogue(s) and as tools to investigate the role of these paralogues in disease.
EXPERIMENTAL SECTION
Chemical Synthesis
1H NMR spectra were recorded on a Bruker 500 or 600 MHz instrument. Chemical shifts are reported in δ values in ppm downfield from TMS as the internal standard. 1H NMR data are reported as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, br = broad, m = multiplet), coupling constant (Hz), integration. High resolution mass spectra were recorded on a Waters LCT Premier system. Low resolution mass spectra were obtained on a Waters Acquity Ultra Performance LC with electrospray ionization and SQ detector. High-performance liquid chromatography (HPLC) analyses were performed on a Waters Autopurification system with PDA, MicroMass ZQ, and ELSD detector, and a reversed phase column (Waters X-Bridge C18, 4.6mm × 150 mm, 5 µm). The purity of FP probe compounds was determined by HPLC using a gradient of: (a) H2O+ 0.1% TFA and (b)CH3CN+ 0.1% TFA, 5–95% b over 13 min at 1.2 mL/min and UV detection at λ 220 and 254 nm and were >95% pure. All reactions were performed under argon protection. FITC was purchased from Aldrich. 12 and 16a were prepared as previously described.49
2-(4-(6-Amino-8-((6-iodobenzo[d][1,3]dioxol-5-yl)thio)-9H-purin-9-yl)butyl)isoindoline-1,3-dione (14b)
13 (200 mg, 0.484 mmol) was dissolved in DMF (8 mL). Cs2CO3 (466 mg, 1.43 mmol) and N-(4-bromobutyl)phthalimide (683 mg, 2.42 mmol) were added, and the mixture was sonicated for 30 min. Cs2CO3 (31.5 mg, 0.097 mmol) was added, and the mixture was again sonicated for 30 min. This was repeated two more times for a total reaction time of 2 h. DMF was removed, and the resulting residue was purified by preparatory TLC (CH2Cl2:MeOH:AcOH, 15:1:0.5) to give 134 mg (45%) of 14b. 1H NMR (500 MHz, CDCl3) δ 8.18 (s, 1H), 7.84 (dd, J = 5.5, 3.1 Hz, 2H), 7.72 (dd, J = 5.5, 3.1 Hz, 2H), 7.22 (s, 1H), 6.89 (s, 1H), 6.76 (br s, 2H), 5.99 (s, 2H), 4.23 (t, J = 7.1 Hz, 2H), 3.69 (t, J = 7.0 Hz, 2H), 1.67–1.83 (m, 4H). MS (ESI) m/z 615.2 [M + H]+.
9-(4-Aminobutyl)-8-((6-iodobenzo[d][1,3]dioxol-5-yl)thio)-9Hpurin-6-amine (15b)
To a suspension of 14b (38.9 mg, 0.063 mmol) in 2mL of MeOH/CH2Cl2 (7:1 mL) was added hydrazine hydrate (46 µL, 0.950 mmol), and the mixture was stirred at rt for 12 h. Solvent was removed under reduced pressure, and the resulting residue was purified by preparatory TLC (CH2Cl2:MeOH-NH3 (7N), 10:1) to give 18 mg (59%) of 15b. 1H NMR (500 MHz, CDCl3/MeOH-d4) δ 8.22 (s, 1H), 7.38 (s, 1H), 7.04 (s, 1H), 6.05 (s, 2H), 4.23 (t, J = 7.4 Hz, 2H), 2.78 (t, J = 7.1 Hz, 2H), 1.82–1.91 (m, 2H), 1.55–1.63 (m, 2H). MS (ESI) m/z 485.0 [M + H]+.
PU-C4-FITC (16b)
15b (9.7 mg, 0.020 mmol), FITC (8.57 mg (0.022 mmol), and Et3N (0.1 mL) in DMF (0.2 mL) was stirred for 3 h at rt. The reaction mixture was directly purified by HPLC to give 5.2 mg (30%) of 16b. 1H NMR (600 MHz, MeOH-d4) δ 8.22 (s, 1H), 8.00 (s, 1H), 7.61 (d, J = 7.6 Hz, 1H), 7.37 (s, 1H), 7.19 (s, 1H), 7.06 (d, J = 8.2 Hz, 1H), 6.58–6.67 (m, 4H), 6.48 (dd, J = 8.7, 2.0 Hz, 2H), 5.97 (s, 2H), 4.30 (t, J = 7.0 Hz, 2H), 3.58 (br s, 2H), 1.90–2.00 (m, 2H), 1.61–1.70 (m, 2H). HRMS (ESI) m/z [M + H]+ calcd for C37H29IN7O7S2, 874.0615; found 874.0610. HPLC Rt = 9.57.
