Abstract
Mixed-lineage leukemia (MLL) maintains the expression of cellular memory genes during development, while leukemic MLL fusion proteins aberrantly maintain expression of hematopoietic stem cell program genes such as HOXA9 to cause leukemia. However, the molecular mechanism of gene activation is unclear. Here we show that only two functional modules are necessary and sufficient for target recognition: those that bind to non-methylated CpGs and di-/tri-methylated histone H3 lysine 36 (H3K36me2/3). An artificial protein composed of the two targeting modules and an interaction domain for AF4-family coactivators can functionally substitute for MLL fusion proteins. Because H3K36me2/3 markers are indicative of active transcription, MLL fusion proteins target previously active CpG-rich genes and activate transcription by recruiting coactivators thereto. Our results indicate that such chromatin context-dependent gene activation is the fundamental mechanism by which MLL fusion proteins maintain the expression of the cellular memory/hematopoietic stem cell program genes.
INTRODUCTION
The MLL gene encodes an epigenetic regulator that maintains HOX gene expression during embryogenesis (1). HOX genes are so-called cellular memory genes because their expression is maintained throughout the development. In the hematopoietic lineage, the MLL protein (also known as HRX, MLL1 and KMT2A) activates the transcription of posterior HOXA genes (e.g. HOXA7-A10) (2,3). Posterior HOXA genes are hematopoietic stem cell (HSC) program genes (4) that promote the self-renewal of HSCs/immature progenitors (5). In normal hematopoiesis, their expression is maintained by MLL in the HSC/immature progenitor compartments, which diminishes as cells differentiate. Chromosomal translocation generates MLL fusion genes, whose products constitutively activate the posterior HOXA genes, which results in aberrant self-renewal of hematopoietic progenitors, leading to leukemia (6). However, the precise molecular mechanism by which MLL and MLL fusion proteins activate their target genes remains unclear. MLL fusion proteins exert their oncogenic functions as a complex with the lens epithelium-derived growth factor (LEDGF) (also known as PSIP1) (7). Disruption of Psip1 in mice causes homeotic skeletal transformation, a characteristic phenotype caused by dysregulation of Hox gene expression (8). LEDGF also facilitates the specific integration of the HIV genome into transcriptionally active regions, presumably by tethering the HIV genome/integrase complex with transcriptionally active chromatin (9,10). In leukemia, MLL frequently fuses with the ALL1-fused gene from chromosome 4 (AF4)- and eleven-nineteen leukemia (ENL)-family genes, whose protein products are the components of a transcriptional coactivator, termed AEP (the AF4 family/ENL family/PTEFb complex) (11–14). MLL fusion proteins constitutively recruit AEP components to activate transcription through direct interaction (e.g. MLL-ENL) or by an as-yet-uncharacterized indirect mechanism (e.g. MLL-AF6) (14). These previous studies postulate that MLL fusion proteins recognize their target genes through the MLL portion and constitutively recruit the AEP coactivator through the fusion partner portion.
Here, we report that MLL fusion proteins recognize a specific chromatin context to activate cellular memory genes. Our structure/function analyses demonstrate that MLL fusion proteins recognize their target genes through two functional domains: the PWWP domain of LEDGF and the CXXC domain of MLL that specifically bind to H3K36me2/3 and non-methylated CpGs, respectively. Because H3K36me2 and H3K36me3 are generally linked to gene activation (15,16), a previously transcribed gene containing non-methylated CpGs in its promoter is subjected to transcriptional activation by MLL fusion proteins. These studies provide a novel chromatin context-dependent gene activation mechanism by which MLL fusion proteins maintain cellular memory.
MATERIALS AND METHODS
Cell culture
The human leukemia cell line ML-2 was cultured in RPMI 1640 medium supplemented with 10% fetal calf serum and non-essential amino acids. 293T, plat-E and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and non-essential amino acids.
Vector construction
The pMSCV-neo MLL-ENL, PME (7) and Bcl2-ires-E2A-HLF (17) vectors are described elsewhere. Various MLL fusion constructs were generated by restriction enzyme digestion or polymerase chain reaction (PCR)-based mutagenesis. The cDNAs were cloned into the pMSCV neo vector (for virus production) (Clontech, Mountain View, CA) or the pCMV5 vector (for transient expression). The expression vectors for FLAG-tagged GAL4 fusion proteins were constructed by PCR using pM (Clontech) as template and cloned into the pCMV5 vector. Various LEDGF constructs were generated by PCR-based mutagenesis and cloned into the pcDNA4 HisMax vector (Invitrogen, Carlsbad, CA).
Western blotting
Western blotting was performed as described elsewhere (14). The antibodies used in this study have been listed in Supplementary Table S2.
Reverse transcription-PCR
RNAs were prepared using the RNeasy kit (Qiagen, Valencia, CA) and reverse-transcribed with the Superscript III first strand cDNA synthesis kit with oligo(dT) primers (Invitrogen). Expression of PCE was confirmed by regular PCR using the primer pair 5′-AGAATTCGATATCGGAAACATGACTCGCGATTTCAAACC-3′/5′-TCACGTGTTCGCGATGCGACGGGCTTTCGTGGAGGAG-3′. Quantitative PCR (qPCR) for other genes was performed as described previously (14) using TaqMan probes [Gapdh (Mm99999915_g1), Hoxa7 (Mm00657963_m1), Hoxa9 (Mm00439364_m1), Hoxa10 (Mm00433966_m1), Cbx5 (Mm00483092_m1), Myb (Mm00501741_m1) and Hmgb3 (Mm01377544_gH) (Applied Biosystems)]. The expression levels, normalized to Gapdh, were estimated using a standard curve and the relative quantification method, as described in ABI User Bulletin #2.
Virus production
The ecotropic retrovirus was produced using plat-E packaging cells (18). Supernatant medium containing the virus was harvested 24–48 h after transfection and used for viral transduction.
Myeloid progenitor transformation assay
The myeloid progenitor transformation assay was performed using cells harvested from the femurs and tibiae of C57BL/6 mice. C-kit-positive cells were enriched using magnetic beads conjugated with anti-c-kit antibody (Miltenyi Biotech, Bergisch Gladbach, Germany), transduced with recombinant retrovirus by spinoculation (19) and plated in methylcellulose medium (Iscove’s modified Dulbecco’s medium, 20% fetal bovine serum, 1.6% methylcellulose, 100 µM β-mercaptoethanol) containing stem cell factor, interleukin-3 and granulocyte macrophage colony-stimulating factor (10 ng/ml). The colony-forming units per 104 plated cells at the third and fourth rounds were quantified after 5–7 days of culture.
Subcellular fractionation and nucleosome co-immunoprecipitation analysis
Subcellular fractions of 293T cells were obtained by CSK buffer extraction (20) and MNase treatment. 293T cells cultured in a 10-cm dish were resuspended in 1 ml of CSK buffer [100 mM NaCl, 10 mM PIPES (pH 6.8), 3 mM MgCl2, 1 mM EGTA (pH 7.6), 0.3 M sucrose, 0.5% Triton X-100, 5 mM sodium butyrate, 0.5 mM DTT, EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland) and 2 mM vanadyl ribonucleoside complexes (Sigma, St. Louis, MO)], incubated on ice for 5 min and then centrifuged (400g, 4°C, 4 min). The supernatant (the soluble fraction) was transferred to a new tube, and the pellet was resuspended in 1 ml of MNase buffer [50 mM Tris-HCl (pH 7.5), 4 mM MgCl2, 1 mM CaCl2, 0.3 M sucrose, 5 mM sodium butyrate, 0.5 mM DTT and protease inhibitor cocktail]. One unit of MNase (Sigma) was added to the suspension, and the mixture was incubated at 37°C for 10–12 min to obtain mononucleosomes. The MNase reaction was stopped by adding EDTA (pH 8.0) at a final concentration of 20 mM. The reaction mixture was centrifuged (13 000 rpm, 4°C, 5 min) to separate the supernatant (the nucleosome fraction) and the pellet. The pellet was resuspended in elution buffer (1% sodium dodecyl sulfate, 50 mM NaHCO3). The nucleosome fractions were subjected to immunoprecipitation (IP) using specific antibodies; washed five times with MNase buffer with 20 mM EDTA and analyzed by western blotting, Oriole staining (Bio-Rad, Hercules, CA), SYBR Green staining (Lonza, Basel, Switzerland) and mass spectrometry. Optionally, the precipitates were washed with MNase buffer three times, treated with DNase I (Qiagen) for 10 min at 37°C and washed five times with MNase buffer with 20 mM EDTA. The precipitates were analyzed by western blotting and SYBR Green staining.
