Abstract
Hemophores from Pseudomonas aeruginosa (HasAp), Serratia marcescens (HasAsm), and Yersinia pestis (HasAyp) bind hemin between two loops. One of the loops harbors conserved axial ligand Tyr75 (Y75 loop) in all three structures, whereas the second loop (H32 loop) contains axial ligand His32 in HasAp and HasAsm, but a noncoordinating Gln32 in HasAyp. Binding of hemin to the Y75 loop of HasAp or HasAsm causes a large rearrangement of the H32 loop that allows His32 coordination. The Q32 loop in apo-HasAyp is already in the closed conformation, such that binding of hemin to the conserved Y75 loop occurs with minimal structural rearrangement and without coordinative interaction with the Q32 loop. In this study, structural and spectroscopic investigations of the hemophore HasAp were conducted to probe (i) the role of the conserved Tyr75 loop in hemin binding and (ii) the proposed requirement of the His83–Tyr75 hydrogen bond to allow the coordination of hemin by Tyr75. High-resolution crystal structures of H83A holo-HasAp obtained at pH 6.5 (0.89 Å) and pH 5.4 (1.25 Å) show that Tyr75 remains coordinated to the heme iron, and that a water molecule can substitute for Nδ of His83 to interact with the Oη atom of Tyr75, likely stabilizing the Tyr75–Fe interaction. Nuclear magnetic resonance spectroscopy revealed that in apo-Y75A and apo-H83A HasAp, the Y75 loop is disordered, and that disorder propagates to nearby elements of secondary structure, suggesting that His83 Nδ–Tyr75 Oη interaction is important to the organization of the Y75 loop in apo-HasA. Kinetic analysis of hemin loading conducted via stopped-flow UV–vis and rapid-freeze-quench resonance Raman shows that both mutants load hemin with biphasic kinetic parameters that are not significantly dissimilar from those previously observed for wild-type HasAp. When the structural and kinetic data are taken together, a tentative model emerges, which suggests that HasA hemophores utilize hydrophobic, π–π stacking, and van der Waals interactions to load hemin efficiently, while axial ligation likely functions to slow hemin release, thus allowing the hemophore to meet the challenge of capturing hemin under inhospitable conditions and delivering it selectively to its cognate receptor.
Iron-containing proteins are integral components of fundamental physiological processes such as DNA synthesis, gene regulation, respiration, oxygen transport, and degradation of xenobiotics.1,2 The relative availability of iron in the host environment is an important determinant of virulence, which is why iron bioavailability is associated with sepsis,3 and an innate response of the immune system is to deploy iron-binding proteins such as lactoferrin and transferrin to decrease free iron concentrations to vanishingly low levels (≈10–18 M) in plasma.4 In spite of the iron-limiting conditions imposed by the host, pathogenic bacteria can establish infections, which is a manifestation of the molecular mechanisms that have evolved in pathogens to procure iron from their host. Approximately 95% of all iron in mammals is found in hemoglobin heme.5 Consequently, hemoglobin and other host proteins that scavenge hemoglobin or heme from blood plasma, such as hemopexin, haptoglobin, and serum albumin, are a source of iron for pathogenic bacteria.6 Indeed, pathogenic bacteria are able to overcome the extremely low levels of free iron within their mammalian hosts by deploying several iron and heme iron acquisition systems.7−9 Some Gram-negative bacteria deploy the has (heme acquisition system), which is characterized by the secretion of the hemophore HasA to bind and sequester extracellular heme and deliver it to a specific outer membrane receptor, HasR, for subsequent internalization.7,10,11