2-(6-(6-Amino-8-((6-iodobenzo[d][1,3]dioxol-5-yl)thio)-9H-purin-9-yl)hexyl)isoindoline-1,3-dione (14c)
13 (200 mg, 0.484 mmol) was dissolved in DMF (8 mL). Cs2CO3 (466 mg, 1.43 mmol) and N-(6-bromohexyl)phthalimide (751 mg, 2.42 mmol) were added, and the mixture was sonicated for 2 h. Solvent was removed under reduced pressure, and the resulting residue was purified by preparatory TLC (CH2Cl2:MeOH:AcOH, 15:1:0.5) to give 100 mg (32%) of 14c. 1H NMR (500 MHz, CDCl3) δ 8.26 (s, 1H), 7.83 (dd, J = 5.4, 3.1 Hz, 2H), 7.70 (dd, J = 5.4, 3.0 Hz, 2H), 7.26 (s, 1H), 6.87 (s, 1H), 6.36 (br s, 2H), 5.96 (s, 2H), 4.18 (t, J = 7.5 Hz, 2H), 3.66 (t, J = 7.2 Hz, 2H), 1.70–1.79 (m, 2H), 1.60–1.68 (m, 2H), 1.32–1.43 (m, 4H). MS (ESI) m/z 643.2 [M + H]+.
9-(6-Aminohexyl)-8-((6-iodobenzo[d][1,3]dioxol-5-yl)thio)-9Hpurin-6-amine (15c)
To a suspension of 14c (97 mg, 0.1511 mmol) in 4 mL of MeOH/CH2Cl2 (7:1 mL) was added hydrazine hydrate (110 µL, 2.27 mmol), and the mixture was stirred at rt for 12 h. Solvent was removed under reduced pressure, and the resulting residue was purified by preparatory TLC (CH2Cl2:MeOH-NH3 (7N), 10:1) to give 47 mg (61%) of 15c. 1H NMR (500 MHz, CDCl3) δ 8.32 (s, 1H), 7.31 (s, 1H), 6.90 (s, 1H), 5.99 (s, 2H), 5.84 (br s, 2H), 4.20 (t, J = 7.5 Hz, 2H), 2.67 (t, J = 6.5 Hz, 2H), 1.72–1.84 (m, 2H), 1.31–1.45 (m, 6H). MS (ESI) m/z 513.0 [M + H]+.
PU-C6-FITC (16c)
15c (9.7 mg, 0.01894 mmol), FITC (8.11 mg, 0.0208 mmol), and Et3N (0.1 mL) in DMF (0.2 mL) was stirred for 3 h at rt. The reaction mixture was directly purified by HPLC to give 8.0 mg (47%) of 16c. 1H NMR (600 MHz, MeOH-d4) δ 8.23 (s, 1H), 8.09 (s, 1H), 7.65 (d, J = 7.9 Hz, 1H), 7.35 (s, 1H), 7.16 (s, 1H), 7.08 (d, J = 8.3 Hz, 1H), 6.71 (d, J = 8.8 Hz, 2H), 6.67 (d, J = 2.2 Hz, 2H), 6.53 (dd, J = 8.8, 2.2 Hz, 2H), 5.96 (s, 2H), 4.24 (t, J = 7.1 Hz, 2H), 3.50 (br s, 2H), 1.79–1.88 (m, 2H), 1.52–1.61 (m, 2H), 1.31–1.42 (m, 4H). HRMS (ESI) m/z [M + H]+ calcd for C39H33IN7O7S2, 902.0928; found 902.0939. HPLC Rt = 10.02.
2-(8-(6-Amino-8-((6-iodobenzo[d][1,3]dioxol-5-yl)thio)-9H-purin-9-yl)octyl)isoindoline-1,3-dione (14d)
13 (200 mg, 0.484 mmol) was dissolved in DMF (8 mL). Cs2CO3 (466 mg, 1.43 mmol) and N-(8-bromooctyl)phthalimide (819 mg, 2.42 mmol) were added, and the mixture was sonicated for 1.5 h. Solvent was removed under reduced pressure, and the resulting residue was purified by preparatory TLC (CH2Cl2:MeOH:AcOH, 15:1:0.5) to give 120 mg (34%) of 14d. 1H NMR (500 MHz, CDCl3) δ 8.29 (s, 1H), 7.84 (dd, J = 5.5, 3.1 Hz, 2H), 7.70 (dd, J = 5.5, 3.1 Hz, 2H), 7.28 (s, 1H), 6.87 (s, 1H), 6.29 (br s, 2H), 5.96 (s, 2H), 4.18 (t, J = 7.5 Hz, 2H), 3.67 (t, J = 7.3 Hz, 2H), 1.62–1.77 (m, 4H), 1.25–1.36 (m, 8H). MS (ESI) m/z 671.3 [M + H]+.
