Abstract
The Arabidopsis thaliana plasma membrane proton ATPase genes, AHA1 and AHA2, are the two most highly expressed isoforms of an 11 gene family and are collectively essential for embryo development. We report the translational fusion of a tandem affinity-purification tag to the 5′ end of the AHA1 open reading frame in a genomic clone. Stable expression of TAP-tagged AHA1 in Arabidopsis rescues the embryonic lethal phenotype of endogenous double aha1/aha2 knockdowns. Western blots of SDS-PAGE and Blue Native gels show enrichment of AHA1 in plasma membrane fractions and indicate a hexameric quaternary structure. TAP-tagged AHA1 rescue lines exhibited reduced vertical root growth. Analysis of the plasma membrane and soluble proteomes identified several plasma membrane-localized proteins with alterred abundance in TAP-tagged AHA1 rescue lines compared to wild type. Using affinity-purification mass spectrometry, we uniquely identified two additional AHA isoforms, AHA9 and AHA11, which copurified with TAP-tagged AHA1. In conclusion, we have generated transgenic Arabidopsis lines in which a TAP-tagged AHA1 transgene has complemented all essential endogenous AHA1 and AHA2 functions and have shown that these plants can be used to purify AHA1 protein and to identify in planta interacting proteins by mass spectrometry.
The Arabidopsis thaliana H+-ATPase genes, AHA1 and AHA2, belong to a protein family of eleven 100 kDa, plasma membrane-localized P-type ATPases. AHA proteins belong to the electrogenic class of P-type ATPases responsible for generating primary active membrane potentials, which also includes the Ca2+-ATPase of the sarcoplasmic reticulum and the Na+/K+-ATPase of mammalian cells.1 Specifically, AHAs link ATP catalysis to the active transport of protons out of the cell, thereby establishing a transmembrane proton motive force (PMF) that drives many secondary transport processes.2−4 The PMF is composed of both a pH gradient and an electrical membrane potential that can reach in excess of −250 mV in roots.5 AHAs are highly expressed in cell types specialized for active transport, including phloem tissue, guard cells, and epidermal root hairs, and are major consumers of cellular ATP.4,6−8
Of the 11 genes encoding functional proteins, AHA1 and AHA2 are the two most highly expressed isoforms, are abundant in all tissues and developmental stages based on EST expression data, and are estimated to make up to 80% of all AHA activity.5,9,10 The plasma membrane proton pumps are hypothesized to play roles in the control of tip growth by generating localized proton fluxes11,12 and are highly regulated to control guard cell opening and closure in response to biotic and abiotic stimuli.13−16 Using reverse-genetic analysis, roles for AHA3 in pollen development, AHA4 in salt stress, and AHA10 in proanthocyanidin and vacuole biosynthesis have been reported.17−19 However, the essential, yet genetically redundant, nature of AHA1 and AHA2 has complicated reverse-genetic approaches to elucidate specific cellular functions for these two proteins.20 To create a more facile system for investigating AHA1 specific functions, we developed transgenic endogenous double aha1/aha2 knockdown lines rescued with a tandem affinity-purification (TAP)-tagged AHA1, thereby replacing ∼70% of the AHA protein in crude seedling and vegetative extracts with a fusion protein capable of facilitating isoform-specific methods including purification, biochemical analysis, and mass spectrometry.
An increasing interest in defining protein-interaction networks has been greatly facilitated by emerging proteomic, genomic, and imaging technologies.21−23 An especially significant contribution to the identification of protein interactions is the growing access to modern mass spectrometry (MS) resources and the ability to perform quantitative experiments.24 Recently, two large Arabidopsis protein-interaction networks have been published including a yeast two-hybrid approach that identified 6200 interactions among 2700 proteins25 and a split ubiquitin approach focused on membrane proteins that revealed 541 interactions between 239 proteins.26 No AHA1 or AHA2 protein interactions were identified in the yeast two-hybrid interactome database, and only two high-confidence candidates, both annotated as leucine-rich repeat kinases, were generated by the split ubiquitin approach. Furthermore, proteins previously published to interact with the plasma membrane proton pumps were not identified in these interactome studies.27−29,14,30,31
Affinity purification of epitope-tagged proteins combined with mass spectrometry-based proteomic analysis has become a widely established method for the characterization of protein complexes, and tandem affinity-purification tags have been successfully used to investigate protein complexes in plants via these affinity-purification mass spectrometry (AP–MS) approaches.32−36,23 Affinity-purification mass spectrometry methods have also been combined with chemical cross-linking for the purification of low-abundance plasma membrane protein complexes from plants.36
Herein, we report the functional rescue of aha1/aha2 double-knockout plants with a TAP-tagged AHA1 genomic transgene (Ntapi:gAHA1). We observed reduced vertical root growth in Ntapi:gAHA1 transgenic rescue lines, but no other phenotypic consequences in overall plant growth was apparent under standard laboratory growth conditions. To investigate the basis of the short-root phenotype and to assess the validity of using these plants for future AHA1 functional studies, we used a metabolically labeled mass spectrometry approach to characterize proteins showing increased or decreased abundance in Ntapi:gAHA1 compared to wild-type plants. We used affinity purification followed by tandem mass spectrometry to identify AHA1-interacting protein candidates by Ntapi:AHA1 copurification and investigated the effect of in vivo elicitation on copurifying proteins. The Ntapi:gAHA1 transgenic plants reported here will provide an important tool for AHA1 isoform-specific biochemical analysis, identification of new protein interactions, and verification of interactions previously identified using heterologous techniques.
Experimental Procedures
Plant Materials and Growth Conditions
Mutants (ecotype Columbia) carrying homozygous T-DNA insertions in AHA1 (aha1–6, SALK016325) and heterozygous insertions in AHA2 (aha2–4, SALK082786) were created by crossing as described previously.20 Seeds were germinated on plates containing half-strength Murashige & Skoog (M&S) salts, 1% (w/v) sucrose, and 0.7% (w/v) agar. Seedlings were transferred to soil/perlite mixture and grown under constant light at 21 °C.
Root Growth Assays
Root growth asssays were conducted as described previously.20,37
DNA Extraction and Plant Genotyping
Plant genomic DNA was extracted using the method of Krysan et al. with the elimination of the phenol/chloroform step.38 The presence of aha1–6 and aha2–4 T-DNA insertion or wild-type alleles was determined by PCR using allele-specific primers spanning the T-DNA and AHA1 or AHA2 genomic junctions (T-DNA LB: TCAAACAGGATTTTCGCCTGCT; S016325 LP: CGTCTCAACAAAAGTCTCTTTCA; S016325 RP: CGAAAGATCAACCTCGTGAGT; S082786 LP: ATGTTCATTGCAAAGGTGGTG; and S082786 RP: CCCATTAGCTCGTGGTTATTG).20
Cloning and Transformation of TAP-Tagged AHA1
Cloning of a 9.9 kb genomic region containing AHA1 with 3377 bp of upstream and 1100 bp of downstream DNA (gAHA1) was described previously.20 A unique SalI restriction-enzyme site was created at the 5′ end of the AHA1 open reading frame using the QuikChange II XL kit (Stratagene) and the following mutagenic primers:
Forward: GATTTCTTCTGGGTGAAGATGTCGACTCTCGAAGATATCAAGAACGAGA
Reverse: TCTCGTTCTTGATATCTTCGAGAGTCGACATCTTCACCCAGAAGAAATC
The gAHA1 fragment was subcloned into the Agrobacterium transformation vector pGreenII0179 at the SacI and XhoI sites.39 The N-terminal TAPi tag was amplified with a high-fidelity polymerase from the gateway cloning vector ntapi.289.gw32 using the following primers designed to append SalI restriction overhangs:
Forward: AAGTCGACCGTGGTGGACAACAAGTTCAA
Reverse: TTGTCGACGAAGAAGATCCTCCTCCTCCCAGTGCGCCGCTG
Adenosine overhangs were added with Taq polymerase, and the fragment was cloned into pCR2.1. Ntapi SalI restriction fragments were cloned into the unique AHA1 SalI site. Constructs were confirmed by sequencing and used to transform aha1–6/aha1–6;AHA2/aha2–4 plants via Agrobacterium-mediated floral dip, strain GV3101 carrying pSOUP.40 Transgenic seeds were selected on hygromycin media (half-strength M&S, 1% sucrose, 0.7% agar, and 25 μg/mL of hygromycin). Hygromycin-resistant (HygR) aha1/aha1;aha2/aha2 plants complemented by the presence of the N-terminal Ntapi:gAHA1 construct were isolated in the T2 population by PCR genotyping.
Plasma Membrane Preparation
Arabidopsis seedlings were grown for 9 days in half-strength M&S liquid media as described previously.41 Nine day old seedlings were extracted in 2× (v/w) homogenization buffer (230 mM sorbitol, 50 mM Tris-HCl, pH 7.5, 10 mM KCl, 1 mM potassium metabisulfite, 1 mM PMSF, 10 mM sodium fluoride, 2 mM sodium pyrophosphate, 1 mM ammonium molybdate, and complete EDTA-free protease inhibitor cocktail (Roche)). Microsomes were collected as previously described.20 The microsomal pellet was resuspended in resuspension buffer (330 mM sorbitol, 5 mM KCl, and 5 mM K2HPO4, pH 7.8) and homogenized in a Teflon homogenizer, and plasma membranes were enriched by phase partition as described previously.42 Protein concentrations were determined with Bradford assays.