HasA hemophores from Serratia marcescens (HasAsm),12,13Pseudomonas aeruginosa (HasAp),14,15 and Yersinia pestis (HasAyp)16 have been structurally characterized in their heme-free (apo) and heme-bound (holo) forms. The structures of HasAp, HasAyp, and HasAsm are composed of an “α-helix wall” packed against a “β-sheet wall” and two extended loops that bridge the α-helix and β-sheet walls. The heme iron of HasAp and HasAsm is coordinated by Tyr75 and His32, with each axial ligand harbored in distinct loops, termed the Y75 and H32 loops, respectively (Figure 1A). This combination of axial ligands has been observed only in hemoglobin M Saskatoon,17 hemoglobins from invertebrates,18,19 and heme d1 in cytochrome cd1 reductase,20 whereas examples of heme proteins that utilize Tyr as the only protein-provided ligand, which include hemoglobins M17,21 methemalbumin,22 catalase,23 HasAyp,16 Isd hemophores secreted by Gram-positive organisms,2 and heme-binding periplasmic binding proteins,24,25 are relatively more abundant. The main difference between apo- and holo-HasAp (or HasAsm) is a large reorganization of the H32 loop, which relocates the His32 side chain approximately 30 Å (Figure 1B). The role played by His32 in heme binding has been investigated using the H32A mutant of HasAp.14,26 The X-ray crystal structure of the holo form revealed a noncrystallographic dimer linked by cofacial interaction of two heme molecules, which are coordinated by Tyr75; each subunit is nearly identical in structure to the apoprotein, including the open conformation of the H32 loop. These observations suggested that heme loads onto the Y75 loop while the H32 loop is in the open (apo-like) conformation. Subsequent closure of the H32 loop in wild-type (wt) HasAp excludes water from the heme environment and provides the sixth ligand to the heme.14 This idea found additional support in the structure of monomeric H32A holo-HasAp in complex with imidazole, which revealed the heme iron coordinated by Tyr75 and an exogenous imidazole. While the H32 loop is not completely defined in the imidazole complex, segments with clearly discernible electron density indicate a closed loop conformation similar to that seen in wt holo-HasAp. Moreover, the plane of the coordinated imidazole in H32A holo-HasAp is nearly superimposable with the His32 imidazole plane in wt HasAp. Solution nuclear magnetic resonance (NMR) studies of H32A holo-HasAp and its imidazole complex provided corroborating evidence that loading of hemin on the Y75 loop triggers closure of the H32 loop, even in the absence of the His32 side chain.14
Figure 1.
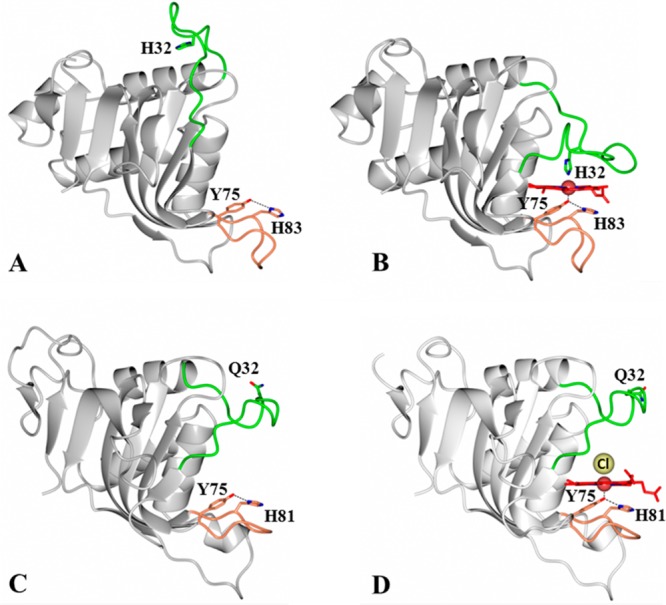
HasA hemophores display structural conservation of the proximal loop (coral) but divergence in the structure and function of the distal loop (green). Structures of (A) apo-HasAp [Protein Data Bank (PDB) entry 3MOK] and (B) holo-HasAp (PDB entry 3ELL) illustrate the large conformational rearrangement of the distal loop (green) caused by heme binding, which results in heme axial ligation by His32 and Tyr75. In contrast, the structures of (C) apo-HasAyp (PDB entry 4JER) and (D) holo-HasAyp (PDB entry 4JET) illustrate the minimal reorganization of the distal loop upon heme binding and the coordination of heme by only one protein-provided ligand, Tyr75. The Cl– ion that coordinates to the distal site is shown as a yellow sphere.