Fractionation-assisted native chromatin immunoprecipitation
293T cells cultured in a 10-cm dish or ML-2 cells (2 × 107) were suspended in 1 ml of CSK buffer to remove chromatin-unbound materials. The pellet was then resuspended in 1 ml of MNase buffer and mixed with 0.2–1 U of MNase for 5–10 min at 37°C to obtain chromatin with DNA lengths of 150–3000 base pairs (bp). The MNase reaction was stopped by adding EDTA (pH 8.0) at a final concentration of 20 mM, and then the mixture was placed on ice. Lysis buffer (1 ml) [10% glycerol, 20 mM sodium phosphate (pH 7.0), 250 mM NaCl, 30 mM sodium pyrophosphate, 0.1% Nonidet P-40, 5 mM EDTA, 10 mM NaF and protease inhibitor cocktail] (21) was added to solubilize the proteins, and the mixture was spun down to remove insoluble material. The supernatant was then subjected to IP. Approximately 1 µg of the primary antibodies listed in Supplementary Table S2 was added to 400 µl of chromatin suspension, and the mixture was incubated for 4–6 h at 4°C. Ten microliters of protein-G magnetic beads (Invitrogen) was added to each sample, and the mixture was incubated for 2 h with rotation. The beads were washed five times with 500 µl of a 1:1 mixture of MNase buffer containing 20 mM EDTA and lysis buffer. For histone modifications, incubation and washing were performed with higher NaCl concentrations (plus 400 mM) to strip chromatin-binding proteins. DNAs were harvested from the precipitates by dissolving in elution buffer followed by phenol/chloroform/iso-amyl alcohol extraction and ethanol precipitation. Deep sequencing of the precipitated DNAs was performed using the Illumina genome analyzer IIx at the Joint Usage/Research Center (RIRBM), Hiroshima University. qPCR was performed on the precipitated DNAs using the custom-made primer sets described in Supplementary Table S3. The value relative to the input was determined using a standard curve and the relative quantification method as described in the ABI User Bulletin #2. Optionally, co-precipitated proteins were harvested by dissolving the immunoprecipitates in elution buffer, mixed with an equal volume of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer and subjected to western blotting.
Liquid chromatography-tandem mass spectrometry analysis
The method for trypsin digestion of protein has been described previously (22). Tandem mass spectrometry analysis was performed using an LTQ Orbitrap ELITE ETD mass spectrometer (Thermo Fisher Scientific). The methods used for liquid chromatography-tandem mass spectrometry (LC-MS/MS) were slightly modified from those described previously (23). The mass spectrometer was operated in data-dependent acquisition mode in which MS acquisition with a mass range of m/z 400–1000 was automatically switched to MS/MS acquisition under the control of Xcalibur software. The top four precursor ions in the MS scan were selected by Orbitrap, with resolution R = 240 000, and those in the subsequent MS/MS scans, with an ion trap in automated gain control (AGC) mode, where AGC values were 1 × 106 and 1.00 × 104 for full MS and MS/MS, respectively. For fragmentation, electron transfer dissociation was used.
In vivo leukemogenesis assay
C-kit-positive cells (2 × 105) prepared from mouse femurs and tibiae were transduced with retrovirus by spinoculation and intravenously transplanted into sublethally irradiated (two doses of 500 rad in 2 days) C57BL/6 mice. Moribund mice were sacrificed, and the spleen cells were either subjected to cytospin preparation followed by May-Grunwald/Giemsa staining or temporarily cultured in methylcellulose medium in the presence of G418 (1 mg/ml) to remove untransformed cells, and then subjected to secondary transplantation or reverse transcription-PCR (RT-PCR) analysis.
CpG island recovery assay
CpG island recovery assays for non-methylated CpGs (CIRA) and methylated CpGs (MIRA) were performed using the Unmethyl Collector kit and Methyl Collector Ultra kit, respectively (Active Motif, Carlsbad, CA). Deep sequencing after CIRA and MIRA was carried out at the Joint Usage/Research Center (RIRBM), Hiroshima University.
Transactivation assay
Transactivation assays using the pFR-luc reporter (Clontech) were performed as described elsewhere (14). Relative luciferase activities were normalized to Renilla luciferase activity and expressed in terms of the average values and standard deviations (SDs) of triplicate determinations relative to the GAL4 DNA binding domain controls.
RESULTS
Murine leukemia models define the major functional modules required for leukemic transformation by MLL-ENL
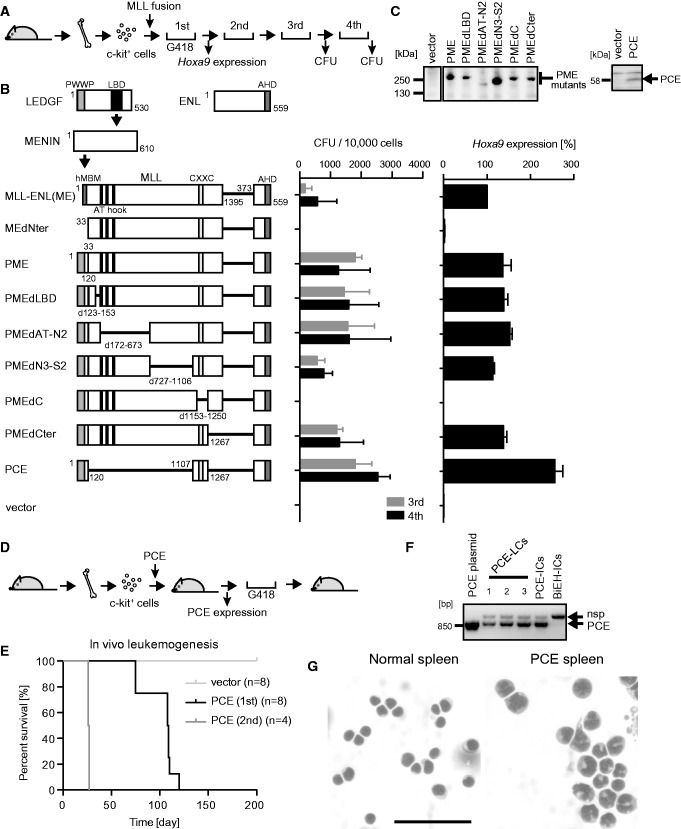
MLL fusion proteins form a trimeric complex with menin and LEDGF through the MLL portion (7). Because MLL fusion proteins associate with LEDGF through menin as a mediator, an MLL-ENL mutant (MEdNter) lacking the high-affinity menin-binding motif failed to transform hematopoietic progenitors in a myeloid progenitor transformation assay (Figure 1A), in which successful transformation is represented by vigorous colony-forming activity in the third and fourth rounds of plating, and elevated expression of Hoxa9 in first-round colonies (Figure 1B). An artificial MLL-ENL fusion protein (PME), in which high-affinity menin-binding motif is replaced by the PWWP domain of LEDGF, transformed myeloid progenitors despite its inability to form the trimeric complex (7). Therefore, the PWWP domain is the single functional module within menin and LEDGF that is required for MLL-ENL-dependent transformation.
Figure 1.
Major functional domains required for leukemogenesis by MLL-ENL. (A) Experimental scheme for the myeloid progenitor transformation assay. (B) Transforming ability of various MLL-ENL (ME) mutants. The schematic structures of LEDGF, MENIN, ENL and various ME mutants are shown, with the key functional modules indicated (left). P: the PWWP domain. C: the CXXC domain. E: the ENL portion included in MLL-ENL. The numbers of colony-forming units (CFUs) at the third and fourth rounds of replating are shown with error bars (SD of >3 independent experiments) (middle). Hoxa9 expression is presented relative to the value of ME (arbitrarily set at 100%) with error bars (SD of triplicate PCRs) (right). (C) Protein expression of the PWWP-MLL-ENL (PME) and PCE mutants in the packaging cells. The PME and PCE proteins were visualized using the anti-ENL antibody. Molecular standards are shown on the left. (D) Experimental scheme for the in vivo leukemogenesis assay. (E) Survival of the transplanted animals in the in vivo leukemogenesis assay. Light gray, vector; black, first PCE transplantation; dark gray, second PCE transplantation. n, number of animals analyzed. (F) Expression of PCE in the PCE-induced leukemia cells (PCE-LCs). Leukemic cells derived from moribund mice were cultured ex vivo in the presence of G418 and analyzed by RT-PCR. The PCE plasmid, PCE-immortalized cells (PCE-ICs) and Bcl2/E2A-HLF-immortalized cells (BiEH-ICs) were included for comparison. nsp, non-specific band. (G) Morphology of PCE-LCs in the spleen. The splenic cells harvested from a PCE-leukemic mouse were stained by the May-Grunwald/Giemsa staining method. Normal spleen is included for comparison. Scale bar, 50 µm.