9-(8-Aminooctyl)-8-((6-iodobenzo[d][1,3]dioxol-5-yl)thio)-9Hpurin-6-amine (15d)
To a suspension of 14d (90.1 mg, 0.1345 mmol) in 4 mL of MeOH/CH2Cl2 (7:1 mL) was added hydrazine hydrate (98 µL, 2.017 mmol), and the mixture was stirred at rt for 12 h. Solvent was removed under reduced pressure, and the resulting residue was purified by preparatory TLC (CH2Cl2:MeOH-NH3 (7N), 10:1) to give 25 mg (34%) of 15d. 1HNMR(500 MHz, CDCl3) δ 8.33 (s, 1H), 7.31 (s, 1H), 6.90 (s, 1H), 5.99 (s, 2H), 5.72 (br s, 2H), 4.20 (t, J = 7.5 Hz, 2H), 2.66 (t, J = 7.1 Hz, 2H), 1.70–1.80 (m, 2H), 1.36–1.45 (m, 2H), 1.21–1.35 (m, 8H). MS (ESI) m/z 541.1 [M + H]+.
PU-C8-FITC (16d)
15d (15.0 mg, 0.028 mmol), FITC (11.9 mg, 0.031 mmol), and Et3N (0.1 mL) in DMF (0.2 mL) was stirred for 4 h at rt. The reaction mixture was directly purified by HPLC to give 16.9 mg (66%) of 16d. 1H NMR (600 MHz, MeOH-d4) δ 8.22 (s, 1H), 8.11 (s, 1H), 7.68 (d, J = 7.8 Hz, 1H), 7.34 (s, 1H), 7.12 (s, 1H), 7.09 (d, J = 8.2 Hz, 1H), 6.72 (d, J = 8.7 Hz, 2H), 6.67 (d, J = 2.0 Hz, 2H), 6.53 (dd, J = 8.7, 2.0 Hz, 2H), 5.96 (s, 2H), 4.20 (t, J = 7.1 Hz, 2H), 3.50 (br s, 2H), 1.74–1.81 (m, 2H), 1.52–1.59 (m, 2H), 1.23–1.35 (m, 8H). HRMS (ESI) m/z [M + H]+ calcd for C41H37IN7O7S2, 930.1241; found 930.1231. HPLC Rt = 10.60.
Biological Evaluation
Reagents
Hsp90α was purchased from (Stressgen no. SPP776). Recombinant Hsp90β was purchased from Enzo Life Sciences (ADI-SPP-777). Canine Grp94 was generated as previously reported.52 Recombinant human Trap-1 was purchased from Enzo Life Sciences (ADI-SPP-848). The human breast cancer cell line SKBr3 was obtained from the American Type Culture Collection (Manassas, VA). Cells were cultured in DME HG:F-12 medium supplemented with 10% fetal bovine serum, NEAA, 1% penicillin, and streptomycin. Cells were collected and frozen to rupture the membranes and then dissolved in binding buffer with added protease and phosphatase inhibitors to form the lysate. Lysates were stored at −80 °C before use. Total protein content was determined using the bicinchoninic acid assay kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer’s instructions.
Hsp90 Saturation Binding Assay
The Hsp90 FP saturation assays were performed on an Analyst GT instrument (Molecular Devices, Sunnyvale, CA) and carried out in black 96-well microplates (Corning no. 3650) in a total volume of 100 µL in each well. A stock of 10 µM 12 or 16a–d was prepared in DMSO and diluted with Felts buffer (20 mM Hepes (K), pH 7.3, 50 mM KCl, 2 mM DTT, 5 mM MgCl2, 20 mM Na2MoO4, and 0.01% NP40 with 0.1 mg/mL BGG). To determine the equilibrium binding of 12 or 16a–d, increasing amounts of Hsp90α, Hsp90β, Grp94, or Trap-1 (up to 250 nM), or SKBr3 lysate (up to 50 µg of total protein) were incubated with 3 nM of 12 or 16a–d. The assay plate was incubated on a shaker at 4 °C for the indicated times, and the FP values in mP were measured. The assay window was calculated as the difference between the FP value recorded for the bound fluorescent tracer and the FP value recorded for the free fluorescent tracer (defined as mP − mPf).
Hsp90 Competition Assay
The Hsp90 FP competition assays were performed on an Analyst GT instrument (Molecular Devices, Sunnyvale, CA) and carried out in black 96-well microplates (Corning no. 3650) in a total volume of 100 µL in each well. A stock of 10 µM16a was prepared in DMSO and diluted with Felts buffer (20 mM Hepes (K), pH 7.3, 50mMKCl, 2 mM DTT, 5 mM MgCl2, 20mMNa2MoO4, and 0.01% NP40 with 0.1 mg/mL BGG). To each well was added 3 nM fluorescent 16a, protein (25 nMHsp90α, 25 nMHsp90β, 25 nMGrp94, 30 nM Trap-1) or SKBr3 lysate (4.5 µg total protein), and tested inhibitor (initial stock in DMSO) in a final volume of 100 µL of Felts buffer. Compounds were added in triplicate wells. For each assay, background wells (buffer only), probe controls (free, probe only), and bound probe controls (probe in the presence of protein or SKBr3 lysate) were included on each assay plate. The assay plate was incubated on a shaker at 4 °C for 24 h, and the FP values in mP were measured. The fraction of probe bound to Hsp90 was correlated to the mP value and plotted against values of competitor concentrations. The inhibitor concentration at which 50% of bound probe was displaced was obtained by fitting the data. All experimental data were analyzed using SOFTmax Pro 4.3.1 and plotted using Prism4.0 (GraphPad Software Inc., SanDiego,CA).