Kinetic studies of binding of heme to wt apo-HasAp using rapid mixing techniques coupled to electronic absorption, electron paramagnetic resonance (EPR), and resonance Raman spectroscopic methods identified two distinct kinetic phases.26 The spectroscopic changes in the first phase, which is complete within 20 ms, are indicative of efficient loading of heme onto the Y75 loop, while the slower phase, which lasts ≈1 s at 4 °C, reflects closure of the H32 loop and coordination of the heme iron by His32; accordingly, the H32A variant exhibits only the first millisecond phase corresponding to the loading of hemin onto the Y75 loop.26 Taken together, our structural and kinetic investigations clearly demonstrate that hemin loads onto the Y75 loop within a few milliseconds, where it is coordinated by Y75, and triggers closure of the H32 loop and coordination of the heme on a time scale of seconds. A recent study conducted with the H32A mutant of HasAsm produced a similar model for heme loading.27
Amino acid sequence alignments of HasA proteins from Yersinia species show conservation of Tyr75 and His83 in the Y75 loop, but no conservation of His32.16 Structural characterization of HasA from Y. pestis, apo- and holo-HasAyp, where residue 32 is a glutamine, led to three important observations. (i) The Gln32-bearing loop adopts a closed conformation in apo-HasAyp (Figure 1C). (ii) Tyr75 is the only endogenous heme ligand in holo-HasAyp. (iii) Binding of heme to apo-HasAyp occurs on a submillisecond time scale with minimal structural reorganization (Figure 1D).16 This study of HasAyp highlights variations in the coordination state of hemin in HasA hemophores, but it also underscores a strong structural conservation of the proximal loop, which includes the invariant Tyr75 ligand and its hydrogen bond partner, His81 (Figure 1). The function of this hydrogen bond, which is thought to enhance the phenolate character of Tyr75, has been proposed to be essential for proximal hemin coordination by Tyr75.15,28,29 A low-resolution crystal structure of H83A holo-HasAsm has been reported, but poor electron density did not permit the determination of whether Tyr75 remains coordinated to the heme iron in H83A holo-HasAsm.30 Resonance Raman and NMR spectroscopic studies, however, led the same authors to suggest that in the absence of His83 the Tyr75–Fe bond is severed. Thus, the H83A mutant of HasAsm was described as a five-coordinate species, with His32 as the only axial ligand at pH 5.2, His32 and water as axial ligands at neutral pH, and His32 and a hydroxide ligand at basic pH.30 A different study proposed that in the absence of Tyr75, His83 likely acts as a heme iron ligand,31,32 but direct experimental support for this notion is lacking.
Given the structural conservation of the Y75 loop in HasA hemophores, and the proposed requirement for the His83–Tyr75 unit for coordination of hemin by Tyr75, we conducted a structural and spectroscopic characterization of the Y75A and H83A HasAp mutants. Our high-resolution crystal structures of H83A HasAp clearly demonstrate that in the absence of His83, the Tyr75 ligand remains coordinated to the heme iron at neutral pH and at pH 5.4. These structures also show that in the absence of the His83 side chain, a water molecule can substitute for the missing Nδ atom of His83 to interact with the Oη atom of Tyr75 and aid its coordination to the ferric heme iron. NMR spectroscopic studies conducted with Y75A and H83A apo-HasAp demonstrate that disruption of the Tyr75–His83 hydrogen bond induces significant conformational disorder in the Y75 loop, which propagates into neighboring elements of secondary structure. Stopped-flow UV–vis absorption and rapid-freeze-quench resonance Raman (RFQ-RR) analyses of the binding of hemin to Y75A and H83A apo-HasAp show biphasic kinetics comparable to those observed in wt HasAp. These results suggest that the importance of hemin iron axial ligation in the process of the acquisition of hemin by HasAp should not be overestimated and point toward noncovalent interactions as factors dominating heme capture and the formation of holo-HasAp.
Experimental Procedures
Site-Directed Mutagenesis
Y75A and H83A HasAp mutants were constructed in the background of the gene encoding the truncated form of HasAp missing the last 21 C′-terminal residues.15 Primers were synthesized by Integrated DNA Technologies, Inc., and were used without further purification in conjunction with the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Primers used to install the Y75A mutation were 5′-GGCGGCGATCTGCATGCGACCCTGTTT-3′ and 5′-GCTAAACAGGGTCGCATGCAGATCGCCGCC-3′ and those use for the H83A mutation 5′-CTGTTTAGCAACCCGAGCGCGACCCTGTGGGGC-3′ and 5′-GCCCCACAGGGTCGCGCTCGGGTTGCTAAACAG-3′, where the underlined codons represent target substitutions. Each gene construct was transformed into XL1-Blue competent cells (Stratagene) for amplification, and the DNA sequence was verified by SeqWright (Houston, TX). The recombinant DNA plasmid with the correct sequence was transformed into Escherichia coli BL21-GOLD(DE3) competent cells for subsequent protein expression.