Protein Purification
Plasma membrane protein (100–500 μg) was solubilized with 10:1 (w/w) n-dodecyl-β-d-maltoside in resuspension buffer for 1 h at 4 °C. Unsolubilized material was pelleted in a benchtop microcentrifuge at full speed for 30 min at 4 °C. Solubilized upper-phase protein (0.1 mg, 100–200 μL total volume) was incubated for 1 h at 4 °C with 15 μL of rabbit IgG agarose resin (Sigma, A9294) or 10 μL of IgG Sepharose 6 Fast Flow resin (GE Life Sciences, 17-0969-01) equilibrated in resuspension buffer with 0.1% (w/v) DDM. Resin was pelleted at 0.8 rcf for 1 min, and supernatant was discarded. Resin was washed two times at 4 °C for 10 min with a 100× volume of resuspension buffer containing 1% NP40 followed by a final wash in a 100× volume of resuspension buffer containing 1% NP40 and 0.01% SDS. Resin was pelleted for 1 min at 0.8 rcf between each wash, and supernatant was discarded. Protein was eluted overnight at 4 °C with 30 units AcTEV protease (Invitrogen).
SDS-PAGE
SDS-PAGE analysis was performed on Novex 10% Tris-glycine gels (Invitrogen) using 1× Novex Tris-Glycine SDS running buffer or on NuPAGE 4–12% Bis-Tris gels using 1× MOPS running buffer at 80 V for 3 to 4 h. Protein samples were diluted 1:1 in Laemmli sample buffer with 5% β-mercaptoethanol (BioRad). For western blot, gels were incubated at room temperature for 20 min in 100 mL of equilibration buffer (2× NuPAGE transfer buffer, 10% methanol, and 1:1000 NuPAGE antioxidant). Proteins were transferred to PVDF using the iBlot dry blotting system (Invitrogen) using program P3 for 12 min. Immunoblotting was performed according standard protocols using HRP-conjugated rabbit anti-goat IgG for colorometric visualization or rabbit anti-CBP (Calmodulin-binding protein, Genscript) followed by IRDye goat anti-rabbit (LI-COR) for membranes visualized with a LI-COR Odyssey infrared scanner.
Blue Native PAGE
Three micrograms of plasma membrane protein was combined with 1% digitonin in membrane-solubilization buffer (50 mM Tris-HCl, 150 mM NaCl, and 1 mM EDTA, pH 7.5) as previously described.43 NativePAGE sample buffer (Invitrogen) was added to the supernatant to a final concentration of 1×, and NativePAGE 5% G-250 sample additive was added to a final concentration of 0.06%. Samples were run on NativePAGE Novex 3–12% Bis-Tris gels at 150 V for 20 min with dark blue cathode buffer followed by 45 min with light blue cathode buffer. Gels were washed for 20 min in equilibration buffer (80 mL of ddH20, 10 mL of 20× transfer buffer, 10 mL of methanol, and 100 μL of NuPage antioxidant), transferred to Immobilon-FL membranes (Millipore) using an iBlot dry blotting system (Invitrogen). Membrane was allowed to dry 30 min. Immunoblotting was performed according to a standard protocol using rabbit anti-CBP and IRDye goat anti-rabbit (LI-COR). Membranes were visualized with a LI-COR Odyssey infrared-imaging system.
Metabolic Labeling and Mass Spectrometry
Seedlings for metabolically labeled mass spectrometry experiments were grown in 1× M&S micronutrient solution supplemented with 1.5 mM CaCl2, 0.75 mM MgSO4, 0.625 mM KH2PO4, 2.3 mM MES salts, and 29.2 mM sucrose. Natural abundance (14N) or heavy (15N) NH4NO3 ( 0.825 g) and 14N or 15N KNO3 (0.96 g) were added to 1 L of media, and the pH was adjusted to 5.7 with KOH. Fourteen milligrams of seeds was grown in 75 mL of sterile media supplemented with ampicillin. Seeds were stratified in the dark for 2 nights at 4 °C and then grown under constant light with gentle shaking for 9 days. Seedlings were spun-dry and weighed, and equal fresh weights of heavy and light seedlings were combined. Microsome and plasma membrane purification proceeded as above. Methanol/chloroform extraction was performed on plasma membrane and soluble fractions as previously described.44 Protein was suspended in 8 M urea/50 mM NH4HCO3 with gentle sonication and diluted to 1 M urea using 50 mM NH4HCO3. A BCA protein assay was performed to determine protein concentration. Aliquots of samples were reduced with 5 mM DTT at ∼50 °C for 45 min, alkylated with 15 mM iodoacetamide at room temperature for 45 min, and digested overnight at 37 °C with a 50:50 mixture of trypsin/lys-C (total protein/enzyme ratio 100:1). Samples were acidified with 0.3% TFA and desalted using Waters tC18 sep-pak columns according to the manufacturer’s instructions. After drying the samples, peptides were solubilized in LC–MS quality 0.1% formic acid. Approximately 5 μg of samples was loaded for analysis using an Agilent 1100 LC system inline with an LTQ Orbitrap XL. Peptides were separated over an in-house packed reversed-phase analytical column (Magic-C18, 200 Å, 3 μm, Michrom Bioresources, Inc.) with a postloading mobile phase elution gradient of 0–40% B over 3 h (A, 0.1% formic acid; B, 95% CH3CN and 0.1% formic acid) at 200 nL/min. Data-dependent analysis was performed in the LTQ Orbitrap XL using FTMS preview mode and full MS1 scan acquisition at 100 000 resolving power. The top five ions, excluding unassigned and +1 charge states, were repeatedly isolated with a 2.5 Da window for CID analysis in the linear ion trap using a normalized collision energy of 35.0 and activation Q and time of 0.25 and 30 ms, respectively. Dynamic exclusion was used for 40 s with a repeat count of 1 throughout the duration of analysis. The TAIR9 proteome database was searched using Mascot v2.2, allowing one missed tryptic cleavage, +2 and +3 charge states, a peptide tolerance of 20 ppm, and MS/MS ion tolerance of 0.6 Da. Carbamidomethylation was set as a fixed modification, and phosphorylated S/T/Y, deamidated N/Q, and oxidated M were set as variable modifications. A reverse database was appended and concurrently searched. Data was filtered to a 1% false discovery rate using in-house developed software (available at http://www.biotech.wisc.edu/sussmanlab/home), and ratio calculations were performed using Census.45 Every sample was manually median-normalized prior to analysis.
Quantitative Analysis of Metabolically Labeled Proteomes
Peptides with Mascot scores below 30 or peak correlations below 0.8 were discarded from analysis. Peptide ratios were averaged to achieve a single ratio for every protein identified. Proteins showing 14N/15N and reciprocal ratios of ≥1.2 or ≤0.8 identified in each experimental pair were subjected to manual verification. Peptides were blasted against the TAIR10 protein database (www.arabidopsis.org) to determine uniqueness, and nonunique peptides were discarded. Reciprocal protein ratios for proteins identified by three or more peptides in 14N/15N and 15N/14N tissue pairs were tested for statistical significance using a two-sample, two-tailed student’s t test with p ≤ 0.05. Protein-abundance changes were strengthened when the protein was identified in both biological replicates and the direction of change in both replicates agreed. Proteins showing opposite abundance changes in reciprocal pairs were discarded.
Mass Spectrometry of IgG-Purified Proteins
IgG-purified, TEV-eluted protein samples were TCA/acetone-precipitated, and pellets were suspended in 7.4 μL of 8 M urea, 50 mM NH4HCO3 (pH 8.5), and 1 mM Tris-HCl for 5 min and subsequently diluted to 30 μL with 1.25 μL of 25 mM DTT, 2.5 μL MeOH, 18.75 μL of 25 mM NH4HCO3 (pH 8.5) for reduction and alkylation. Samples were incubated at 50 °C for 15 min and cooled on ice to room temperature. IAA (1.5 μL of 55 mM) was added, and samples were incubated at room temperature in the dark for 15 min. The reaction was quenched with 4 μL of 25 mM DTT. Eight microliters of trypsin (10 ng/μL Trypsin Gold, Promega Corp., in 25 mM NH4HCO3) and 6.5 μL of 25 mM NH4HCO3 (pH 8.5) was added to a final volume of 50 μL. Digestion proceeded for 2 h at 42 °C followed by an additional 5 μL of trypsin for a final enzyme/substrate ratio of 1:40. Digestion proceeded overnight at 37 °C. Digestions were quenched with 2.5% TFA to 0.3% final concentration, and 8 μL (∼700 ng) was loaded for nanoLC–MS/MS analysis. Microsomal and two-phase plasma membrane-enriched fractions were delipidated with methanol/chloroform separation prior to digestion. One-hundred micrograms of microsomal protein and 10 μg of plasma membrane-enriched fractions were precipitated in methanol/chloroform/water (4:1:1), and the protein pellet at the organic and aqueous interphase was washed in neat methanol. The protein was dried then digested as above in a 100 μL total volume with a 1:40 ratio of trypsin/substrate. Peptides were analyzed by nanoLC–MS/MS using the Agilent 1100 nanoflow system (Agilent Technologies) connected to a linear hybrid ion trap-orbitrap mass spectrometer (LTQ-Orbitrap XL, Thermo Fisher Scientific) equipped with a nanoelectrospray ion source. HPLC was performed on in in-house fabricated 15 cm C18 column packed with MAGIC C18AQ 3 μm particles (Michrom BioResources, Inc.) in a laser-pulled tip (P-2000, Sutter Instrument) using 360 μm × 75 μm fused silica tubing. Sample loading and desalting were done at 15 μL/min using a trapping column in line with the autosampler (Zorbax 300SB-C18, 5 μM, 5 × 0.3 mm, Agilent Technologies). Peptide elution solvent A was 0.1% formic acid in water and solvent B was 0.1% formic acid and 95% acetonitrile in water. The flow rate was 200 nL/min. The column was loaded, desalted, and equilibrated for 20 min in 1% solvent B followed by an increase to 40% solvent B over 195 min, ramped to 60% solvent B over 20 min, increased to 100% solvent B in 5 min, and held at 100% solvent B for an additional 3 min. The column was re-equilibrated to 1% B for 30 min. MS/MS data acquisition was gathered in a data-dependent mode as follows: MS survey scans from m/z 300 to 2000 were collected in centroid mode at a resolving power of 100 000. MS/MS spectra were collected on the five most abundant signals in each survey scan. Dynamic exclusion for 40 s of precursors up to 0.55 m/z below and 1.05 m/z above previously selected precursors was employed to increase the dynamic range and to maximize peptide identifications. Singly charged ions and ions for which the charge state could not be assigned were rejected from consideration for MS/MS.