Molecular Modeling
All the compounds were constructed using the fragment dictionary of Maestro (version 8.5). Ligands were further prepared using Ligprep (version 2.2) utility provided by Schrödinger LLC. The X-ray crystal structure of Hsp90α (PDB ID: 2FWZ, 2VCI, 3QDD, and 3D0B), Hsp90β (PDB ID: 3NMQ), Grp94 (PDB ID: 2EXL and 3O2F), and homology model of Trap-152 were optimized for subsequent grid generation and docking using the default parameters in Protein Preparation Wizard provided by Schröinger LLC. Grids were then prepared using the Receptor Grid Generation tool in Glide (version 5.0).63,64 Next, the extra precision (XP) Glide docking method was used to dock compounds flexibly into the ATP binding site of receptors according to previously reported procedure.65
ACKNOWLEDGMENTS
G.C. is funded by Leukemia and Lymphoma Society, Breast Cancer Research Fund, 1U01 AG032969-01A1, 1R21 CA158609-01, 1R21 AI090501, R01 CA172546-01A1, and 1R01 CA155226-01. T.T. is funded by Susan G. Komen for the Cure (KG091313) and the Department of Defense, Breast Cancer Research Program (PDFBC093421). P.D.P. is supported by funds and resources from St. John’s University. D.G. is funded by 2R01-CA095130. We also thank Dr. George Sukenick and Dr. Hui Liu of the NMR Analytical Core Facility at MSKCC for expert mass spectral analysis.
ABBREVIATIONS USED
- Hsp90
heat shock protein 90
- Grp94
glucose-regulated protein 94
- Trap-1
TNF receptor-associated protein 1
- HER2
human epidermal growth factor receptor 2
- ER
estrogen receptor
- HIF1α
hypoxia-inducible factor 1-alpha
- NBD
N-terminal nucleotide-binding domain
- FP
fluorescence polarization
- FAM
carboxyfluorescein
- 2D-FIDA
two-dimensional fluorescence intensity distribution analysis
- FITC
fluorescein isothiocyanate
- mP
milli-polarization
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Jhaveri K, Taldone T, Modi S, Chiosis G. Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. Biochim. Biophys. Acta. 2012;1823:742–755. doi: 10.1016/j.bbamcr.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biamonte MA, Van de Water R, Arndt JW, Scannevin RH, Perret D, Lee WC. Heat shock protein 90: inhibitors in clinical trials. J. Med. Chem. 2010;53:3–17. doi: 10.1021/jm9004708. [DOI] [PubMed] [Google Scholar]
- 3.Chiosis G, Dickey CA, Johnson JL. A global view of Hsp90 functions. Nat. Struct. Mol. Biol. 2013;20:1–4. doi: 10.1038/nsmb.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 5.Isaacs JS, Xu W, Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell. 2003;3:213–217. doi: 10.1016/s1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 6.Marzec M, Eletto D, Argon Y. GRP94: an HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim. Biophys. Acta. 2012;1823:774–787. doi: 10.1016/j.bbamcr.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLaughlin M, Vandenbroeck K. The endoplasmic reticulum protein folding factory and its chaperones: new targets for drug discovery? Br. J. Pharmacol. 2011;162:328–345. doi: 10.1111/j.1476-5381.2010.01064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang BH, Plescia J, Dohi T, Rosa J, Doxsey SJ, Altieri DC. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell. 2007;131:257–270. doi: 10.1016/j.cell.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 10.Costantino E, Maddalena F, Calise S, Piscazzi A, Tirino V, Fersini A, Ambrosi A, Neri V, Esposito F, Landriscina M. TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 2009;279:39–46. doi: 10.1016/j.canlet.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 11.Restall IJ, Lorimer IA. Induction of premature senescence by hsp90 inhibition in small cell lung cancer. PLoS One. 2010;5:e11076. doi: 10.1371/journal.pone.0011076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodina A, Vilenchik M, Moulick K, Aguirre J, Kim J, Chiang A, Litz J, Clement CC, Kang Y, She Y, Wu N, Felts S, Wipf P, Massague J, Jiang X, Brodsky JL, Krystal GW, Chiosis G. Selective compounds define Hsp90 as a major inhibitor of apoptosis in small-cell lung cancer. Nat. Chem. Biol. 2007;3:498–507. doi: 10.1038/nchembio.2007.10. [DOI] [PubMed] [Google Scholar]