Protein Expression and Purification
Expression, purification, and reconstitution of Y75A and H83A HasAp with heme were conducted using previously described methods.14 The oligomeric state of the mutant hemophores was determined with the aid of a gel filtration column (Superdex 75 prep grade, GE Healthcare), calibrated with cytochrome c (12.4 kDa), chymotrypsin (25 kDa), ovalbumin (44 kDa), and albumin (67 kDa) standards, in 50 mM Tris-HCl (pH 7.5), 100 mM KCl buffer. The preparation of wt, Y75A, and H83A HasAp uniformly labeled with 15N ([U-15N]HasAp, [U-15N]Y75A HasAp, and [U-15N]H83A HasAp) or labeled with both 15N and 13C (wt [U-13C,U-15N]HasAp, Y75A [U-13C,U-15N]HasAp, and H83A [U-13C,U-15N]HasAp) was performed as previously described for wt and H32A HasAp.14
Crystallization and Data Collection
HasAp mutants in 100 mM sodium phosphate (pH 7.8) (Y75A) or 100 mM Tris (pH 7.6) (H83A) were concentrated to 15 mg/mL and screened for crystal growth at 20 °C using Compact Jr. (Emerald biosystems) sitting drop vapor diffusion plates. Equal volumes of protein and crystallization solution were equilibrated against a 100 μL reservoir volume. Crystals of Y75A HasAp were obtained within 24 h from Crystal Screen 2 (Hampton Research) condition #32 [1.6 M (NH4)2SO4, 100 mM HEPES (pH 7.5), and 200 mM NaCl]. Crystals were transferred to a solution containing an 80% crystallization solution and 20% glycerol before being frozen in liquid nitrogen for data collection. Crystals of H83A HasAp were obtained from three distinct conditions. (1) Wizard II (Emerald biosystems) condition #11 [10% 2-propanol, 100 mM sodium cacodylate (pH 6.5), and 200 mM zinc acetate] produced nondiffracting small crystals that grew as clusters of small needles in approximately 24 h at 20 °C. Refinement of the crystallization conditions was conducted using the Hampton Additive screen (Hampton Research). Plate-shaped clusters that diffracted to high resolution were obtained from 10% 2-propanol, 100 mM sodium cacodylate (pH 6.5), 200 mM zinc acetate, and 3% dioxane. These crystals were transferred to a solution containing a 75% crystallization solution and 25% ethylene glycol before being frozen in liquid nitrogen for data collection. (2) Crystals were obtained in approximately 3 days at 20 °C from Hampton Crystal Screen 2 condition #23 [1.6 M (NH4)2SO4, 100 mM MES (pH 6.5), and 10% dioxane]. Crystals were transferred to a solution containing an 80% crystallization solution and 20% ethylene glycol before being frozen in liquid nitrogen for data collection. (3) Crystals were obtained in approximately 3 days at 20 °C from 1.6 M (NH4)2SO4, 100 mM MES (pH 5.4), and 10% dioxane. Samples were transferred to a solution containing an 80% crystallization solution and 20% ethylene glycol before being frozen in liquid nitrogen for data collection. High-resolution diffraction data from all crystals were acquired at Advanced Photon Source IMCA-CAT beamline 17ID using a Dectris Pilatus 6M pixel array detector.
Structure Solution and Refinement
Diffraction intensities were integrated and scaled using XDS33 and Scala,34 respectively, and the Laue class was checked using Pointless.35 Coordinates from the structure of wt HasAp [Protein Data Bank (PDB) entry 3ELL] were used for molecular replacement with Molrep36 searching for a single molecule in the asymmetric unit for all data sets. Structural refinement of positional and anisotropic atomic displacement parameters and manual model building for both mutants were performed with Phenix37 and Coot,38 respectively. Structure validation was conducted using Molprobity,39 and figures were prepared using CCP4mg40 and PyMol (http://www.pymol.org). Data collection and refinement statistics are summarized in Table 1.