Quantitative Analysis
Label-free raw data files were converted to Mascot generic format (mgf) through initial open-source mzXML format conversion using the Trans-Proteomic Pipeline (TPP) software suite version 4.4. Mgf files were searched against the Arabidopsis thaliana TAIR9 database with decoy reverse entries and common contaminants were included for a total of 27 279 Arabidopsis protein-coding genes and 62 523 total entries using in-house Mascot search engine 2.2.07 (Matrix Science) with variable methionine oxidation and asparagine and glutamine deamidation. Peptide mass tolerance was set at 20 ppm with fragment mass at 0.8 Da. Protein annotations, identification significance, and spectral-based quantification were done using Scaffold software (version 3.6.3, Proteome Software Inc., Portland, OR.) Protein identifications were accepted if they were established with greater than 95% probability and contained at least two peptides using a 1% false discovery rate. Protein probabilities were assigned using Protein Prophet.46 Proteins containing nonunique peptides that could not be assigned on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Metabolically labeled raw data was acquired, converted, and searched as above. Mascot search results were transformed to pepXML format using TPP software with an in-house-generated false-discovery-rate filter. FDR for peptide identification was set at 10% for for low-quality MS/MS spectra to be allowed and available for manual inspection if desired. Relative quantification of 14N/15N peptide ratios was performed using TPP tools and Census software (Scripps Research Institute).
Results
Complementation with a TAP-Tagged AHA1 Transgene
Our lab has previously shown that wild-type AHA1 or AHA2 transgenes successfully rescue aha1/aha1;aha2/aha2 double-knockdown embryo lethality.20 We therefore wished to test whether an AHA1 transgene fused to a tandem affinity-purification (TAP) tag could also rescue embryonic lethality. A TAP-tag construct optimized for use in plants containing two tandem protein A domains separated from a calmodulin-binding protein domain (CBP) by a TEV protease site was ligated to the 5′ end of a genomic AHA1 open reading frame32 (Supporting Information Figure S1).
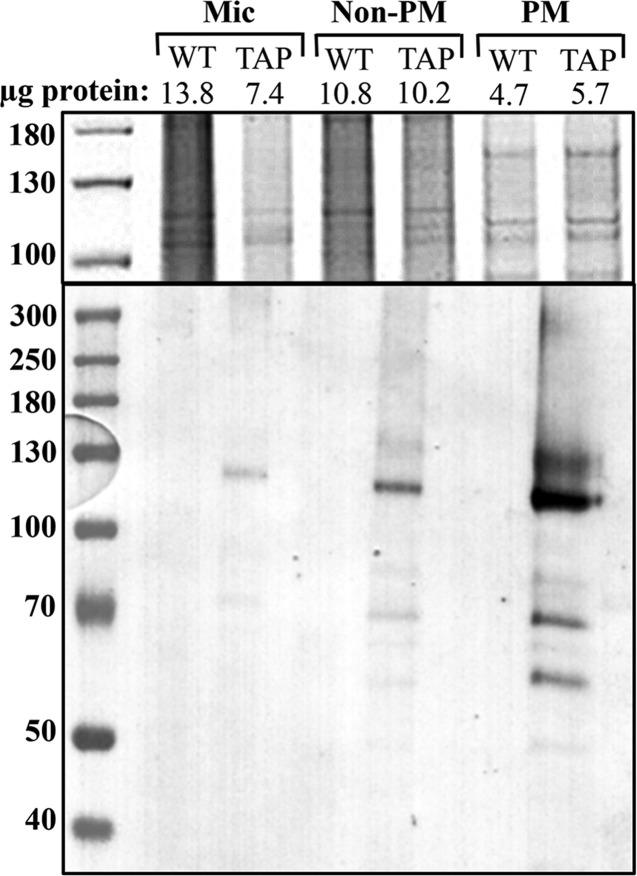
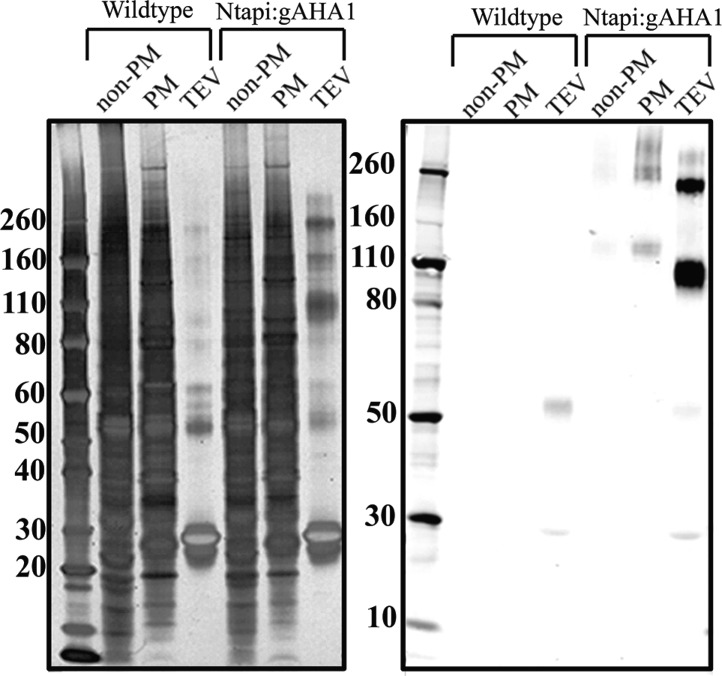
Arabidopsis (Col-0, aha1/aha1;AHA2/aha2) were transformed with the Ntapi:gAHA1 clone via Agrobacterium-mediated floral dip, and successful transformation events were selected on hygromycin media.40,47 Plants were allowed to self, and PCR genotyping of the T2 population identified aha1/aha1;aha2/aha2 double-knockdown plants carrying the Ntapi:gAHA1 transgene. We recovered two independent rescue lines expressing Ntapi:gAHA1 (Ntapi:gAHA1a and Ntapi:gAHA1b; TAPa and TAPb). A band of approximately 125 kDa was detected in western blots from denaturing SDS protein gels of Ntapi:gAHA1 microsomal fractions probed for protein A. Following two-phase partitioning, the majority of protein A staining enriches in the plasma membrane phase (Figure 1). In addition to the major band between the 110 and 130 kDa markers, we commonly observe the slightly higher and smaller ∼70–80 kDa bands on western blots. Smaller bands may be due to partial proteolysis of the C-terminal regulatory domain, whereas larger bands may be due to alterred migration resulting from bound lipid or detergent molecules.
Figure 1.
Detection of Ntapi:AHA1 in total microsomes (Mic), plasma membrane (PM), and intracellular membranes (non-PM). Size markers are in kDa. Upper panel is coomassie stained, and lower panel is a western blot probed with HRP-conjugated rabbit anti-goat IgG.
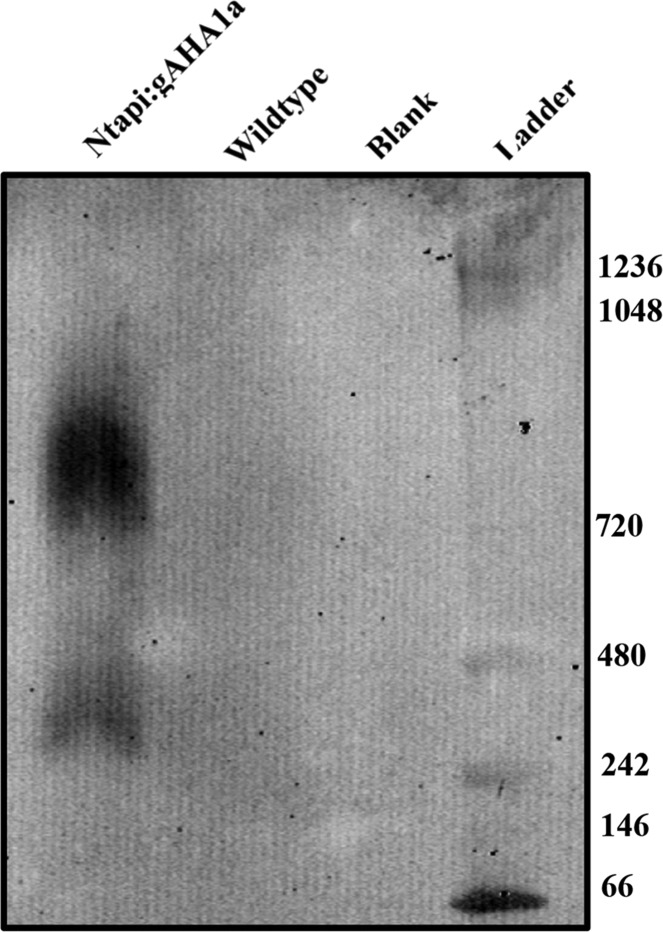
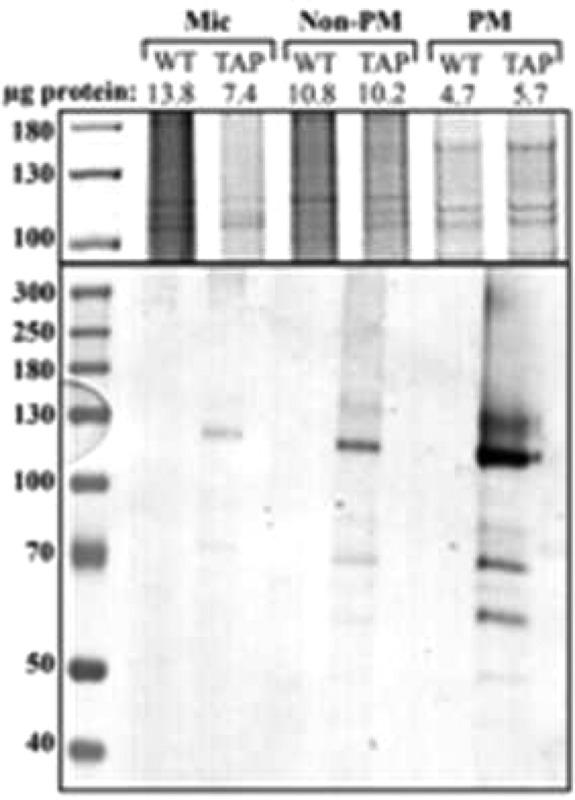
Previous studies of the Nicotiana plumbaginifolia plasma membrane proton pump, PMA2, indicate that the protein is present in the membrane as hexameric complexes, especially after activation by fusicoccin and subsequent 14-3-3 protein binding.48,49 To determine the molecular weight of nondenatured Ntapi:AHA1, purified plasma membranes were run on blue native gels and probed with rabbit anti-CBP antibody. We observed two bands migrating close to the 242 and 720 kDa markers. Proteins may migrate higher than expected in blue native gels based on detergents and dyes used or bound lipids; thus, these bands likely correspond to dimeric and hexameric complexes observed previously (Figure 2).48
Figure 2.