To identify the structural requirements of MLL fusion proteins besides the PWWP domain, we examined the transforming ability of a series of PME mutants with various internal deletions in the MLL gene. A mutant lacking the CXXC domain (PMEdC) failed to transform myeloid progenitors (Figure 1B and C). Conversely, a mutant (PCE) consisting only of the PWWP domain, the CXXC domain and the ENL portion activated Hoxa9 expression and immortalized myeloid progenitors. The PCE mutant induced leukemia in recipient mice within 3–4 months in the in vivo leukemogenesis assay (Figure 1D–F). The PCE-induced leukemic blasts had monocytic features (Figure 1G) and elevated expression of posterior Hoxa genes and leukemia stem cell signature genes including Myb, Hmgb3 and Cbx5 (Supplementary Figure S1), which are major characteristics of MLL-associated myeloid leukemias (24). The leukemic blasts recapitulated the same disease in secondary recipients with a much shorter latency period (Figure 1E). These results clearly show that PCE is as functional as MLL-ENL for leukemic transformation. Thus, the PWWP domain, the CXXC domain and the ENL portion are the major structural elements required for MLL-associated leukemogenesis.
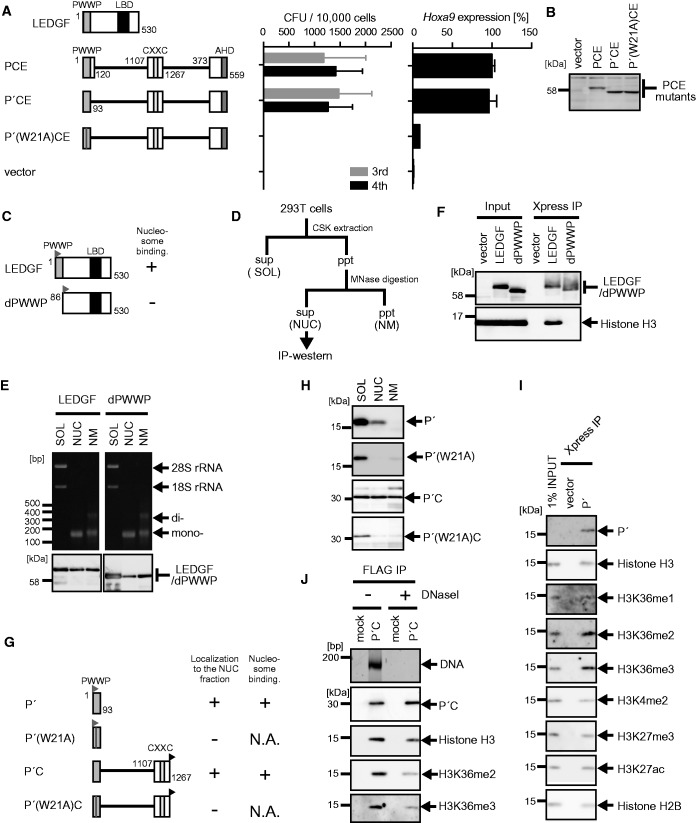
Binding to a specific nucleosome through the PWWP domain is essential for MLL fusion-dependent gene activation
To further analyze the function of the PWWP domain, we next tested a series of artificial PCE derivatives with various alterations in the PWWP domain for the transforming ability of myeloid progenitors (Figure 2A and B). A mutant (P′CE) with the minimum structure of the PWWP domain exhibited this transformation ability, whereas an alanine substitution of the evolutionarily conserved tryptophan (W21) resulted in a loss of the transformation activity. The PWWP domain of LEDGF has been implicated in chromatin binding (25,26). To examine its chromatin-binding capacity, we transiently expressed LEDGF and its mutant (dPWWP) lacking the PWWP domain in 293T cells (Figure 2C), and we divided the cellular components into three subfractions (Figure 2D and Supplementary Figure S2A). In this procedure, (i) chromatin-unbound soluble materials were extracted by cytoskeletal (CSK) buffer (the soluble fraction); (ii) chromatin and its associated factors were eluted by DNA fragmentation with micrococcal nuclease (MNase) (the nucleosome fraction); and (iii) insoluble materials after MNase treatment were solubilized by denaturing detergent (the nuclear matrix fraction). Nearly all the nucleosomes in the nucleosome fraction were mononucleosomes based on their DNA length (∼150 bp) (Figure 2E, top panels). LEDGF substantially localized to the nucleosome fraction (Figure 2E, bottom panels) and was able to pull down mononucleosomes (Figure 2F), whereas dPWWP mainly localized to the soluble fraction (Figure 2E) and failed to pull down nucleosomes (Figure 2F). The PWWP domain (P′) by itself localized in the nucleosome fraction (Figure 2G and H) and pulled down endogenous nucleosomes (Figure 2I). Previously, one group showed that LEDGF had specifically associated with the histone H3 peptide containing either H3K36me2 or H3K36me3 (25), while another group showed that the recombinant PWWP domain of LEDGF directly binds to H3K36me3 but not to H3K36me2 in vitro (26), suggesting that the peptide pull-down assay tends to overrepresent H3K36me3–PWWP interaction, depending on the assay conditions. Our nucleosome co-immunoprecipitation (co-IP) assay, which can detect protein–nucleosome interaction under physiological conditions, showed that the nucleosomes co-precipitated with P′ contained high amounts of not only H3K36me3 but also H3K36me2 (Figure 2I), indicating that the PWWP domain of LEDGF is capable of binding to either H3K36me2 or H3K36me3 in vivo. In accordance with these results, an artificial protein (P′C) composed of the PWWP domain and the CXXC domain was substantially localized in the nucleosome fraction (Figures 2G and H) and co-precipitated with mononucleosomes and DNAs (Supplementary Figure S2B and C). The W21A substitution mutants of P′ and P′C exclusively localized to the soluble fraction (Figures 2G and H) and therefore were not used for the nucleosome co-IP analysis (Supplementary Figures S2D and E). These results indicate that the PWWP domain, and not the CXXC domain, is mainly responsible for stable association with chromatin. The purified P′C–nucleosome complex contained no other proteins but histones at the stoichiometric level (Supplementary Figure S2B). Complete removal of the DNAs by DNase I treatment did not abolish the P′C–nucleosome interaction (Figure 2J) or P′–nucleosome interaction (Supplementary Figure S2F). Thus, the PWWP domain directly associates with histones. Taken together, these findings show that the binding ability of the PWWP domain to specific histones is required for MLL fusion-dependent gene activation.
Figure 2.