- 13.Duerfeldt AS, Peterson LB, Maynard JC, Ng CL, Eletto D, Ostrovsky O, Shinogle HE, Moore DS, Argon Y, Nicchitta CV, Blagg BS. Development of a Grp94 inhibitor. J. Am. Chem. Soc. 2012;134:9796–9804. doi: 10.1021/ja303477g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang BH, Plescia J, Song HY, Meli M, Colombo G, Beebe K, Scroggins B, Neckers L, Altieri DC. Combinatorial drug design targeting multiple cancer signaling networks controlled by mitochondrial Hsp90. J. Clin. Invest. 2009;119:454–464. doi: 10.1172/JCI37613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein Hsp90–pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeBoer C, Meulman PA, Wnuk RJ, Peterson DH. Geldanamycin, a new antibiotic. J. Antibiot. 1970;23:442–447. doi: 10.7164/antibiotics.23.442. [DOI] [PubMed] [Google Scholar]
- 17.Schulte TW, Neckers LM. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother. Pharmacol. 1998;42:273–279. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- 18.Kasibhatla SR, Hong K, Biamonte MA, Busch DJ, Karjian PL, Sensintaffar JL, Kamal A, Lough RE, Brekken J, Lundgren K, Grecko R, Timony GA, Ran Y, Mansfield R, Fritz LC, Ulm E, Burrows FJ, Boehm MF. Rationally designed high-affinity 2-amino-6-halopurine heat shock protein 90 inhibitors that exhibit potent antitumor activity. J. Med. Chem. 2007;50:2767–2778. doi: 10.1021/jm050752+. [DOI] [PubMed] [Google Scholar]
- 19.He H, Zatorska D, Kim J, Aguirre J, Llauger L, She Y, Wu N, Immormino RM, Gewirth DT, Chiosis G. Identification of potent water soluble purine-scaffold inhibitors of the heat shock protein. J. Med. Chem. 2006;49:381–390. doi: 10.1021/jm0508078. [DOI] [PubMed] [Google Scholar]
- 20.Bao R, Lai C-J, Qu H, Wang D, Yin L, Zifcak B, Atoyan R, Wang J, Samson M, Forrester J, DellaRocca S, Xu G-X, Tao X, Zhai H-X, Cai X, Qian C. CUDC-305, a novel synthetic HSP90 inhibitor with unique pharmacologic properties for cancer therapy. Clin. Cancer Res. 2009;15:4046–4057. doi: 10.1158/1078-0432.CCR-09-0152. [DOI] [PubMed] [Google Scholar]
- 21.Huang KH, Veal JM, Fadden RP, Rice JW, Eaves J, Strachan J-P, Barabasz AF, Foley BE, Barta TE, Ma W, Silinski MA, Hu M, Partridge JM, Scott A, DuBois LG, Freed T, Steed PM, Ommen AJ, Smith ED, Hughes PF, Woodward AR, Hanson GJ, McCall WS, Markworth CJ, Hinkley L, Jenks M, Geng L, Lewis M, Otto J, Pronk B, Verleysen K, Hall SE. Discovery of novel 2-aminobenzamide inhibitors of heat shock protein 90 as potent, selective and orally active antitumor agents. J. Med. Chem. 2009;52:4288–4305. doi: 10.1021/jm900230j. [DOI] [PubMed] [Google Scholar]
- 22.Brough PA, Aherne W, Barril X, Borgognoni J, Boxall K, Cansfield JE, Cheung K-MJ, Collins I, Davies NGM, Drysdale MJ, Dymock B, Eccles SA, Finch H, Fink A, Hayes A, Howes R, Hubbard RE, James K, Jordan AM, Lockie A, Martins V, Massey A, Matthews TP, McDonald E, Northfield CJ, Pearl LH, Prodromou C, Ray S, Raynaud FI, Roughley SD, Sharp SY, Surgenor A, Walmsley DL, Webb P, Wood M, Workman P, Wright L. 4,5-Diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer. J. Med. Chem. 2008;51:196–218. doi: 10.1021/jm701018h. [DOI] [PubMed] [Google Scholar]
- 23.Ying W, Du Z, Sun L, Foley KP, Proia DA, Blackman RK, Zhou D, Inoue T, Tatsuta N, Sang J, Ye S, Acquaviva J, Ogawa LS, Wada Y, Barsoum J, Koya K. Ganetespib, a unique triazolone-containing Hsp90 inhibitor, exhibits potent antitumor activity and a superior safety profile for cancer therapy. Mol. Cancer Ther. 2012;11:475–484. doi: 10.1158/1535-7163.MCT-11-0755. [DOI] [PubMed] [Google Scholar]