Three micrograms of Ntapi:gAHA1 or wild-type plasma membrane protein was run on blue native gels. Proteins were transferred to membranes and probed with rabbit anti-CBP antibodies.
The efficiency of aha1/aha2 double-knockdown rescue by Ntapi:gAHA1 was assessed by comparing the genotypes of aha1–6/aha1–6;AHA2/aha2–4;Ntapi:gAHA1a progeny to the ratios expected on the basis of Mendelian segregation for either a single or two unlinked insertional events (Table 1). The presence of multiple insertions and complex transgenic loci following Agrobacterium-mediated transformation is common.50,51 Progeny ratios differed significantly from both expected scenarios based on chi-squared analysis but more closely resembled a scenario involving two unlinked transgene insertional events. A reduction in the number of rescued progeny from the expected number was observed and could be due to altered expression, translation, trafficking, or biochemical activity of the fusion protein. A similar segregation analysis of Ntapi:gAHA1b progeny was not performed because the genotype of the transformed seedling was aha1–6;aha1–6/aha2–4;aha2–4.
Table 1. Progeny of aha1–6/aha1–6;AHA2/aha2–4 Parental Plant Carrying the Ntapi:gAHA1 Transgene.
| genotype | plants obsa | experiment Ab | experiment Bc |
|---|---|---|---|
| AHA2/aha2–4 | 8 | 18.33 | 4.58 |
| AHA2/AHA2 | 5 | 9.17 | 2.29 |
| aha2–4/aha2–4 | 0 | 0 | 0 |
| AHA2/aha2–4;Ntapi:gAHA1 | 55 | 41.25 | 51.56 |
| AHA2/AHA2;Ntapi:gAHA1 | 31 | 20.625 | 25.78 |
| aha2–4/aha2–4;Ntapi:gAHA1 | 11 | 20.625 | 25.78 |
| total | 110 | 110 | 110 |
| p = 0.0002d | p = 0.004 |
Number of plants observed by PCR genotyping.
Number of plants expected by Mendelian segregation with a single transgene insertion.
Number of plants expected by Mendelian segregation of two unliked transgene insertional events.
Result of chi-squared goodness of fit tests between observed and expected genotypes using 4 degrees of freedom.
Ntapi:gAHA1 Plants Exhibit Reduced Root Length
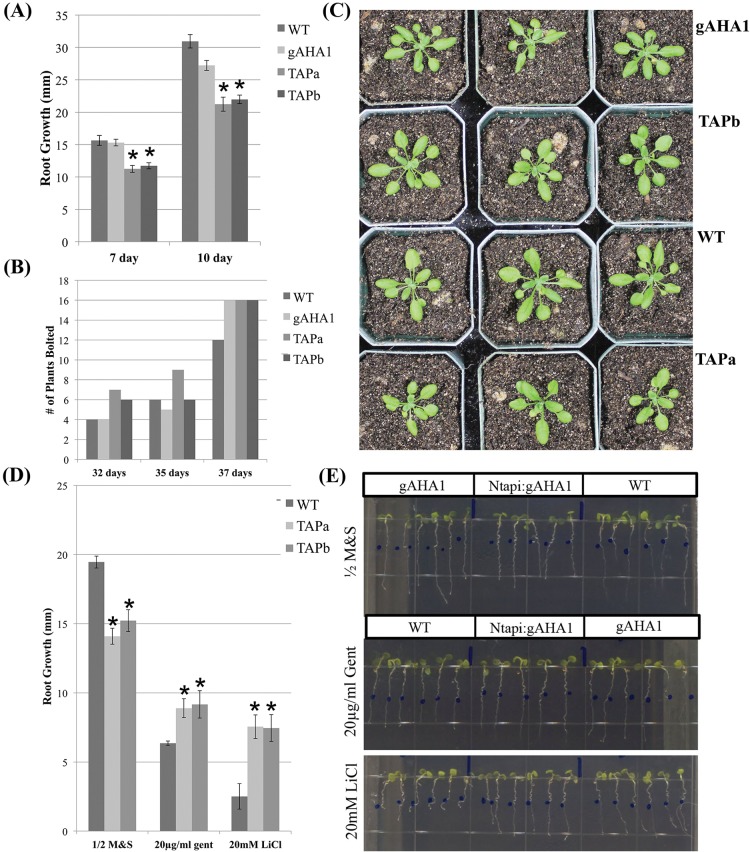
When grown vertically on standard half-strength M&S with 1% (w/v) sucrose nutrient media, we observed approximately 20% reduced root length in Ntapi:gAHA1 plants compared to wild-type or aha1/aha2 double knockdown mutants rescued with a wild-type AHA1 transgene (gAHA1) (Figure 3A). We looked for additional growth changes such as the time to first bolting (Figure 3B) and plant growth in soil (Figure 3C), but we did not observe readily apparent differences. Work in our lab has previously shown that aha2 single-knockdown mutants exhibit root-growth phenotypes when seedlings are grown under stress conditions (high external pH, high external potassium, and toxic cations) that alter or rely on the proton motive force.20,37 To determine effects of the TAP-tagged AHA1 protein on the proton motive force, we grew Ntapi:gAHA1 plants in the presence of gentamicin and lithium chloride. Under these conditions, aha2 knockdowns show enhanced root growth compared to wild-type plants because of the reduced proton motive force that energizes the reduced uptake of these toxic compounds. Ntapi:gAHA1 lines show similar root-growth phenotypes to aha2 single knockdowns rescued with a gAHA1 transgene (Figure 3D).
Figure 3.
Growth of Ntapi:gAHA1 plants. (A) Vertical root growth (mm) of plants on half-strength M&S with 1% (w/v) sucrose media was measured at 7 and 10 days. Bars indicate standard error, and two-tailed, two-sample Student t tests were used to determine statistical difference (p < 0.005, n ≥ 12). (B) Sixteen seedling of each genotype were transferred to soil and monitored visually for bloting. (C) Representative experiment shown. Vegetative growth of three representative plants for each genotype at 32 days. (D) Vertical root growth (mm) of 4 day old wild-type, Ntapi:gAHA1a, and Ntapi:gAHA1b seedlings transferred to stress media for 5 days. Bars indicate standard error, and Student t tests were performed as above (p < 0.05, n = 7). (E) Vertical root growth (mm) of 4 day old wild-type, Ntapi:gAHA1a, and gAHA1 seedlings transferred to stress media for 5 days.
Proteome Analysis of Ntapi:gAHA1 Plants
With the ultimate goal of using Ntapi:gAHA1 plants for the in planta physiological investigation of AHA1, we wished to establish how closely transgenic lines resembled wild-type plants beyond merely visible phenotypes. To this end, we compared the plasma membrane and soluble proteomes of Ntapi:gAHA1 plants to wild-type and gAHA1 rescue lines using a metabolically labeled mass spectrometry approach to determine whether expressing Ntapi:AHA1 had proteomic consequences. Equal wet weights of seedlings grown in natural-abundance (14N) and isotopically heavy nitrogen (15N) liquid media were mixed prior to protein extraction. Subsequent downstream sample processing and mass spectrometry analysis was identical for paired tissues, and the relative abundance of a peptide from the original tissue can be determined by calculating the ratio of 14N/15N peptide intensity.52 A ratio of 1.0 indicates no difference in peptide abundance between metabolically labeled tissue pairs; a ratio greater than 1.0 indicates increased peptide abundance from 14N-grown tissue compared to 15N-grown tissue; and a ratio less than 1.0 indicates decreased peptide abundance in 14N-grown tissue
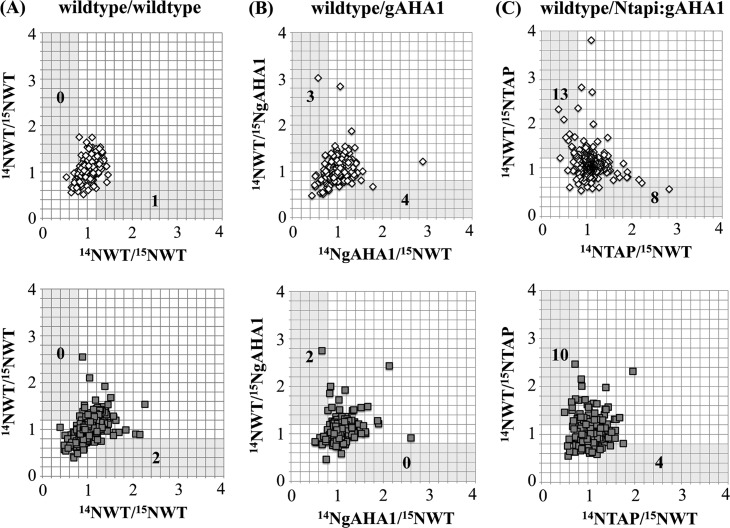
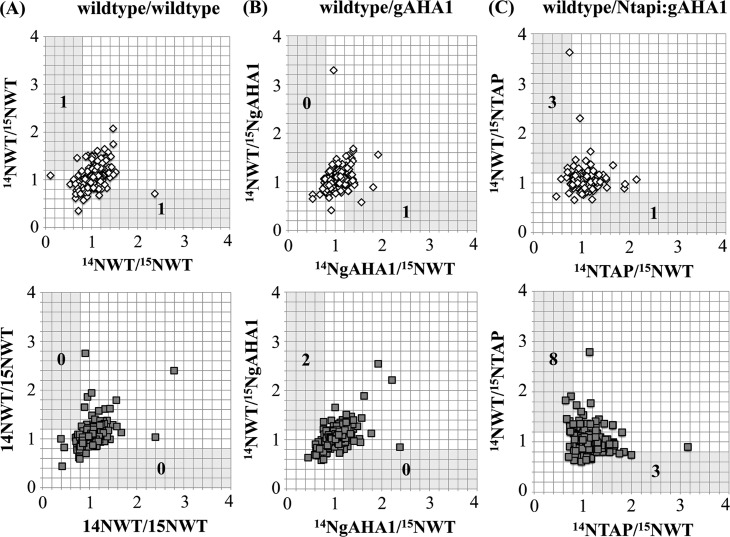
We generated 14N/15N and reciprocal 15N/14N tissue pairs of liquid-grown, 9 day old wild-type/wild-type, wild-type/gAHA1, and wild-type/Ntapi:gAHA1a plants and performed biological duplicates for each experimental pair. Proteomic analysis of both soluble and plasma membrane proteomes was performed. The number of unique peptides and proteins identified by tandem mass spectrometry from each metabolically labeled tissue pair is summarized in Supporting Information Tables S1 and S2. Approximately half of the protein identifications in all samples were based on a single peptide and were discarded from further analysis (Supporting Information Table S3). For proteins identified by more than one peptide, peptide ratios were averaged to achieve a single 14N/15N protein ratio. Protein ratios for plasma membrane (Figure 4) and soluble (Figure 5) fractions were plotted as the ratios from 14N/15N to reciprocal 15N/14N genotype pairs.