Nucleosome binding by the PWWP domain is essential for MLL-ENL-dependent transformation. (A) Transforming ability of PCE mutants containing various mutations within the PWWP domain. P′: the minimum structure of the PWWP domain. The schematic structures of LEDGF and various PCE mutants are shown (left). The CFUs at the third and fourth rounds of replating are shown with error bars (SD of >3 independent experiments) (middle). Hoxa9 expression in the first-round colonies is expressed relative to that of PCE (arbitrarily set at 100%) with error bars (SD of triplicate PCRs) (right). (B) Protein expression of the PCE mutants in the packaging cells. The PCE mutant proteins were visualized using the anti-ENL antibody. (C) The schematic structures of LEDGF and its mutant lacking the PWWP domain (dPWWP). An Xpress tag (gray flag) is fused to the N-terminal end of the LEDGF proteins. Their ability to associate with nucleosomes is summarized. (D) Experimental scheme of subcellular fractionation. (E) Distribution of DNAs/RNAs (top) and LEDGF proteins (bottom) in the three subfractions. 293T cells transiently expressing the indicated proteins were subfractionated into the soluble fraction (SOL), the nucleosome fraction (NUC) and the nuclear matrix fraction (NM). The LEDGF proteins were visualized using anti-Xpress antibody. (F) Nucleosomes associated with LEDGF and dPWWP. Nucleosome co-IP assay was performed with the nucleosome fractions of 293T cells transiently expressing the LEDGF proteins. The LEDGF mutant proteins and histone H3 proteins were visualized using anti-Xpress and histone H3 antibodies, respectively. (G) The schematic structures of the P′ and the P′C mutants. An Xpress tag (gray flag) is fused to the N-terminal end of P′ mutants. A FLAG tag (black flag) is fused to the C-terminal end of the P′C mutants. The localization to the nucleosome fraction of each mutant and its ability to bind nucleosomes are summarized. N.A., not applicable. (H) Subcellular distribution of the P′ and P′C mutants. Experiments were performed as in (E). The P′ mutants were visualized by using the anti-Xpress antibody. The P′C mutants were visualized by using the anti-FLAG antibody. (I) Nucleosomes associated with P′. P′ and its associating nucleosomes were co-purified by co-IP from the NUC fraction of 293T cells transiently expressing P′ as in (F). Nucleosomes were visualized by antibodies specific for mono-methylated histone H3 lysine 36 (H3K36me1), H3K36me2 and H3K36me3 or by anti-histone H2B and -H3 antibodies. Di-methylated histone H3 lysine 4 (H3K4me2), tri-methylated histone H3 lysine 27 (H3K27me3) and acetylated histone H3 lysine 27 (H3K27ac) were also analyzed for comparison. (J) The role of DNAs in P′C–nucleosome interaction. P′C and its associating nucleosomes were co-purified by co-IP using the anti-FLAG antibody from the NUC fraction of 293T cells expressing P′C. The precipitates were treated with or without DNase I (300 kU/ml) for 10 min and washed to remove unbound material. DNAs co-precipitated with P′C were visualized by SYBR Green staining. Nucleosomes were visualized by antibodies specific for H3K36me2 and H3K36me3 modification or by the anti-histone-H3 antibody.
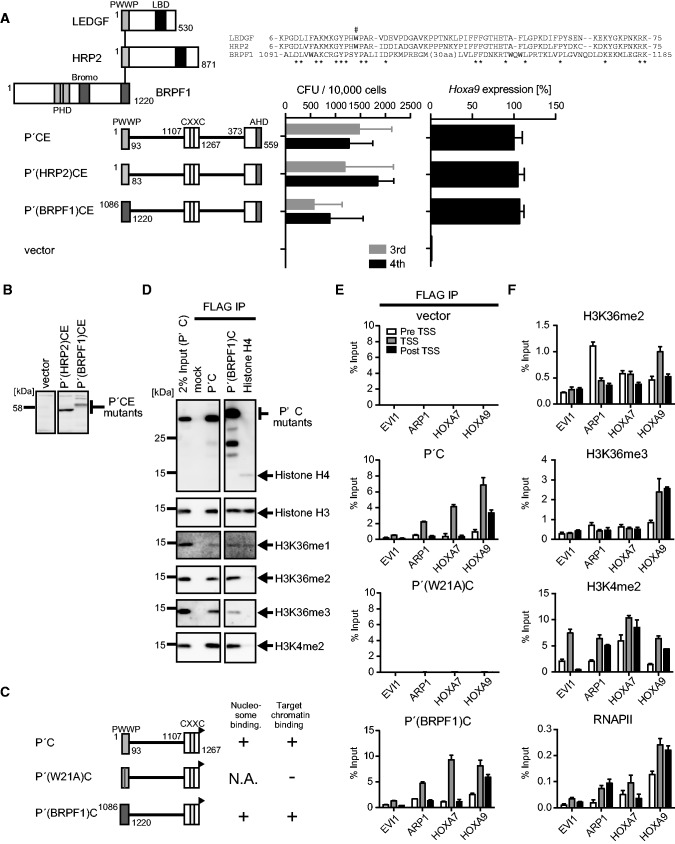
The PWWP domain and the CXXC domain target transcriptionally active promoters containing either H3K36me2 or H3K36me3
We next examined the functional similarities among various PWWP domains. HRP2 is a close homolog of LEDGF and has a highly homologous PWWP domain (Figure 3A). The bromodomain and PHD finger-containing 1 (BRPF1) protein is an associated factor of the MOZ histone acetyltransferase (27) and has a less conserved PWWP domain, which has been recently shown to directly bind to histone H3 peptide containing H3K36me2 or H3K36me3 in vitro (28). To investigate the functional differences and similarities of the PWWP domains of LEDGF, HRP2 and BRPF1, we tested whether these PWWP domains are interchangeable in the myeloid progenitor transformation assay. The P′CE mutants whose PWWP domain is replaced by those of HRP2 and LEDGF successfully activated Hoxa9 expression and immortalized myeloid progenitors (Figure 3A and B). Nucleosome co-IP assay with the P′C mutants containing the PWWP domains of LEDGF and BRPF1 specifically enriched nucleosomes containing H3K36me2 more efficiently than those containing H3K36me3 (Figure 3C and D). Consistent with these results, mass spectrometry analysis indicated that two-thirds of P′C-bound nucleosomes contained the H3K36me2 marker, while the H3K36me3 marker was minor compared with P′-bound nucleosomes (Supplementary Figure S3). In addition, chromatin modifications characteristic to active promoters such as di-methylated histone H3 lysine 4 (H3K4me2) (Figures 2I and 3D), acetylated histone H3 lysine 9 (H3K9ac) and acetylated histone H3 lysine 27 (H3K27ac) (Supplementary Figure S3) were enriched in P′C-bound nucleosomes, suggesting that the CXXC domain confers affinity toward active promoters. To determine the targeting ability of these artificial proteins to known MLL target genes in vivo, we performed chromatin immunoprecipitation (ChIP) followed by qPCR on 293T cells transiently expressing those P′C mutants (Figure 3C and E). To this end, we devised a highly sensitive native ChIP protocol by altering the aforementioned nucleosome IP protocol. In this ChIP method, we resuspended the cells in CSK buffer to remove the chromatin-unbound materials and treated the cells with MNase to digest the DNAs to achieve the average length of 500–1000 bp. The highly solubilized chromatin fraction was extracted with lysis buffer and then subjected to IP. We designated this protocol fractionation-assisted native ChIP, but hereafter it is referred to as ChIP. Ecotropic viral integration site 1 (EVI1), paired-like homeodomain transcription factor 2 (PITX2), HOXA7 and HOXA9 were previously reported as potential MLL target genes (14,28–30). Although the chromatin environment in 293T cells might be different from that in leukemia cells, the P′C mutants, but not P′(W21A)C, were localized at the transcription start sites (TSSs) of PITX2, HOXA7 and HOXA9, but not EVI1 (Figure 3E). These genes showed enrichment of H3K4me2 near the TSS, consistent with the observation that P′C-bound nucleosomes contain high levels of H3K4me2 modification (Figure 3D–F). H3K36me2 was present in all of the P′C mutant-occupied loci, while H3K36me3 was only found at the HOXA9 locus (Figure 3F), supporting the notion that the PWWP domain binds either H3K36me2 or H3K36me3 in vivo. The presence of RNA polymerase II (RNAPII) indicated that PITX2, HOXA7 and HOXA9, but not EVI1, were transcriptionally active in 293T cells (Figure 3F). Thus, the PWWP domain and the CXXC domain collaboratively target the promoter-proximal regions of transcriptionally active genes containing either H3K36me2 or H3K36me3.
Figure 3.