Table 1. X-ray Data Collection and Refinement Statistics.
| Y75A HasAp | H83Amono HasAp | H83Aortho HasAp | H83ApH5.4 HasAp | |
|---|---|---|---|---|
| Data Collection | ||||
| unit cell parameters | a = 34.44 Å, b = 46.73 Å, c = 101.04 Å | a = 34.90 Å, b = 66.23 Å, c = 41.00 Å, β = 97.1° | a = 34.38 Å, b = 46.99 Å, c = 100.90 Å | a = 34.43 Å, b = 46.78 Å, c = 101.81 Å |
| space group | P212121 | P21 | P212121 | P212121 |
| resolution (Å)a | 40.00–0.95 (1.00–0.95) | 66.23–1.32 (1.39–1.32) | 100.90–0.89 (0.94–0.89) | 101.81–1.25 (1.32–1.25) |
| wavelength (Å) | 1.0000 | 1.0000 | 0.9184 | 1.0000 |
| temperature (K) | 100 | 100 | 100 | 100 |
| no. of observed reflections | 901848 | 151394 | 714733 | 296237 |
| no. of unique reflections | 97317 | 43290 | 120441 | 46491 |
| ⟨I/σ(I)⟩a | 18.7 (2.6) | 7.7 (2.2) | 18.0 (2.9) | 12.1 (2.5) |
| completeness (%)a | 94.1 (80.0) | 99.6 (99.8) | 95.8 (87.2) | 100 (100) |
| multiplicitya | 9.3 (5.6) | 3.5 (3.5) | 5.9 (3.7) | 6.4 (6.3) |
| Rmerge (%)a,b | 5.5 (60.1) | 9.5 (68.1) | 5.0 (38.7) | 9.2 (80.6) |
| Rmeas (%)a,d | 5.8 (66.3) | 11.2 (80.5) | 5.4 (44.7) | 10.0 (88.1) |
| Rpim (%)a,d | 1.7 (27.3) | 5.9 (42.3) | 2.1 (21.8) | 3.9 (35.1) |
| Refinement | ||||
| resolution (Å) | 34.30–0.95 | 34.63–1.32 | 32.54–0.89 | 42.51–1.25 |
| no. of reflections (working/test) | 92362/4861 | 41067/2180 | 114343/6000 | 44065/2350 |
| Rfactor/Rfree (%)c | 13.9/14.6 | 14.3/16.8 | 12.4/13.4 | 15.3/18.2 |
| no. of atoms | ||||
| protein | 1357 | 1356 | 1436 | 1360 |
| heme | 43 | 43 | 43 | 43 |
| water | 322 | 215 | 235 | 213 |
| model quality | ||||
| root-mean-square deviation | ||||
| bond lengths (Å) | 0.012 | 0.010 | 0.015 | 0.011 |
| bond angles (deg) | 1.456 | 1.356 | 1.688 | 1.370 |
| average B factor (Å2) | ||||
| all atoms | 12.4 | 13.5 | 8.9 | 12.7 |
| protein | 10.3 | 11.8 | 7.3 | 10.9 |
| heme | 8.0 | 9.2 | 6.5 | 8.6 |
| zinc | – | 16.3 | – | – |
| ethylene glycol | – | 23.3 | 20.0 | 19.9 |
| formate | 7.4 | – | – | – |
| water | 22.2 | 24.5 | 18.9 | 24.4 |
| coordinate error, maximum likelihood (Å) | 0.07 | 0.16 | 0.13 | 0.30 |
| Ramachandran plot (%) | ||||
| most favored | 98.4 | 98.9 | 97.1 | 98.4 |
| additionally allowed | 1.6 | 1.1 | 2.4 | 1.6 |
Values in parentheses are for the highest-resolution shell.
Rmerge = ∑hkl∑i|Ii(hkl) – ⟨I(hkl)⟩|/∑hkl∑iIi(hkl), where Ii(hkl) is the intensity measured for the ith reflection and ⟨I(hkl)⟩ is the average intensity of all reflections with indices hkl.
Rfactor = ∑hkl||Fobs(hkl)| – |Fcalc(hkl)||/∑hkl|Fobs(hkl)|. Rfree is calculated in an identical manner using 5% of the randomly selected reflections that were not included in the refinement.
Y75A HasAp
Following initial refinement, prominent difference (Fobs – Fcalc) electron density was observed near the heme on the side that would normally be occupied by Tyr75. Three peaks were present in this region that were at an angle of approximately 120° and separated by approximately 1.3 Å. Refinement with an ethanol molecule at this site resulted in positive Fobs – Fcalc electron density at the C2 atom of ethanol, indicating an underestimation of electrons, whereas placement of a formate ion did not yield any residual electron density. Similar electron density was observed from four diffraction data sets ranging from 0.95 to 1.30 Å resolution obtained from four unique crystallization conditions: Hampton Crystal Screen 2 condition #23 described above, Hampton Crystal Screen 2 condition #32 [1.6 M ammonium sulfate, 0.1 M HEPES (pH 7.5), and 0.1 M sodium chloride], Hampton Crystal Screen 2 condition #42 [1.5 M ammonium sulfate, 0.1 M Tris (pH 8.5), and 12% (v/v) glycerol], and Hampton Crystal Screen 1 condition #38 [1.4 M sodium citrate tribasic dehydrate and 0.1 M HEPES (pH 7.5)]. Herein, we report only the 0.95 Å resolution data obtained with Crystal screen 2 condition #23. Because the electron density was not consistent with any of the crystallization components, protein storage buffer, or purification buffers, it is likely that the formate ion was present in the heme used in the reconstitution of the apoprotein.