Figure 4.
Reciprocal plasma membrane protein ratios in metabolically labeled tissue pairs. Wild-type (WT), wild-type AHA1-rescued (gAHA1), and Ntapi:gAHA1-rescued plants were grown in natural-abudance (14N) and heavy isotope (15N) nitrogen media. Equal fresh weights of (A) wild-type/wild-type, (B) wild-type/gAHA1, and (C) wild-type/Ntapi:gAHA1 genotype pairs from two biologically replicated experiments (upper and lower plots) were mixed prior to homogenization, sample processing, and MS analysis. Protein ratios (14N/15N) were calculated by averaging median-normalized peptide ratios determined by Census, with a minimum MASCOT score of 30. Calculated protein ratios for reciprocally labeled tissue pairs were plotted for proteins identified by two or more peptides in both samples. The number of proteins showing reciprocal ratios of ≥1.2 and ≤0.8 in each sample are indicated.
Figure 5.
Reciprocal soluble protein ratios in metabolically labeled tissue pairs. Wild-type (WT), wild-type AHA1-rescued (gAHA1), and Ntapi:gAHA1-rescued plants were grown in natural-abudance (14N) and heavy isotope (15N) nitrogen media. Equal fresh weights of (A) wild-type/wild-type, (B) wild-type/gAHA1, and (C) wild-type/Ntapi:gAHA1 genotype pairs from two biologically replicated experiments (upper and lower plots) were mixed prior to homogenization, sample processing, and MS analysis. Protein ratios (14N/15N) were calculated by averaging median-normalized peptide ratios determined by Census, with a minimum MASCOT score of 30. Protein ratios for reciprocally labeled tissue pairs were plotted for proteins identified by two or more peptides in both samples. The number of proteins showing reciprcal ratios of ≥1.2 and ≤0.8 in each sample are indicated.
Following manual verification of proteins showing 14N/15N and reciprocal ratios of ≥1.2 or ≤0.8, no proteins met all of the required criteria (see Experimental Procedures) in 14Nwild-type/15Nwild-type and reciprocal plasma membrane or soluble protein fractions. In wild-type/gAHA1 tissue pairs, only AHA1 itself showed statistically significant and reciprocal abundance changes in both biological replicates (Table 2). Despite the reduced levels of AHA1 protein at the membrane, gAHA1-rescue lines show no observable phenotypes under standard laboratory growth conditions. In wild-type/Ntapi:gAHA1 tissue pairs, we identified nine plasma membrane proteins that were differentially abundant in all three reciprocal experiments with at least a 20% abundance change in one experiment (Table 3). The majority of proteins identified show decreased abundance in Ntapi:gAHA1 seedlings compared to wild-type, but two, the phosphate transporter ATPT2 and AHA1 itself, show increased abundance. The known cellular roles of identified proteins include membrane transport (ATPT2), cell wall structure (SVL2), vesicle trafficking (PATL1), and microtubule structure and organization (DREPP).53−58
Table 2. Manually Verified Protein Abundance Changes in gAHA1 vs Wild-Type Plasma Membrane Fractions.
| experiment
1 |
experiment
2 |
||||
|---|---|---|---|---|---|
| accession | annotation | 14NgAHA1/ 15NWT | 14NWT/ 15NgAHA1 | 14NgAHA1/ 15NWT | 14NWT/ 15NgAHA1 |
| AT2G18960 | AHA1 | 0.20 (5)a | 5.64 (6) | 0.15 (3) | 4.67 (8) |
| p = 0.0003b | p = 0.001 | ||||
| AT5G44610 | MAP18 | 1.33 (3) | 0.74 (2) | ||
The number of unique peptides whose ratios were averaged to calculate the total protein ratio are indicated in parentheses.
Two sample, two-tailed Student t tests were used to determine statistical significance of reciprocal protein ratios if three or more unique peptides were detected in each sample.
Table 3. Manually Verified Protein Abundance Changes in Ntapi:gAHA1 vs Wild-Type Plasma Membrane Fractions.
| experiment
1 |
experiment
2 |
experiment
3 |
|||||
|---|---|---|---|---|---|---|---|
| accession | annotation | 14NTAPa/15NWT | 15NWT/14NTAPa | 14NTAPa/15NWT | 15NWT/14NTAPa | 14NTAPb/15NWT | 15NWT/14NTAPb |
| AT2G18960 | AHA1 | 1.06 (6)a | 0.79 (9) | 1.93 (6) | 0.62 (7) | 1.34 (9) | 0.70 (5) |
| p = 0.01b | p = 1 × 10–5 | p = 1 × 10–4 | |||||
| AT2G38940 | ATPT2 | 1.69 (4) | 0.88 (3) | 0.76 (2) | 1.08 (4) | 0.69 | |
| p = 0.05 | |||||||
| AT3G01290 | band 7 | 1.53 (6) | 0.69 (8) | 1.17 (4) | 1.05 (6) | 1.74 (7) | 0.47 (4) |
| p = 6 × 10–9 | p = 2 × 10–6 | ||||||
| AT1G66970 | SVL2 | 0.84 (7) | 1.37 (10) | 0.66 (8) | 1.57 (8) | 0.80 (8) | 0.95 (8) |
| p = 2 × 10–5 | p = 4 × 10–7 | ||||||
| AT1G72150 | PATL1 | 0.94 (7) | 1.55 (7) | 0.72 (6) | 2.47 (8) | 0.78 (10) | 1.07 (11) |
| p = 1 × 10–5 | p = 6 × 10–6 | p = 0.03 | |||||
| AT3G16450 | jacalin family | 0.96 | 2.20 (4) | 0.64 | 0.75 (4) | 1.38 (4) | |
| p = 1 × 10–4 | |||||||
| AT3G16460 | jacalin family | 0.65 (5) | 1.36 (8) | 0.91 (6) | 1.35 (7) | 0.80 (11) | 1.45 (16) |
| p = 5 × 10–6 | p = 8 × 10–4 | p = 2 × 10–11 | |||||
| AT3G05900 | neurofilament protein-related | 0.64 (10) | 1.21 (11) | 0.91 (4) | 1.29 (4) | 0.70 (8) | 1.44 (10) |
| AT4G20260 | DREPP family | 0.95 (6) | 1.24 (8) | 0.80 (3) | 1.71 (6) | 0.93 (7) | 1.15 (5) |
| p = 0.009 | p = 0.008 | p = 0.006 | |||||
The number of unique peptides whose ratios were averaged to calculate the total protein ratio are indicated in parentheses.
Two sample, two-tailed Student t tests were used to determine statistical significance of reciprocal protein ratios if three or more unique peptides were detected in each sample. A third experiment was performed comparing Ntapi:gAHA1b and wildtype plants to distinguish between effects specific to Ntapi:AHA1 expression versus transformation artifacts.
TAP-Tag-Mediated Purification of AHA1
With the goal of using Ntapi:gAHA1 plants to facilitate efficient isoform-specific purification of AHA1 from Arabidopsis tissue, we investigated the ability to specifically purify Ntapi:AHA1 with IgG purification resin. We developed a modified tandem affinity-purification protocol to purify Ntapi:AHA1 using only the first affinity-purification step to limit the possibility of losing protein interactors with increased sample handling.60 Purification of Ntapi:AHA1 from plasma membrane fractions by IgG resin is demonstrated by western blot in Figure 6. Including 0.1% SDS in the IgG resin washing buffer decreases nonspecifically bound proteins while retaining high Ntapi:AHA1 antibody reactivity in eluted fractions. Using normalized spectral counts as a relative quantification of the abundance of Ntapi:AHA1 in membrane and purified samples, the protein enrichment from upper-phase fractions by IgG resin is approximately 3.5-fold under mild washing conditions (Table 4). Addition of 150 mM NaCl further reduces MS identification of contaminating proteins but also results in loss of Ntapi:AHA1 staining in western blots of TEV-eluted fractions (Supporting Information Figure S2). We therefore routinely use washing conditions containing 0.1% SDS with no additional salts for reducing background protein contamination while retaining specifically bound Ntapi:AHA1.