The PWWP domain and the CXXC domain target active promoters containing H3K36me2/3 in vivo. (A) Transforming ability of P′CE mutants containing three different PWWP domains. The schematic structures of LEDGF, HRP2, BRPF1 and various P′CE mutants are shown (left). Sequence alignment of the three PWWP domains is shown. Asterisk, conserved residue. The tryptophan residue mutated in Figure 2A is indicated by #. The CFUs at the third and fourth rounds of replating are shown with error bars (SD of >3 independent experiments) (middle). Hoxa9 expression in the first-round colonies is expressed relative to that of P′CE (arbitrarily set at 100%) with error bars (SD of triplicate PCRs) (right). (B) Protein expression of the P′CE mutants carrying non-LEDGF PWWP domains in the packaging cells. The P′CE mutant proteins were visualized using the anti-ENL antibody. (C) The schematic structures of various P′C mutants. A FLAG tag (black flag) is fused to the C-terminal end of the P′C mutants. Their abilities to bind nucleosomes and target promoters are summarized. (D) Nucleosomes associated with the P′C mutants. The P′C mutants and their associating nucleosomes were co-purified by co-IP using the anti-FLAG antibody from the NUC fraction of 293T cells transiently expressing the P′C mutants. Nucleosomes were visualized by antibodies specific for histone H3 or the indicated modifications. Nucleosomes co-purified with FLAG-tagged histone H4 were also analyzed for comparison. (E) Chromatin targeting ability of the P′C mutants. Genomic localization of the P′C mutants was analyzed by ChIP using the anti-FLAG antibody in 293T cells transiently expressing the P′C mutants. The precipitated DNAs were analyzed by qPCR using specific probes for pre-TSS (−1.0 to −0.5 kb of TSS), TSS (0 to +0.5 kb of TSS) and post-TSS (+1.0 to 1.5 kb of TSS) of the indicated genes. (F) Epigenetic status of MLL target genes in 293T cells. ChIP analysis was performed on 293T cells using antibodies specific for H3K36me2, H3K36me3, H3K4me2 and RNAPII. The precipitated DNAs were analyzed by qPCR as in (E).
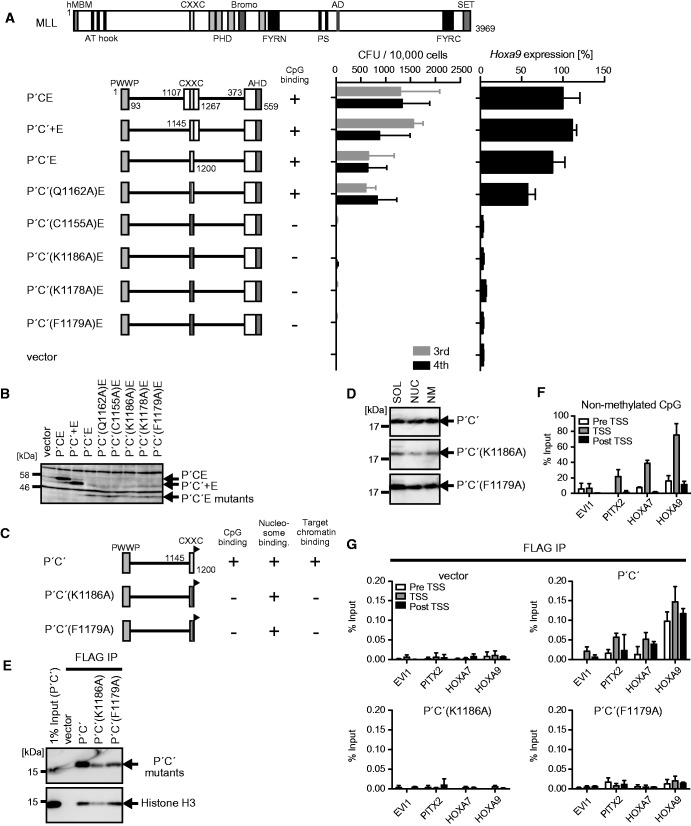
The chromatin context comprising H3K36me2/3 and non-methylated CpGs is targeted by MLL fusion proteins
Next, we examined the structural requirements of the CXXC domain. A deletion mutant (P′C′E) that lacks the sequences flanking the CXXC domain successfully transformed myeloid progenitors (Figure 4A and B). Therefore, the CXXC domain, but not its flanking sequences, is required for the proper targeting of MLL-ENL. The CXXC domain of MLL directly binds to non-methylated CpGs (31–34). Point mutations that have been proved to abolish the binding ability to non-methylated CpGs (C1155A and K1186A) (31) resulted in a loss of transforming activity (Figure 4A and B), while the Q1162A substitution, which has no effect on non-methylated CpG-specific binding, did not compromise transformation. Mutations in the KFGG motif (K1178A and K1179A) that severely attenuate the binding ability to non-methylated CpGs (31) also abolished transforming activity. The artificial protein (P′C′) composed of the minimum structures of the PWWP domain and the CXXC domain localized in the nucleosome fraction (Figure 4C and D) and associated with nucleosomes (Figure 4E). The P′C′ mutants carrying CpG binding-deficient mutations retained the nucleosome binding activity because they had an intact PWWP domain (Figure 4C–E). To investigate the CpG methylation status of the MLL target genes, we performed CIRA using the recombinant CXXC domain protein followed by qPCR, which showed that PITX2, HOXA7 and HOXA9 loci contained ample amounts of non-methylated CpGs near the TSSs in 293T cells (Figure 4F). ChIP-qPCR analysis of 293T cells transiently expressing these P′C′ mutants showed that only the wild-type control, not the CpG binding-deficient mutants, was able to associate with the promoters of MLL target genes (Figure 4G). Thus, binding to non-methylated CpGs through the CXXC domain is essential for the proper targeting of MLL fusion proteins.
Figure 4.
MLL-ENL recognizes the chromatin context with H3K36me2/3 and non-methylated CpGs. (A) Transforming ability of P′CE mutants with various mutations within the CXXC domain. The schematic structures of MLL and various P′CE mutants are shown (left). C′: the minimum structure of the CXXC domain; C′+, the minimum structure of the CXXC domain and its C-terminal flanking region. Their ability to bind to non-methylated CpGs is summarized. The CFUs at the third and fourth rounds of replating are shown with error bars (SD of >3 independent experiments) (middle). Hoxa9 expression in the first-round colonies is expressed relative to the value of P′CE (arbitrarily set at 100%) with error bars (SD of triplicate PCRs) (right). (B) Protein expression of the P′CE mutants in the packaging cells. The P′CE mutant proteins were visualized by the anti-ENL antibody. (C) The schematic structures of various P′C′ mutants. A FLAG tag (black flag) is fused to the C-terminal end of the P′C′ mutants. Their abilities to bind non-methylated CpGs, nucleosomes and target promoters are summarized. (D) Subcellular distribution of the P′C′ mutants. Experiments were performed as in Figure 2E. The P′C′ mutants were visualized by the anti-FLAG antibody. (E) Nucleosomes associated with the P′C′ mutants. The P′C mutants and their associating nucleosomes were analyzed as in Figure 2F. (F) Distribution of non-methylated CpGs at the MLL target genes. Genomic DNAs of 293T cells were analyzed by CIRA. The precipitated DNAs were analyzed by qPCR as in Figure 3E. (G) Chromatin targeting ability of the P′C mutants. Genomic localization of the P′C′ mutants was analyzed by ChIP as in Figure 3E.
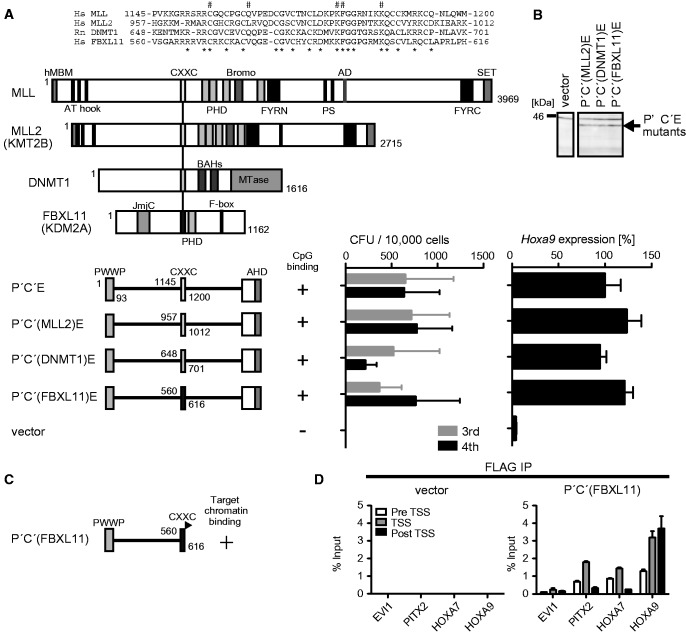
There are dozens of CXXC domain-containing proteins in mammals. To examine the functional similarity of different CXXC domains, we generated several domain swap mutants of P′C′E whose CXXC domain is replaced by other CXXC domains (Figure 5A and B). The CXXC domains of MLL2 (also known as HRX2, MLL4 and KMT2B) (35), DNA (cytosine-5)-methyltransferase 1 (DNMT1) (36) and F-box and leucine-rich repeat protein 11 (FBXL11; also known as KDM2A) (37) were previously shown to bind to non-methylated CpGs. P′C′E mutants carrying these three non-MLL CXXC domains were competent for leukemic transformation (Figure 5A and B). Furthermore, the P′C′ mutant with the CXXC domain of FBXL11 (Figure 5C and Supplementary Figure S4) was able to associate with MLL target genes in 293T cells (Figure 5D). These results indicate that the three different CXXC domains are functionally equivalent in terms of MLL-dependent gene activation, confirming the notion that the binding ability to non-methylated CpGs, a common function of those three CXXC domains, is required for targeting of MLL fusion proteins.