- 24.McLaughlin SH, Ventouras LA, Lobbezoo B, Jackson SE. Independent ATPase activity of Hsp90 subunits creates a flexible assembly platform. J. Mol. Biol. 2004;344:813–826. doi: 10.1016/j.jmb.2004.09.055. [DOI] [PubMed] [Google Scholar]
- 25.Richter K, Soroka J, Skalniak L, Leskovar A, Hessling M, Reinstein J, Buchner J. Conserved conformational changes in the ATPase cycle of human hsp90. J. Biol. Chem. 2008;283:17757–17765. doi: 10.1074/jbc.M800540200. [DOI] [PubMed] [Google Scholar]
- 26.Frey S, Leskovar A, Reinstein J, Buchner J. The ATPase cycle of the endoplasmic chaperone Grp94. J. Biol. Chem. 2007;282:35612–35620. doi: 10.1074/jbc.M704647200. [DOI] [PubMed] [Google Scholar]
- 27.Rosser MF, Nicchitta CV. Ligand interactions in the adenosine nucleotide-binding domain of the Hsp90 chaperone, GRP94. I. Evidence for allosteric regulation of ligand binding. J. Biol. Chem. 2000;275:22798–22805. doi: 10.1074/jbc.M001477200. [DOI] [PubMed] [Google Scholar]
- 28.Leskovar A, Wegele H, Werbeck ND, Buchner J, Reinstein J. The ATPase Cycle of the Mitochondrial Hsp90 Analog Trap1. J. Biol. Chem. 2008;283:11677–11688. doi: 10.1074/jbc.M709516200. [DOI] [PubMed] [Google Scholar]
- 29.Llauger-Bufi L, Felts SJ, Huezo H, Rosen N, Chiosis G. Synthesis of novel fluorescent probes for the molecular chaperone Hsp90. Bioorg. Med. Chem. Lett. 2003;13:3975–3978. doi: 10.1016/j.bmcl.2003.08.065. [DOI] [PubMed] [Google Scholar]
- 30.Kim J, Felts S, Llauger L, He H, Huezo H, Rosen N, Chiosis G. Development of a fluorescence polarization assay for the molecular chaperone Hsp90. J. Biomol. Screening. 2004;9:375–381. doi: 10.1177/1087057104265995. [DOI] [PubMed] [Google Scholar]
- 31.Du Y, Moulick K, Rodina A, Aguirre J, Felts S, Dingledine R, Fu H, Chiosis G. High-throughput screening fluorescence polarization assay for tumor-specific Hsp90. J. Biomol. Screening. 2007;12:915–924. doi: 10.1177/1087057107306067. [DOI] [PubMed] [Google Scholar]
- 32.Howes R, Barril X, Dymock BW, Grant K, Northfield CJ, Robertson AG, Surgenor A, Wayne J, Wright L, James K, Matthews T, Cheung KM, McDonald E, Workman P, Drysdale MJ. A fluorescence polarization assay for inhibitors of Hsp90. Anal. Biochem. 2006;350:202–213. doi: 10.1016/j.ab.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 33.Moulick K, Clement CC, Aguirre J, Kim J, Kang Y, Felts S, Chiosis G. Synthesis of a red-shifted fluorescence polarization probe for Hsp90. Bioorg. Med. Chem. Lett. 2006;16:4515–4518. doi: 10.1016/j.bmcl.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 34.Lea WA, Simeonov A. Fluorescence Polarization Assays in Small Molecule Screening. Expert Opin. Drug Discovery. 2011;6:17–32. doi: 10.1517/17460441.2011.537322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jameson DM, Ross JA. Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev. 2010;110:2685–2708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayer MP. Gymnastics of molecular chaperones. Mol. Cell. 2010;39:321–331. doi: 10.1016/j.molcel.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 37.Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90–geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 38.Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 39.Immormino RM, Kang Y, Chiosis G, Gewirth DT. Structural and quantum chemical studies of 8-aryl-sulfanyl adenine class Hsp90 inhibitors. J. Med. Chem. 2006;49:4953–4960. doi: 10.1021/jm060297x. [DOI] [PubMed] [Google Scholar]
- 40.Brough PA, Barril X, Borgognoni J, Chene P, Davies NGM, Davis B, Drysdale MJ, Dymock B, Eccles SA, Garcia-Echeverria C, Fromont C, Hayes A, Hubbard RE, Jordan AM, Jensen MR, Massey A, Merrett A, Padfield A, Parsons R, Radimerski T, Raynaud FI, Robertson A, Roughley SD, Schoepfer J, Simmonite H, Sharp SY, Surgenor A, Valenti M, Walls S, Webb P, Wood M, Workman P, Wright L. Combining hit identification strategies: fragment-based and in silico approaches to orally active 2-aminothieno[2,3-d]pyrimidine inhibitors of the Hsp90 molecular chaperone. J. Med. Chem. 2009;52:4794–4809. doi: 10.1021/jm900357y. [DOI] [PubMed] [Google Scholar]