Figure 6.
Purification of Ntapi:AHA1 visulized by western blot. One-hundred micrograms of solubilized plasma membrane protein was incubated with IgG purification resin, washed three times with 0.1% SDS in resuspension buffer followed by three times with AcTEV, and eluted overnight with AcTEV protease. Eluted proteins were acetone-precipitated and resuspended directly in Laemmli sample buffer with reducing agent. One microgram of nonplasma membrane (non-PM) and plasma membrane (PM) wild-type or Ntapi:gAHA1 protein and half of TEV-eluted proteins were loaded on 4–12% NuPAGE gels. The left panel is silver stained for total protein, and the right panel is a western blot probed with rabbit anti-CBP followed by LI-COR IRDye goat anti-rabbit antibody. Size-marker values are in kDa.
Table 4. Successive Purification of Ntapi:AHA1 from Liquid-Grown Seedlings.
| step | fraction | spectral Ctsa | NSAFb | fold enrichmentc |
|---|---|---|---|---|
| 1 | microsomes | 75 | 0.0012 | |
| 2 | non-PM membranes | 63 | 0.0010 | |
| 3 | plasma membranes | 273 | 0.0051 | 4.25 |
| 4 | IgG bound, TEV-eluted (mild washd) | 99 | 0.0175 | 14.6 |
| 5 | IgG bound, TEV-eluted (stringent washe) | 100 | 0.5639 | 470 |
Spectral counts of a representative sample as determined by “number of assigned spectra” in Scaffold.
Normalized spectral abundance factor calculated according to the method of Zhang et al.59
Enrichment of Ntapi:AHA1 from microsomal samples as determined by increasing NSAF values.
Twice with 100× volume 1% NP40, once with 100× volume 1% NP40, 0.01% SDS.
Three times with 100× volume 0.1% SDS, 150 mM NaCl.
Identification of Ntapi:AHA1-Copurifying Proteins
After establishing specific IgG purification of Ntapi:AHA1, we pursued the identification of Ntapi:AHA1-copurifying proteins using an affinity-purification mass spectrometry approach. Ntapi:gAHA1 and wild-type plants were grown in 14N and 15N liquid media and combined prior to homogenization as described previously. More than three technical purification replicates were performed on 14NNtapi:gAHA1/15Nwild-type and reciprocal 14Nwild-type/15NNtapi:gAHA1 tissue pairs. Eluted proteins were subjected to tandem mass spectrometry analysis, and proteins showing reciprocal 14N/15N ratios deviating from 1.0 between wild-type and Ntapi:gAHA1 seedlings were subjected to manual analysis. Four proteins showed increased purification from Ntapi:gAHA1 plants compared to wild-type including two additional AHA isoforms (Table 5). The enrichment of AHA9 and AHA11 from Ntapi:gAHA1 versus wild-type protein extracts was validated by searching the raw data for peaks corresponding to unique peptides from each isoform (Figure 7). Because of the high degree of sequence identity between isoforms, many other AHA peptides were discarded from our analysis; however, a list of all identified AHA peptides and their 14N/15N ratios are included in the Supporting Information (Table S4).
Table 5. Ntapi:AHA1-Copurifying Proteins in Reciprocal Metabolically Labeled Tissue Pairs.
| accession | annotation | unique peptidesa | 14NTAP/15NWT ratiob | 14NWT/15NTAP ratioc |
|---|---|---|---|---|
| AT2G18960 | AHA1 | 7 | 14.10 | 0.03 |
| AT1G80660 | AHA9 | 1 | 24.34 | 0.04 |
| AT5G62670 | AHA11 | 2 | 1.73 | NDd |
| AT4G10480 | unknown/α-NAC | 2 | 1.35 | 0.71 |
| AT1G62480 | unknown/Ca2+-binding | 2 | 1.32 | 0.72 |
Peptides were BLASTed agains the TAIR10 database to determine uniqueness.
Peptide ratios for IgG-purified, TEV-eluted proteins from 14NNtapi:gAHA1/15NCol-0 WT metabolically labeled, liquid-grown tissue pairs.
Peptide ratios for IgG-purified, TEV-eluted proteins from reciprocally labeled tissue pairs.
Peptide intensity from 14N-grown tissue was too low for detection in raw data by Mascot set up in 14N search mode.
Figure 7.
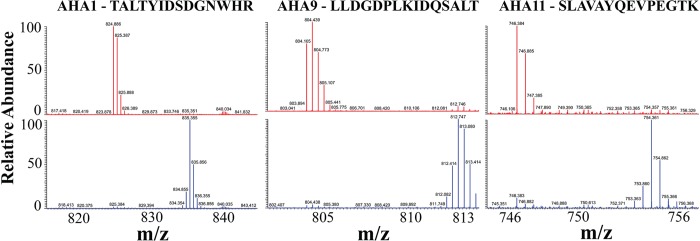
Peptides unique to other AHA isoforms enrich with Ntapi:AHA1 after TAP-tag-mediated purification. Metabolically labeled 14NNtapi:gAHA1/15Nwild-type (red) and reciprocal 14Nwild-type/15NNtapi:gAHA1 (blue) plasma membrane pairs were incubated with IgG resin, washed, and eluted overnight with TEV protease. The raw data was inspected manually for peaks corresponding to the indicated unique peptides from AHA1, AHA9, and AHA11.
Response of Ntapi:AHA1-Copurifying Proteins to in Vivo Treatment
Because of the autoinhibited state of AHA proteins,61 we wished to investigate whether treatment in vivo with compounds known or hypothesized to cause AHA activation would effect identification of NTapi:AHA1-copurifying proteins. The fungal phytotoxin fusicoccin (FC) binds to and stabilizes the AHA phosphorylated C-terminus and 14-3-3 protein complex, causing permanent activation of the pump.62 We hypothesized that treating liquid-grown seedlings with FC would increase mass spectrometry identification of 14-3-3 proteins in IgG-purified eluates, which we failed to identify in nonelicited copurification experiments (Supporting Information Table S5). Nine day old Ntapi:gAHA1 liquid-grown 14N + FC/15N – FC and reciprocally treated pairs were subjected to combined homogenization, purification, and MS analysis as previously described. Treatment of seedlings with 1 μM FC for 30 min increased purification of AHA11 and several 14-3-3 protein isoforms including GRF1, GRF2, and GRF3 (Table 6). Auxin also regulates plasma membrane proton pump activity by phosphorylation, although the protein kinases and/or phosphotases responsible have not been identified.63 Additionally, the inhibitory effect of auxin on root growth is well-established and likely involves regulation of proton pump activity.64 When seedlings were treated with 1 μM indole-3-acetic-acid for 15 min, we observed decreased copurification of the protein kinase PHOT1 (Table 6).
Table 6. Response of Ntapi:AHA1-Copurifying Proteins to in Vivo Treatments.
| accession | annotation | treatment | 14NTAP+/15NTAP–a | 14NTAP–/15NTAP+b |
|---|---|---|---|---|
| AT2G18960 | AHA1 | FCc | 0.93 (8) | 0.98 (6) |
| p = 0.74 | ||||
| IAAd | 1.00 (5) | 1.14 (7) | ||
| p = 0.52 | ||||
| AT5G62670 | AHA11 | FC | 1.10 (3)e | 0.67 (3) |
| p = 0.03f | ||||
| AT4G09000 | GRF1 | FC | 1.41(4) | 0.95 (4) |
| p = 0.006 | ||||
| AT1G78300 | GRF2 | FC | 1.13 (3) | 0.80 (3) |
| p = 0.003 | ||||
| AT5G38480 | GRF3 | FC | 1.25 (4) | 0.76 (5) |
| p = 0.003 | ||||
| AT3G45780 | PHOT1 | IAA | 0.87 (2) | 1.17 (5) |
| AT2G26080 | ATGLDP2 | IAA | 1.23 (3) | 0.94 (3) |
| p = 0.02 | ||||
| AT3G23990 | HSP60 | IAA | 1.21 (4) | 0.81 (2) |
Liquid-grown Ntapi:gAHA1 seedlings: 14N seedlings treated, 15N seedlings mock treated.
Reciprocal experiment: 14N seedlings mock treated, 15N seedlings treated.
Fusicoccin treatment: 1 μM for 30 min.
Indole-3-acetic acid treatment: 1 μM for 15 min.
Number of unique peptides included in protein ratio average are indicated in parentheses.
Statistical significance is based on two-sample, two-tailed Student’s t test.
Discussion
Herein, we describe the functional rescue of aha1/aha2 knockdown embryonic lethality in Arabidopsis by stable transformation with an N-terminal TAP-tagged genomic AHA1 transgene, and we used an affinity-purification mass spectrometry approach to copurify candidate Ntapi:AHA1-interacting proteins. The ability to identify proteins interacting with AHA1 in planta is a particular strength of our approach that is not feasible with heterologous methods used to identify protein interactions, including the yeast two-hybrid and split ubiquitin systems.
While we observed a slight reduction in the number of endogenous double knockdown, transgene-rescued progeny from the expected numbers based on Mendelian segregation, this could be a result of non-WT transgene expression or altered translation, trafficking, or activity of the tagged protein, resulting in some amount of embryonic lethality. Western blots of two-phase separated membranes showed that Ntapi:AHA1 was enriched with plasma membrane fractions compared to nonplasma membrane fractions, as expected. This result alleviated concerns that the TAP tag could be deleteriously affecting proper maturation and trafficking of Ntapi:AHA1 protein, although we cannot rule out the possibility that a portion of the protein may be mistrafficked or misfolded.