Figure 5.
Non-MLL CXXC domains can functionally substitute for the MLL CXXC domain. (A) Transforming ability of P′C′E mutants with various non-MLL CXXC domains. The schematic structures of MLL, MLL2, DNMT1, FBXL11 and various P′C′E mutants are shown (left). Sequence alignment of the CXXC domains of MLL, MLL2, DNMT1 and FBXL11. The mutated residues in Figure 4A are indicated by #. Asterisk, conserved residue. The CFUs at the third and fourth rounds of replating are shown with error bars (SD of >3 independent experiments) (middle). Hoxa9 expression in the first-round colonies is expressed relative to the value of P′C′E (arbitrarily set at 100%) with error bars (SD of triplicate PCRs) (right). (B) Protein expression of the P′C′E mutants carrying non-MLL CXXC domains in the packaging cells. The P′C′E mutant proteins were visualized by the anti-ENL antibody. (C) The schematic structure of the P′C′ mutant carrying the CXXC domain of FBXL11 [P′C′(FBXL11)]. A FLAG tag is fused to the C-terminal end of the CXXC domain (black flag). Its ability to associate with the target promoters is indicated. (D) Chromatin targeting ability of P′C′(FBXL11). Genomic localization of P′C′(FBXL11) was analyzed by ChIP as in Figure 3E.
Taken together, the minimum functional modules required for the targeting of MLL fusion protein are the PWWP domain and the CXXC domain, which bind to nucleosomes with H3K36me2/3 and non-methylated CpGs, respectively. Therefore, the MLL fusion complex targets a chromatin context in which H3K36me2/3 and non-methylated CpGs coexist.
The MLL-AF6 complex localizes at transcriptionally active promoters containing H3K36me2/3 and non-methylated CpGs in vivo
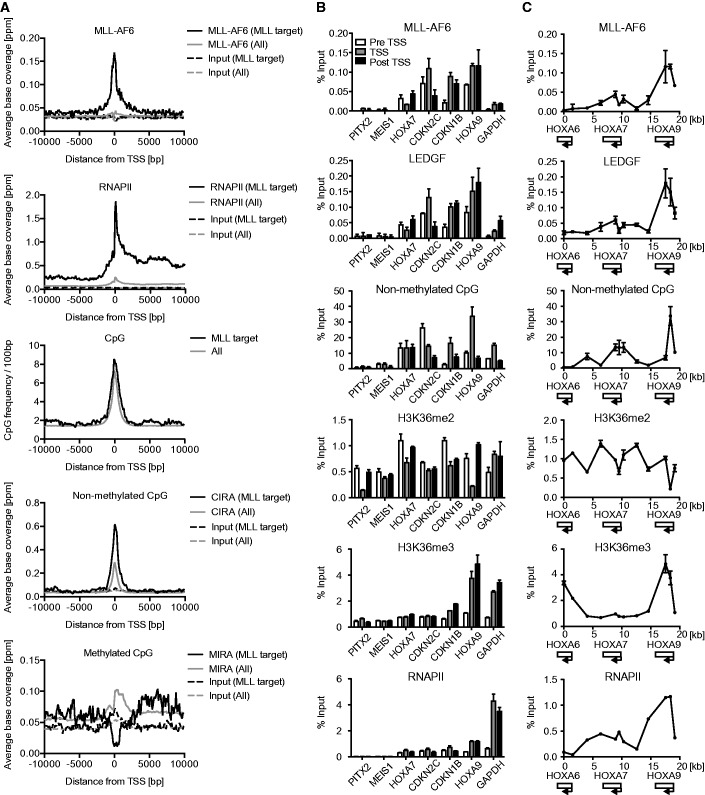
To determine the genomic landscape of the MLL target genes under physiological conditions, we performed a genome-wide ChIP analysis of cells that express an MLL fusion protein endogenously. ML-2 cells express MLL-AF6, but not wild-type MLL (38,39); therefore, the ChIP signal obtained from ML-2 cells using the anti-MLL antibody can be ascribed to the presence of MLL-AF6. Our ChIP analysis followed by deep sequencing (ChIP-seq) using the anti-MLL antibody identified 154 TSSs of 131 MLL-AF6 target genes (Supplementary Table S1) by virtue of enriched ChIP signals of MLL-AF6 near the TSSs. These regions included previously identified MLL target genes such as HOXA9, HOXA10, cyclin-dependent kinase inhibitor 1B (CDKN1B), CDKN2C, transcription factor 4 (TCF4) and runt-related transcription factor 2 (RUNX2) (14,40), and some newly identified genes such as special AT-rich sequence-binding protein-1 (SATB1), ribosomal protein L31 (RPL31) and RPL10A (Supplementary Figure S5A and Supplementary Table S1). Gene ontology analysis of these MLL-AF6 target genes indicates that MLL-AF6 mainly associates with genes involved in gene expression and translation (Supplementary Figure S5B). The average distribution of MLL-AF6 at the MLL target loci indicates that MLL-AF6 localizes near the TSSs (Figure 6A). MLL target loci contain a relatively large amount of CpG sequence near the TSSs. CIRA and MIRA followed by deep sequencing (CIRA-seq and MIRA-seq respectively) showed that MLL target loci are rich with non-methylated CpGs but are devoid of methylated CpGs. MLL-AF6-occupied loci also exhibited an enrichment of RNAPII and H3K36me3 (Figure 6A and Supplementary Figure S5C). Thus, endogenously expressed MLL-AF6 specifically localizes at CpG-rich promoters of transcriptionally active genes.
Figure 6.
Genomic landscape of MLL-AF6 target genes in ML-2 cells. (A) The average distribution of MLL-AF6, RNAPII, CpG dinucleotide, non-methylated CpGs and methylated CpGs at the 154 MLL-AF6-occupied TSSs. The input data and all TSS data are included for comparison. ChIP-seq or CIRA/MIRA-seq tags were clustered into 100-bp bins. On the x axis, the position of the associated TSS is designated as 0 bp. The y axis indicates the normalized ChIP-seq tag count (ppm). The average number of CpG dinucleotide in each bin was also plotted. (B) Genomic localization of MLL-AF6, LEDGF, non-methylated CpGs, H3K36me2, H3K36me3 and RNAPII in ML-2 cells at various gene loci. Genomic localization was determined by ChIP-qPCR or CIRA-qPCR. The precipitated DNAs were analyzed using the specific probes for pre-TSS (−1.0 to −0.5 kb of TSS), TSS (0 to +0.5 kb of TSS) and post-TSS (+1.0 to 1.5 kb of TSS) of the indicated genes. (C) Genomic localization of MLL-AF6, LEDGF, non-methylated CpGs, H3K36me2, H3K36me3 and RNAPII in ML-2 cells at the posterior HOXA loci. Relative locations of the HOXA6, HOXA7 and HOXA9 loci are shown at the bottom.