- 41.Yun TJ, Harning EK, Giza K, Rabah D, Li P, Arndt JW, Luchetti D, Biamonte MA, Shi J, Lundgren K, Manning A, Kehry MR. EC144, A Synthetic Inhibitor of Heat Shock Protein 90, Blocks Innate and Adaptive Immune Responses in Models of Inflammation and Autoimmunity. J. Immunol. 2011;186:563–575. doi: 10.4049/jimmunol.1000222. [DOI] [PubMed] [Google Scholar]
- 42.Barta TE, Veal JM, Rice JW, Partridge JM, Fadden RP, Ma W, Jenks M, Geng L, Hanson GJ, Huang KH, Barabasz AF, Foley BE, Otto J, Hall SE. Discovery of benzamide tetrahydro-4H-carbazol-4-ones as novel small molecule inhibitors of Hsp90. Bioorg. Med. Chem. Lett. 2008;18:3517–3521. doi: 10.1016/j.bmcl.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 43.Immormino RM, Metzger LE, Reardon PN, Dollins DE, Blagg BS, Gewirth DT. Different poses for ligand and chaperone in inhibitor-bound Hsp90 and GRP94: implications for paralog-specific drug design. J. Mol. Biol. 2009;388:1033–1042. doi: 10.1016/j.jmb.2009.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi J, Van de Water R, Hong K, Lamer RB, Weichert KW, Sandoval CM, Kasibhatla SR, Boehm MF, Chao J, Lundgren K, Timple N, Lough R, Ibanez G, Boykin C, Burrows FJ, Kehry MR, Yun TJ, Harning EK, Ambrose C, Thompson J, Bixler SA, Dunah A, Snodgrass-Belt P, Arndt J, Enyedy IJ, Li P, Hong VS, McKenzie A, Biamonte MA. EC144 is a potent inhibitor of the heat shock protein 90. J. Med. Chem. 2012;55:7786–7795. doi: 10.1021/jm300810x. [DOI] [PubMed] [Google Scholar]
- 45.Alexander LD, Partridge JR, Agard DA, McAlpine SR. A small molecule that preferentially binds the closed conformation of Hsp90. Bioorg. Med. Chem. Lett. 2011;21:7068–7071. doi: 10.1016/j.bmcl.2011.09.096. [DOI] [PubMed] [Google Scholar]
- 46.Schilb A, Riou V, Schoepfer J, Ottl J, Müller K, Chene P, Mayr LM, Filipuzzi I. Development and implementation of a highly miniaturized confocal 2D-FIDA-based high-throughput screening assay to search for active site modulators of the human heat shock protein 90beta. J. Biomol. Screening. 2004;9:569–577. doi: 10.1177/1087057104265538. [DOI] [PubMed] [Google Scholar]
- 47.Massey AJ, Schoepfer J, Brough PA, Brueggen J, Chène P, Drysdale MJ, Pfaar U, Radimerski T, Ruetz S, Schweitzer A, Wood M, Garcia-Echeverria C, Jensen MR. Preclinical antitumor activity of the orally available heat shock protein 90 inhibitor NVP-BEP800. Mol. Cancer Ther. 2010;9:906–919. doi: 10.1158/1535-7163.MCT-10-0055. [DOI] [PubMed] [Google Scholar]
- 48.Eccles SA, Massey A, Raynaud FI, Sharp SY, Box G, Valenti M, Patterson L, Brandon AH, Gowan S, Boxall F, Aherne W, Rowlands M, Hayes A, Martins V, Urban F, Boxall K, Prodromou C, Pearl L, James K, Matthews TP, Cheung K-M, Kalusa A, Jones K, McDonald E, Barril X, Brough PA, Cansfield JE, Dymock B, Drysdale MJ, Finch H, Howes R, Hubbard RE, Surgenor A, Webb P, Wood M, Wright L, Workman P. NVP-AUY922: a novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res. 2008;68:2850–2860. doi: 10.1158/0008-5472.CAN-07-5256. [DOI] [PubMed] [Google Scholar]
- 49.Taldone T, Gomes-DaGama EM, Zong H, Sen S, Alpaugh ML, Zatorska D, Alonso-Sabadell R, Guzman ML, Chiosis G. Synthesis of purine-scaffold fluorescent probes for heat shock protein 90 with use in flow cytometry and fluorescence microscopy. Bioorg. Med. Chem. Lett. 2011;21:5347–5352. doi: 10.1016/j.bmcl.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lynch BA, Loiacono KA, Tiong CL, Adams SE, MacNeil IA. A Fluorescence Polarization Based Src-SH2 Binding Assay. Anal. Biochem. 1997;247:77–82. doi: 10.1006/abio.1997.2042. [DOI] [PubMed] [Google Scholar]
- 51.Wu P, Brasseur M, Schindler U. A High-Throughput STAT Binding Assay Using Fluorescence Polarization. Anal. Biochem. 1997;249:29–36. doi: 10.1006/abio.1997.2158. [DOI] [PubMed] [Google Scholar]