Ntapi:gAHA1-rescue lines exhibited minimal observable phenotypic consequences beyond the ∼20% reduced vertical root growth on standard media. However, mass spectrometry revealed a subtle plasma membrane proteomic phenotype of Ntapi:gAHA1 plants. Several proteins showing abundance differences in Ntapi:gAHA1 versus wild-type plants have reported roles in cell wall structure, vesicle trafficking, and cell elongation, and these proteins could be contributing factors to the observed Ntapi:gAHA1 short-root phenotype. SVL2 is related to the plasma membrane anchored, glycerophosphoryl diester phosphodiesterase protein SHV3. SHV3 plays a role in cell wall rigidity, and knockout mutants exhibit ruptured root hairs and swollen guard cells.57 PATL1 is expressed in root tip cells and plays an important role in membrane trafficking and cell plate maturation, both important processes for cell elongation.54,65,66 AtPrx34 is a known cell wall associated peroxidase expressed in root cells that plays a role in cell elongation. Knockout lines of a closely related gene, AtPrx33, have short roots, and double knockouts exhibit an additive effect.56 The DREPP family protein identified in our study, PcaP1, has microtubule destabilizing activity that results in the negative regulation of hypotcotyl elongation.58 A closely related gene, PcaP2, negatively regulates root hair tip growth.67 These two proteins bind to Ca2+/calmodulin complexes; therefore, our Ntapi:AHA1 construct may be interefereing with normal regulation and activity of this protein with respect to calmodulin binding. Together, the altered abundance of these proteins in Ntapi:gAHA1 seedlings may be contributing to reduced vertical root growth. It is also possible that other tip growing systems may be affected. For example, if pollen tube growth of endogenous knockout pollen carrying the Ntapi:gAHA1 transgene is also negatively affected, it may explain the reduced numbers of Ntapi:gAHA1 rescue progeny we observed.
Because of the essential nature of AHA1 and AHA2 and the number of important plant processes in which they are hypothesized to function, we hypothesize that many interacting proteins, including protein kinases important in the regulation of AHA by phosphorylation at multiple sites in the C-terminal regulatory region,68 are yet to be identified. Initial experimentation to identify Ntapi:AHA1-interacting proteins identified two additional copurifying AHA isoforms, AHA9 and AHA11. We and others have shown that AHA proteins form multimeric complexes in plasma membranes, but whether those complexes are homomultimeric or heteromultimeric with respect to isoform composition has not been shown. Our copurification results suggest that these complexes are mixed in their isoform composition and further opens the possibility that the isoform composition of native AHA complexes could play important regulatory or signaling roles.70 These results are in contrast to a recent publication by Bobik et al., which showed that Nicotiana plumbaginifolia H+-ATPase isoforms PMA2 and PMA4 did not oligomerize with each other.69 The different results may be due to differences in experimental design, including the use of natural versus overexpression promoters or differences in the biological activity of H+-ATPase isoforms from Arabidopsis versus Nicotiana. We also observed increased copurification of AHA11 after treatment with the fungal toxin FC, suggesting that the isoform composition of quaternary AHA complexes may be constantly in flux and responsive to the environment. One obvious limitation is that our rescue plants do not express AHA2, which, because of its high expression levels and shared sequence identity with AHA1, we would predict to be the most common isoform to complex with AHA1. In the absence of AHA2, other isoforms may be artifactually incorporating into multimeric AHA complexes.
The autoinhibited state of AHA1 may reduce our ability to identify interacting proteins. We therefore tested if we could expand the identification of copurifying proteins by combining in vivo treatments that are hypothesized or known to regulate AHA activity with metabolic labeling, affinity purification, and mass spectrometry. The dynamic copurification of PHOT1 and 14-3-3 in response to exogenous auxin or fusicoccin suggest that further experiments will reveal similar, and potentially novel, AHA1-interacting proteins in response to specific conditions. A current limitation is the inability to discern whether proteins with differential abundance in purified samples after in vivo elicitation reflect true Ntapi:AHA1-interacting proteins or indicate changes in the abundance of these proteins in plasma membrane fractions prior to purification with IgG resin. We addressed this problem by also analyzing an aliquot of unpurified plasma membrane samples with tandem mass spectormetry; however, the increased sample complexity of nonpurified samples can result in decreased peptide identification and inconclusive results, especially for less abundant proteins. Introducing cross-linking reagents to maintain protein interactions while increasing the denaturing quality of washing steps to remove nonspecifically bound proteins from the purification resin will facilitate the identification of interacting proteins in the future.
Glossary
Abbreviations Used
- AHA
Arabidopsis H+-ATPase
- PMF
proton motive force
- TAP
tandem affinity purification
- M&S
Murashige & Skoog
- AP–MS
affinity-purification mass spectrometry
- CBP
calmodulin-binding protein
Supporting Information Available
Full predicted protein sequence of the Ntapi:AHA1 expressed protein; further information about the metabolically labeled proteome experiments including total protein and peptide identifications; list of all AHA peptides identified in metabolically labeled, nonelicited Ntapi:gAHA1 copurification experiments; list of 14-3-3 general regulatory factor proteins identified in metabolically labeled, nonelicited Ntapi:gAHA1 copurification experiments and their 14N/15N ratios. This material is available free of charge via the Internet at http://pubs.acs.org.
U.S. Department of Energy Office of Basic Energy Sciences grant DEFG02-88ER13938 to Michael R. Sussman. Molecular Biosciences Training Grant, NIH National Research Service Award T32 GM07215 to Rachel Rodrigues (Nee Nelson).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Kühlbrandt W. (2004) Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5, 282–295. [DOI] [PubMed] [Google Scholar]
- Buckhout T. J. (1989) Surcose transport in isolated plasma-membrane vesicles from sugar beet (Beta vulgaris L.) Evidence for an electrogenic sucrose-proton symport. Planta 178, 393–399. [DOI] [PubMed] [Google Scholar]
- Bush D. R. (1993) Proton-coupled sugar and amino acid transporters in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 513–542. [Google Scholar]
- Palmgren M. G. (2001) Plant plasma membrane H+-ATPases: Powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845. [DOI] [PubMed] [Google Scholar]
- Sussman M. H. R. (2012) The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis. Plant Physiol. 158, 1158–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M. R. (1994) Molecular analysis of proteins in the plant plasma membrane. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 211–234. [Google Scholar]
- DeWitt N. D.; Harper J. F.; Sussman M. R. (1991) Evidence for a plasma membrane proton pump in phloem cells of higher plants. Plant J. Cell Mol. Biol. 1, 121–128. [DOI] [PubMed] [Google Scholar]
- DeWitt N. D.; Sussman M. R. (1995) Immunocytological localization of an epitope-tagged plasma membrane proton pump (H+-ATPase) in phloem companion cells. Plant Cell 7, 2053–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M.; Davison T. S.; Henz S. R.; Pape U. J.; Demar M.; Vingron M.; Schölkopf B.; Weigel D.; Lohmann J. U. (2005) A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Winter D.; Vinegar B.; Nahal H.; Ammar R.; Wilson G. V.; Provart N. J. (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718-1–e718-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certal A. C.; Almeida R. B.; Carvalho L. M.; Wong E.; Moreno N.; Michard E.; Carneiro J.; Rodriguéz-Léon J.; Wu H.-M.; Cheung A. Y.; Feijó J. A. (2008) Exclusion of a proton ATPase from the apical membrane is associated with cell polarity and tip growth in Nicotiana tabacum pollen tubes. Plant Cell 20, 614–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijó J. A.; Sainhas J.; Hackett G. R.; Kunkel J. G.; Hepler P. K. (1999) Growing pollen tubes possess a constitutive alkaline band in the clear zone and a growth-dependent acidic tip. J. Cell Biol. 144, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S.; Leonhardt N.; Fenzi F.; Valon C.; Costa M.; Piette L.; Vavasseur A.; Genty B.; Boivin K.; Muller A.; Giraudat J.; Leung J. (2007) Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 26, 3216–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Elmore J. M.; Fuglsang A. T.; Palmgren M. G.; Staskawicz B. J.; Coaker G. (2009) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 7, e1000139-1–e1000139-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y.; Nakamura S.; Takemiya A.; Takahashi Y.; Shimazaki K.-I.; Kinoshita T. (2010) Biochemical characterization of in vitro phosphorylation and dephosphorylation of the plasma membrane H+-ATPase. Plant Cell Physiol. 51, 1186–1196. [DOI] [PubMed] [Google Scholar]
- Kinoshita T.; Hayashi Y. (2011) New insights into the regulation of stomatal opening by blue light and plasma membrane H(+)-ATPase. Int. Rev. Cell Mol. Biol. 289, 89–115. [DOI] [PubMed] [Google Scholar]
- Robertson W. R.; Clark K.; Young J. C.; Sussman M. R. (2004) An Arabidopsis thaliana plasma membrane proton pump is essential for pollen development. Genetics 168, 1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaudant F.; Duby G.; von Stedingk E.; Zhao R.; Morsomme P.; Boutry M. (2007) Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiol. 144, 1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I. R.; Young J. C.; Armstrong G.; Foster N.; Bogenschutz N.; Cordova T.; Peer W. A.; Hazen S. P.; Murphy A. S.; Harper J. F. (2005) A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 102, 2649–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M.; Burch H. L.; Nelson R. B.; Barrett-Wilt G.; Kline K. G.; Mohsin S. B.; Young J. C.; Otegui M. S.; Sussman M. R. (2010) Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 285, 17918–17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S.; Ehrhardt D. W.; Loqué D.; Chen J.; Rhee S. Y.; Frommer W. B. (2008) Molecular and cellular approaches for the detection of protein-protein interactions: Latest techniques and current limitations. Plant J. 53, 610–635. [DOI] [PubMed] [Google Scholar]