ChIP-qPCR and CIRA-qPCR analyses on each gene confirmed that the MLL-AF6 complex localized at the major MLL target loci including HOXA7, CDKN2C, CDKN1B and HOXA9 but was completely absent at the gene loci of PITX2 and MEIS1, which are potentially regulated by MLL (14) but are transcriptionally inactive in this cell line (Figure 6B and C and Supplementary Figure S5D and E). MLL-AF6, menin and LEDGF co-localized near the TSSs of MLL target genes where non-methylated CpGs are abundant (Figure 6B and Supplementary Figure S5D). H3K36me2 was ubiquitously present and relatively enriched at the transcriptionally active loci. The MLL-AF6 complex localized at the promoter-proximal regions of HOXA7 and CDKN2C, which were H3K36me2-rich but H3K36me3-poor (Figure 6B and C). On the other hand, the MLL-AF6 complex was highly enriched near the TSS of HOXA9, where H3K36me2 was scarce but H3K36me3 was abundant (Figure 6B and C and Supplementary Figure S5D and E). These results confirm the notion that the presence of either H3K36me2 or H3K36me3 is required for targeting of the MLL fusion complex. Hence, our data strongly indicate that the MLL-fusion complex targets the promoter-proximal regions containing H3K36me2/3 and non-methylated CpGs under physiological conditions.
It should be noted that the MLL fusion complex was also found by ChIP-qPCR at a housekeeping gene, glyceraldehyde phosphate dehydrogenase (GAPDH), whose promoter-proximal region also contained H3K36me2/3 and non-methylated CpGs near the TSS (Figure 6B and Supplementary Figure S5D). However, the localization of the MLL-AF6 complex was much weaker than that at well-known MLL target genes. These results suggest that the extent of occupation by the MLL fusion complex is influenced not only by the amounts of H3K36me2/3 and non-methylated CpGs but also by other factors or environments that are yet undetermined.
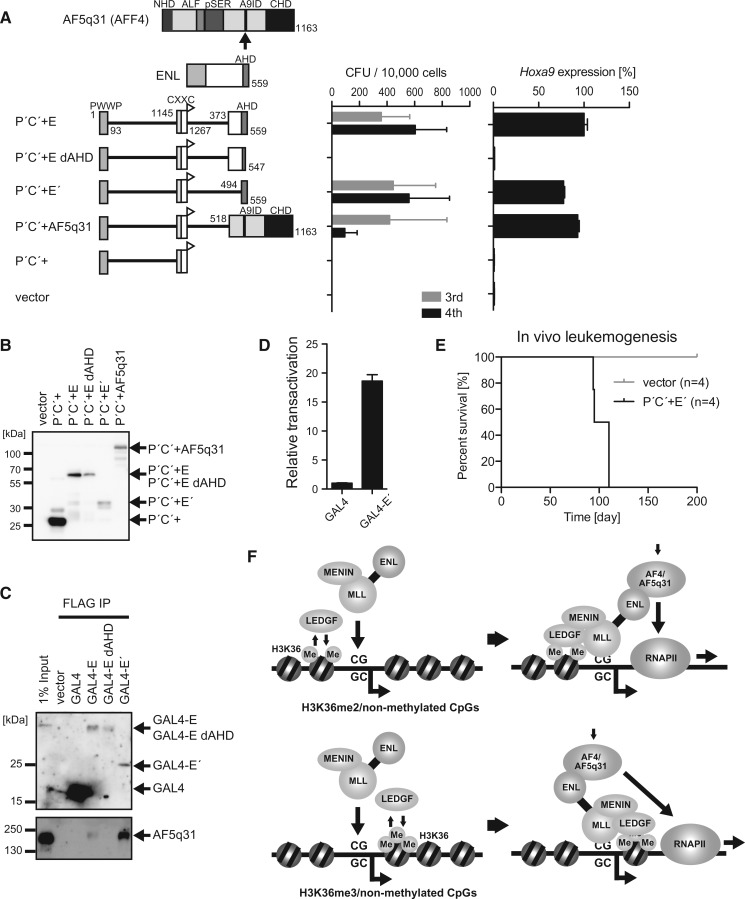
An interaction domain for AF4-family coactivators and the targeting modules are the minimum elements required for MLL-ENL-dependent leukemogenesis
Next, we analyzed the structural requirements of the fusion partner portion. We tested the transforming ability of various fusion partners that were fused with the minimum targeting structure (P′C′+) composed of the PWWP domain and the CXXC domain (the C-terminal flanking sequence of the CXXC domain was included to increase the protein stability). An artificial protein (P′C′+E′) composed of the minimum targeting structure and the ANC1 homology domain (AHD) of ENL activated Hoxa9 expression and transformed myeloid progenitors, whereas a P′C′+E mutant lacking a part of AHD failed to transform myeloid progenitors (Figure 7A and B). AHD is a binding platform for AF4 family transcriptional coactivators (14). Indeed, the GAL4 DNA binding domain fused with AHD (GAL4-E′) associated with AF5q31 (also known as AFF4 and MCEF) (Figure 7C and Supplementary Figure S6), which is a member of the AF4 family, and exhibited transcriptional activation activity (Figure 7D). Removing AHD from P′C′+E′ resulted in the loss of transforming ability (Figure 7A and B), whereas adding back the AF5q31 portion restored the transforming ability, indicating that association with the AF4-family coactivator is essential for MLL-ENL-dependent transformation. In vivo leukemogenesis assay showed that P′C′+E′ induced leukemia in vivo (Figure 7E). Therefore, the PWWP domain, the CXXC domain and AHD are the minimum essential structures required for leukemic transformation. Taken together, these results show that MLL fusion proteins target the promoter-proximal regions of transcriptionally active CpG-rich genes through dual recognition of epigenetic markers and activate transcription by recruiting transcriptional coactivators such as AF4-family proteins (Figure 7F).
Figure 7.
MLL fusion-dependent gene activation is mediated by AF4-family coactivators. (A) Transforming ability of P′C′+ mutants fused with ENL and AF5q31. The schematic structures of ENL, AF5q31 and various P′C′+ mutants are shown (left). An HA tag (white flag) is fused to the C-terminal end of the P′C′+ structure. E dAHD:the ENL portion lacking the ANC1 homology domain (AHD). E′: the minimum transformation domain of ENL composed of AHD. The CFUs at the third and fourth rounds of replating are shown with error bars (SD of >3 independent experiments) (middle). Hoxa9 expression in the first-round colonies is expressed relative to the value of P′C′+E (arbitrarily set at 100%) with error bars (SD of triplicate PCRs) (right). (B) Protein expression of the P′C′+ mutants fused with ENL and AF5q31 in the packaging cells. The P′C′+ mutant proteins were visualized by the anti-HA antibody. (C) Association of AHD and the AF4-family proteins. A series of FLAG-tagged GAL4-ENL fusion proteins (Supplementary Figure S6) were transiently expressed in 293T cells and subjected to ChIP using the anti-FLAG antibody. The precipitates were analyzed by western blotting using the anti-FLAG and anti-AF5q31 antibodies. (D) Transcriptional activation activity of AHD. A FLAG-tagged GAL4 DNA binding domain fused with AHD (GAL4-E′) was transiently expressed in 293T cells with the pFR-luc reporter plasmid (5 GAL4 binding sequences and a TATA box are placed in front of the luciferase gene) and subjected to luciferase assay. Error bars represent the SD of triplicate analyses. (E) Survival of the transplanted animals in the in vivo leukemogenesis assay. Gray, vector; black, P′C′+E. n, number of animals analyzed. (F) Models of MLL-ENL-dependent gene activation.
DISCUSSION
This study demonstrates that MLL fusion proteins target the promoter-proximal regions of previously active genes by simultaneously recognizing H3K36me2/3 and non-methylated CpGs and activate transcription by recruiting transcriptional coactivators. Our structure–function analysis demonstrated that an MLL-ENL mutant composed only of the PWWP domain, the CXXC domain and AHD was able to activate Hoxa9 expression and transform myeloid progenitors. These results imply that the PWWP domain and the CXXC domain are the only structures essentially required for target recognition by MLL fusion proteins. The PWWP domain specifically binds to either H3K36me2 or H3K36me3, both of which are indicators of active transcription. The CXXC domain binds to non-methylated CpGs, which are clustered in the active gene promoters. HOX loci are enriched with CpGs and are therefore preferentially subjected to MLL fusion-dependent gene activation. Our ChIP/CIRA analyses showed that the MLL fusion complex localizes at the promoter-proximal region, where non-methylated CpGs and H3K36me2/3 are present. Thus, dual recognition of the two epigenetic markers is the major determinant for MLL fusion proteins to identify their target genes. AHD is the platform to recruit AF4 family transcriptional coactivators, which confer transcriptional activation activity. The fundamental mechanism of MLL fusion-dependent gene activation is threefold: (i) LEDGF scans the genome and binds to H3K36me2/3 through the PWWP domain. (ii) The MLL fusion/menin complex binds to non-methylated CpGs in the promoter through the CXXC domain and a nearby chromatin-bound LEDGF. These two points of contact allow the MLL fusion complex to stably associate with its target chromatin. (iii) The MLL fusion complex activates transcription by recruiting transcriptional coactivators (Figure 7F). However, it should be noted that these three domains are the minimum elements, not all the elements, involved in MLL fusion-dependent leukemogenesis in the context of a full-length MLL fusion protein. The activity of the MLL fusion complex is likely regulated through intricate regulatory mechanisms. Factors that modulate its function may also play critical roles in MLL fusion-dependent leukemogenesis.