- 52.Patel PD, Yan P, Seidler PM, Patel HJ, Sun W, Yang C, Que NS, Taldone T, Finotti P, Stephani RA, Gewirth DT, Chiosis G. Paralog-selective Hsp90 inhibitors define tumor-specific regulation of HER2. Nat. Chem. Biol. 2013 doi: 10.1038/nchembio.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taldone T, Zatorska D, Patel PD, Zong H, Rodina A, Ahn JH, Moulick K, Guzman ML, Chiosis G. Design, Synthesis and Evaluation of Small Molecule Hsp90 Probes. Bioorg. Med. Chem. 2011;19:2603–2614. doi: 10.1016/j.bmc.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dollins DE, Warren JJ, Immormino RM, Gewirth DT. Structures of GRP94–nucleotide complexes reveal mechanistic differences between the Hsp90 chaperones. Mol. Cell. 2007;28:41–56. doi: 10.1016/j.molcel.2007.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Felts SJ, Owen BA, Nguyen P, Trepel J, Donner DB, Toft DO. The Hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J. Biol. Chem. 2000;275:3305–3312. doi: 10.1074/jbc.275.5.3305. [DOI] [PubMed] [Google Scholar]
- 56.Ge J, Normant E, Porter JR, Ali JA, Dembski MS, Gao Y, Georges AT, Grenier L, Pak RH, Patterson J, Sydor JR, Tibbitts TT, Tong JK, Adams J, Palombella VJ. Design, synthesis, and biological evaluation of hydroquinone derivatives of 17-amino-17-demethoxygeldanamycin as potent, water-soluble inhibitors of Hsp90. J. Med. Chem. 2006;49:4606–4615. doi: 10.1021/jm0603116. [DOI] [PubMed] [Google Scholar]
- 57.Lundgren K, Zhang H, Brekken J, Huser N, Powell RE, Timple N, Busch DJ, Neely L, Sensintaffar JL, Yang Y, McKenzie A, Friedman J, Scannevin R, Kamal A, Hong K, Kasibhatla SR, Boehm MF, Burrows FJ. BIIB021, an orally available, fully synthetic small-molecule inhibitor of the heat shock protein Hsp90. Mol. Cancer Ther. 2009;8:921–929. doi: 10.1158/1535-7163.MCT-08-0758. [DOI] [PubMed] [Google Scholar]
- 58.Caldas-Lopes E, Cerchietti L, Ahn JH, Clement CC, Robles AI, Rodina A, Moulick K, Taldone T, Gozman A, Guo Y, Wu N, de Stanchina E, White J, Gross SS, Ma Y, Varticovski L, Melnick A, Chiosis G. Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8368–8373. doi: 10.1073/pnas.0903392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chandarlapaty S, Sawai A, Ye Q, Scott A, Silinski M, Huang K, Fadden P, Partdrige J, Hall S, Steed P, Norton L, Rosen N, Solit DB. SNX2112, a synthetic heat shock protein 90 inhibitor, has potent antitumor activity against HER kinase-dependent cancers. Clin. Cancer Res. 2008;14:240–248. doi: 10.1158/1078-0432.CCR-07-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Solit DB, Zheng FF, Drobnjak M, Munster PN, Higgins B, Verbel D, Heller G, Tong W, Cardon-Cardo C, Agus DB, Scher HI, Rosen N. 17-Allylamino-17-demethoxygeldanamycin induces the degradation of androgen receptor and HER-2/neu and inhibits the growth of prostate cancer xenografts. Clin. Cancer Res. 2002;8:986–993. [PubMed] [Google Scholar]
- 61.Wright L, Barril X, Dymock B, Sheridan L, Surgenor A, Beswick M, Drysdale M, Collier A, Massey A, Davies N, Fink A, Fromont C, Aherne W, Boxall K, Sharp S, Workman P, Hubbard RE. Structure–activity relationships in purine-based inhibitor binding to Hsp90 isoforms. Chem. Biol. 2004;6:775–785. doi: 10.1016/j.chembiol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 62.Immormino RM, Dollins DE, Shaffer PL, Soldano KL, Walker MA, Gewirth DT. Ligand-induced conformational shift in the N-terminal domain of GRP94, an Hsp90 chaperone. J. Biol. Chem. 2004;279:46162–46171. doi: 10.1074/jbc.M405253200. [DOI] [PubMed] [Google Scholar]
- 63.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 64.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 65.Patel PD, Patel MR, Kaushik-Basu N, Talele TT. 3D QSAR and molecular docking studies of benzimidazole derivatives as hepatitis C virus NS5B polymerase inhibitors. J. Chem. Inf. Model. 2008;48:42–55. doi: 10.1021/ci700266z. [DOI] [PubMed] [Google Scholar]