- Miernyk J. A.; Thelen J. J. (2008) Biochemical approaches for discovering protein-protein interactions. Plant J. Cell Mol. Biol. 53, 597–609. [DOI] [PubMed] [Google Scholar]
- Fukao Y. (2012) Protein-protein interactions in plants. Plant Cell Physiol. 53, 617–625. [DOI] [PubMed] [Google Scholar]
- Gingras A.-C.; Gstaiger M.; Raught B.; Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654. [DOI] [PubMed] [Google Scholar]
- (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Lalonde S.; Obrdlik P.; Noorani Vatani A.; Parsa S. A.; Vilarino C.; Revuelta J. L.; Frommer W. B.; Rhee S. Y. (2012) Uncovering Arabidopsis membrane protein interactome enriched in transporters using mating-based split ubiquitin assays and classification models. Front. Plant Sci. 3, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T.; Fuglsang A. T.; Olsson A.; Brüntrup I. M.; Collinge D. B.; Volkmann D.; Sommarin M.; Palmgren M. G.; Larsson C. (1997) The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 9, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandini P.; Valera M.; Albumi C.; Bonza M. C.; Giacometti S.; Ravera G.; Murgia I.; Soave C.; De Michelis M. I. (2002) A novel interaction partner for the C-terminus of Arabidopsis thaliana plasma membrane H+-ATPase (AHA1 isoform): Site and mechanism of action on H+-ATPase activity differ from those of 14-3-3 proteins. Plant J. Cell Mol. Biol. 31, 487–497. [DOI] [PubMed] [Google Scholar]
- Fuglsang A. T.; Guo Y.; Cuin T. A.; Qiu Q.; Song C.; Kristiansen K. A.; Bych K.; Schulz A.; Shabala S.; Schumaker K. S.; Palmgren M. G.; Zhu J.-K. (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19, 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang I.-F.; Curran A.; Woolsey R.; Quilici D.; Cushman J. C.; Mittler R.; Harmon A.; Harper J. F. (2009) Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 9, 2967–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witthöft J.; Caesar K.; Elgass K.; Huppenberger P.; Kilian J.; Schleifenbaum F.; Oecking C.; Harter K. (2011) The activation of the Arabidopsis P-ATPase 1 by the brassinosteroid receptor BRI1 is independent of threonine 948 phosphorylation. Plant Signaling Behav. 6, 1063–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohila J. S.; Chen M.; Cerny R.; Fromm M. E. (2004) Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. Cell Mol. Biol. 38, 172–181. [DOI] [PubMed] [Google Scholar]
- Rubio V.; Shen Y.; Saijo Y.; Liu Y.; Gusmaroli G.; Dinesh-Kumar S. P.; Deng X. W. (2005) An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. Cell Mol. Biol. 41, 767–778. [DOI] [PubMed] [Google Scholar]
- Chang I.-F. (2006) Mass spectrometry-based proteomic analysis of the epitope-tag affinity purified protein complexes in eukaryotes. Proteomics 6, 6158–6166. [DOI] [PubMed] [Google Scholar]
- Leene J. V.; Stals H.; Eeckhout D.; Persiau G.; Slijke E. V. D.; Isterdael G. V.; Clercq A. D.; Bonnet E.; Laukens K.; Remmerie N.; Henderickx K.; Vijlder T. D.; Abdelkrim A.; Pharazyn A.; Onckelen H. V.; Inzé D.; Witters E.; Jaeger G. D. (2007) A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6, 1226–1238. [DOI] [PubMed] [Google Scholar]
- Qi Y.; Katagiri F. (2009) Purification of low-abundance Arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J. Cell Mol. Biol. 57, 932–944. [DOI] [PubMed] [Google Scholar]
- Haruta M.; Sussman M. R. (2012) The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis thaliana. Plant Physiol. 158, 1158–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan P. J.; Young J. C.; Tax F.; Sussman M. R. (1996) Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci. U.S.A. 93, 8145–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R. P.; Edwards E. A.; Leyland N. R.; Bean S.; Mullineaux P. M. (2000) pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Clough S. J.; Bent A. F. (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Nelson C. J.; Huttlin E. L.; Hegeman A. D.; Harms A. C.; Sussman M. R. (2007) Implications of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics 7, 1279–1292. [DOI] [PubMed] [Google Scholar]
- Larsson C., and Widell S.. Isolation of plant plasma membranes and production of inside-out vesicles, in Aqueous Two-Phase Systems: Methods and Protocols (Hatti-Kaul R., Ed.) pp 159–166, Humana Press, Totowa, NJ. [Google Scholar]
- Lee M. C. S.; Hamamoto S.; Schekman R. (2002) Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J. Biol. Chem. 277, 22395–22401. [DOI] [PubMed] [Google Scholar]
- Wessel D.; Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143. [DOI] [PubMed] [Google Scholar]
- Park S. K.; Venable J. D.; Xu T.; Yates J. R. 3rd (2008) A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5, 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A. I.; Keller A.; Kolker E.; Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Henriques R.; Lin S.-S.; Niu Q.-W.; Chua N.-H. (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Kanczewska J.; Marco S.; Vandermeeren C.; Maudoux O.; Rigaud J.-L.; Boutry M. (2005) Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer. Proc. Natl. Acad. Sci. U.S.A. 102, 11675–11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C.; Marco S.; Jaspert N.; Marcon C.; Schauer N.; Weyand M.; Vandermeeren C.; Duby G.; Boutry M.; Wittinghofer A.; Rigaud J.-L.; Oecking C. (2007) Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol. Cell 25, 427–440. [DOI] [PubMed] [Google Scholar]
- De Buck S.; Podevin N.; Nolf J.; Jacobs A.; Depicker A. (2009) The T-DNA integration pattern in Arabidopsis transformants is highly determined by the transformed target cell. Plant J. 60, 134–145. [DOI] [PubMed] [Google Scholar]
- Oltmanns H.; Frame B.; Lee L.-Y.; Johnson S.; Li B.; Wang K.; Gelvin S. B. (2010) Generation of backbone-free, low transgene copy plants by launching T-DNA from the Agrobacterium chromosome. Plant Physiol. 152, 1158–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.-E.; Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262. [DOI] [PubMed] [Google Scholar]
- Weig A.; Deswarte C.; Chrispeels M. J. (1997) The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol. 114, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman T. K.; Ohol Y. M.; McReynolds L. J.; Luna E. J. (2004) Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol 136, 3080–3094.discussion 3001–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.; Shin H.-S.; Dewbre G. R.; Harrison M. J. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. Cell Mol. Biol. 39, 629–642. [DOI] [PubMed] [Google Scholar]
- Passardi F.; Tognolli M.; De Meyer M.; Penel C.; Dunand C. (2006) Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta 223, 965–974. [DOI] [PubMed] [Google Scholar]
- Hayashi S.; Ishii T.; Matsunaga T.; Tominaga R.; Kuromori T.; Wada T.; Shinozaki K.; Hirayama T. (2008) The glycerophosphoryl diester phosphodiesterase-like proteins SHV3 and its homologs play important roles in cell wall organization. Plant Cell Physiol. 49, 1522–1535. [DOI] [PubMed] [Google Scholar]
- Li J.; Wang X.; Qin T.; Zhang Y.; Liu X.; Sun J.; Zhou Y.; Zhu L.; Zhang Z.; Yuan M.; Mao T. (2011) MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. Plant Cell 23, 4411–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Wen Z.; Washburn M. P.; Florens L. (2010) Refinements to label free proteome quantitation: How to deal with peptides shared by multiple proteins. Anal. Chem. 82, 2272–2281. [DOI] [PubMed] [Google Scholar]
- Puig O.; Caspary F.; Rigaut G.; Rutz B.; Bouveret E.; Bragado-Nilsson E.; Wilm M.; Séraphin B. (2001) The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24, 218–229. [DOI] [PubMed] [Google Scholar]
- Ekberg K.; Palmgren M. G.; Veierskov B.; Buch-Pedersen M. J. (2010) A novel mechanism of P-type ATPase autoinhibition involving both termini of the protein. J. Biol. Chem. 285, 7344–7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtele M.; Jelich-Ottmann C.; Wittinghofer A.; Oecking C. (2003) Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 22, 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K.; Hayashi K.; Kinoshita T. (2012) Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis1. Plant Physiol. 159, 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick C. C.; Goormaghtigh E.; Scarborough G. A. (1987) A hexameric form of the Neurospora crassa plasma membrane H+-ATPase. Arch. Biochem. Biophys. 252, 348–356. [DOI] [PubMed] [Google Scholar]
- Carter C. J.; Bednarek S. Y.; Raikhel N. V. (2004) Membrane trafficking in plants: New discoveries and approaches. Curr. Opin. Plant Biol. 7, 701–707. [DOI] [PubMed] [Google Scholar]
- Peterman T. K.; Sequeira A. S.; Samia J. A.; Lunde E. E. (2006) Molecular cloning and characterization of patellin1, a novel sec14-related protein, from zucchini (Cucurbita pepo). J. Plant Physiol. 163, 1150–1158. [DOI] [PubMed] [Google Scholar]
- Kato M.; Aoyama T.; Maeshima M. (2013) The Ca2+-binding protein PCaP2 located on the plasma membrane is involved in root hair development as a possible signal transducer. Plant J. Cell Mol. Biol. 74, 690–700. [DOI] [PubMed] [Google Scholar]
- Rudashevskaya E. L.; Ye J.; Jensen O. N.; Fuglsang A. T.; Palmgren M. G. (2012) Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments. J. Biol. Chem. 287, 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren M. C.; Soummarin M.; Serrano R.; Larsson C. (1991) Identification of an autoinhibitory domain in the C terminal region of the plant plasma membrane H+-ATPase. J. Biol. Chem. 266, 20740–20745. [PubMed] [Google Scholar]
- Bobik K.; Duby G.; Nizet Y.; Vandermeeren C.; Stiernet P.; Kanczewska J.; Boutry M. (2010) Two widely expressed plasma membrane H+-ATPase isoforms of Nicotiana tabacum are differentially regulated by phosphorylation of their penultimate threonine. Plant J. 62, 291–301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.