Aberrant self-renewal is an essential feature of oncogene-driven proliferation. It requires the continuous expression of a gene set that is required for self-renewal through rounds of cell divisions; however, its mechanisms are largely unclear. Because the PWWP and CXXC domains are sufficient for target recognition, the MLL fusion complex identifies posterior HOXA genes by a chromatin context-dependent mechanism rather than by mediating sequence-specific transcription factors. The H3K36me3 marker is generally deposited into chromatin in a transcriptional elongation-coupled manner by the SETD2 methyltransferase (also known as KMT3A, HYPB and SET2), which associates with actively elongating RNAPII (41–43). The H3K36me2 marker is deposited into chromatin by various SET domain-containing proteins, including NSD2, and is generally linked to transcriptional activation (16). Hence, previously transcribed genes containing H3K36me2/3 and non-methylated CpGs at the promoter-proximal regions are subject to gene activation by MLL fusion proteins. Consequently, the expression of previously transcribed HSC program genes is maintained at high levels through rounds of cell division to cause aberrant self-renewal.
In summary, our study identifies the minimum structural requirements of MLL fusion proteins for HOX gene activation and reveals the chromatin context recognized by them. A surprisingly simple combination of epigenetic markers that defines previously active gene promoters is used in MLL fusion-dependent cellular memory maintenance. Thus, our results provide significant insights into the molecular mechanisms of cellular memory maintenance and aberrant self-renewal caused by MLL fusion proteins.
ACCESSION NUMBERS
We generated ChIP-seq and CIRA/MIRA-seq data in this study. Mass data obtained in this study have been deposited under accession numbers DRA000555, DRA000557–DRA000561 in the DDBJ (DNA Data Bank of Japan) Sequence Read Archive.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Society for the Promotion of Science (KAKENNHI) [23689050, 22118003, 25118511, 25670450 to A.Y., 25870373 to H.O.]. Funding for open access charge: Japan Society for the Promotion of Science (KAKENNHI) [22118003 to A.Y.].
Conflict of interest. A.Y. has research funding from Dainippon Sumitomo Pharma Co., Ltd. The funder had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Chikako Hatanaka, Yukari Jindai, Liu Kehong and Dr. Takahiro Kihara for technical assistance; Dr. Toshio Kitamura for providing plat-E cells; and Drs. Hiroshi Handa and Yuki Yamaguchi for discussion and critical reading of the manuscript.
REFERENCES
- 1.Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl Acad. Sci. USA. 1998;95:10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yagi H, Deguchi K, Aono A, Tani Y, Kishimoto T, Komori T. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood. 1998;92:108–117. [PubMed] [Google Scholar]
- 4.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 5.Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, Humphries K, Sauvageau G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–129. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 6.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutherland HG, Newton K, Brownstein DG, Holmes MC, Kress C, Semple CA, Bickmore WA. Disruption of Ledgf/Psip1 results in perinatal mortality and homeotic skeletal transformations. Mol. Cell. Biol. 2006;26:7201–7210. doi: 10.1128/MCB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, Roeder RG. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc. Natl Acad. Sci. USA. 2011;108:15751–15756. doi: 10.1073/pnas.1111498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell. 2011;44:609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith KS, Rhee JW, Cleary ML. Transformation of bone marrow B-cell progenitors by E2a-Hlf requires coexpression of Bcl-2. Mol. Cell. Biol. 2002;22:7678–7687. doi: 10.1128/MCB.22.21.7678-7687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 19.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoyama A, Kawaguchi Y, Kitabayashi I, Ohki M, Hirai K. The conserved domain CR2 of Epstein-Barr virus nuclear antigen leader protein is responsible not only for nuclear matrix association but also for nuclear localization. Virology. 2001;279:401–413. doi: 10.1006/viro.2000.0715. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujinoki M, Kawamura T, Toda T, Ohtake H, Ishimoda-Takagi T, Shimizu N, Yamaoka S, Okuno M. Identification of 36-kDa flagellar phosphoproteins associated with hamster sperm motility. J. Biochem. 2003;133:361–369. doi: 10.1093/jb/mvg048. [DOI] [PubMed] [Google Scholar]
- 23.Daigo K, Yamaguchi N, Kawamura T, Matsubara K, Jiang S, Ohashi R, Sudou Y, Kodama T, Naito M, Inoue K, et al. The proteomic profile of circulating pentraxin 3 (PTX3) complex in sepsis demonstrates the interaction with azurocidin 1 and other components of neutrophil extracellular traps. Mol. Cell. Proteomics. 2012;11:M111.015073. doi: 10.1074/mcp.M111.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daugaard M, Baude A, Fugger K, Povlsen LK, Beck H, Sorensen CS, Petersen NH, Sorensen PH, Lukas C, Bartek J, et al. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol. 2012;19:803–810. doi: 10.1038/nsmb.2314. [DOI] [PubMed] [Google Scholar]
- 26.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ullah M, Pelletier N, Xiao L, Zhao SP, Wang K, Degerny C, Tahmasebi S, Cayrou C, Doyon Y, Goh SL, et al. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol. Cell. Biol. 2008;28:6828–6843. doi: 10.1128/MCB.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. Biol. 2010;17:617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 29.Arakawa H, Nakamura T, Zhadanov AB, Fidanza V, Yano T, Bullrich F, Shimizu M, Blechman J, Mazo A, Canaani E, et al. Identification and characterization of the ARP1 gene, a target for the human acute leukemia ALL1 gene. Proc. Natl Acad. Sci. USA. 1998;95:4573–4578. doi: 10.1073/pnas.95.8.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W, Kumar AR, Hudson WA, Li Q, Wu B, Staggs RA, Lund EA, Sam TN, Kersey JH. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13:432–440. doi: 10.1016/j.ccr.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, Freund SM, Bycroft M, Warren AJ. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol. Cell. Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 2002;30:958–965. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat. Struct. Mol. Biol. 2010;17:62–68. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bach C, Mueller D, Buhl S, Garcia-Cuellar MP, Slany RK. Alterations of the CxxC domain preclude oncogenic activation of mixed-lineage leukemia 2. Oncogene. 2009;28:815–823. doi: 10.1038/onc.2008.443. [DOI] [PubMed] [Google Scholar]
- 36.Pradhan M, Esteve PO, Chin HG, Samaranayke M, Kim GD, Pradhan S. CXXC domain of human DNMT1 is essential for enzymatic activity. Biochemistry. 2008;47:10000–10009. doi: 10.1021/bi8011725. [DOI] [PubMed] [Google Scholar]
- 37.Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Mol. Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanabe S, Zeleznik-Le NJ, Kobayashi H, Vignon C, Espinosa R, III, LeBeau MM, Thirman MJ, Rowley JD. Analysis of the t(6;11)(q27;q23) in leukemia shows a consistent breakpoint in AF6 in three patients and in the ML-2 cell line. Genes Chromosomes Cancer. 1996;15:206–216. doi: 10.1002/(SICI)1098-2264(199604)15:4<206::AID-GCC2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 40.Wang QF, Wu G, Mi S, He F, Wu J, Dong J, Luo RT, Mattison R, Kaberlein JJ, Prabhakar S, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117:6895–6905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hampsey M, Reinberg D. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- 42.Hu M, Sun XJ, Zhang YL, Kuang Y, Hu CQ, Wu WL, Shen SH, Du TT, Li H, He F, et al. Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc. Natl Acad. Sci. USA. 2010;107:2956–2961. doi: 10.1073/pnas.0915033107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.