H83A HasAp
Condition 1 produced a monoclinic crystal form, henceforth termed H83Amono. Molecular replacement searches in space groups P21 and P2 produced correlation coefficients of 0.62 and 0.30, respectively. Therefore, subsequent structural refinement was conducted in space group P21. Initial structure solution and refinement were conducted using in-house diffraction data collected with Cu Kα radiation. Large peaks greater than 10σ were observed in the difference Fourier maps (Fobs – Fcalc) that were tentatively assigned as zinc ions from the crystallization solution. No anomalous signal was observed at these sites using Cu Kα data. An X-ray fluorescence scan indicated that zinc was indeed present in the crystal. Consequently, data for structural refinement were collected at a wavelength of 1.0000 Å, which still yields an appreciable anomalous signal for Zn ∼ 2.6e–. In addition, diffraction data were collected at a low-energy remote wavelength (λ = 1.28414 Å, 9655 eV), which resulted in the disappearance of the anomalous difference density at the Zn sites but an increase at the heme Fe site. Condition 2 produced an orthorhombic crystal form (H83Aortho), and condition 3 produced crystals at pH 5.4 (H83ApH5.4), which were obtained from attempts to grow crystals in a pH range of 5.0–5.5, to explore the ligation state of Tyr75 under acidic conditions. The structure was determined as described above for H83Aortho.
NMR Spectroscopy
Sequential backbone assignments of wt apo-HasAp were obtained at 32 °C with the aid of two- and three-dimensional NMR experiments [1H–15N HSQC, HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH] conducted in a Varian Unity Inova 600 NMR spectrometer equipped with a triple-resonance probe. Given that the 1H–15N HSQC spectra of Y75A and H83A apo-HasAp are very similar to that of wt apo-HasAp, sequential backbone assignments for the two mutants were obtained at 32 °C with the aid of 1H–15N HSQC, HNCA, and HN(CO)CA experiments conducted in a Bruker Avance 800 spectrometer equipped with a 5 mm TCI 1H/13C/15N cryoprobe. Protein samples for three-dimensional NMR data acquisition were in phosphate buffer (20 mM, 95% H2O, 5% D2O, pH 7.0) at concentrations ranging between 2.5 and 5.0 mM. Two- and three-dimensional NMR spectra were processed using NMRPipe41 and analyzed with Sparky.421H chemical shifts were referenced to the proton reference of DSS at 0 ppm, while 15N and 13C shifts were referenced indirectly using ratios of 0.101329118 and 0.251449530, respectively.43 The effect of temperature on the 1H–15N HSQC spectra of wt and mutant proteins was determined in a 600 MHz Bruker Avance III spectrometer equipped with an inverse H/C/N triple-resonance probe, using samples with concentrations in the range of 1.0–1.7 mM.
RR and EPR Spectroscopy
RR spectra were recorded using a custom McPherson 2061/207 spectrograph (0.67 m with variable gratings) equipped with a Princeton Instruments liquid N2-cooled CCD detector (LN-1100PB). Excitation at 406 nm was provided by a krypton laser (Innova 302, Coherent), and a Kaiser Optical supernotch filter was used to attenuate Rayleigh scattering. Off-Soret excitations were provided by an argon laser (Innova 90, Coherent). Spectra at room temperature were recorded in a 90° scattering geometry on samples mounted on a reciprocating translation stage. Frequencies were calibrated relative to indene and CCl4 and are accurate to ±1 cm–1. CCl4 was also used to check the polarization conditions. The integrity of the RR samples, before and after laser illumination, was confirmed by direct monitoring of their UV–vis spectra in the Raman capillaries. Low-temperature spectra were recorded in a backscattering geometry on samples maintained at ∼105 K in a liquid nitrogen coldfinger. Frequencies were calibrated relative to aspirin and are accurate to ±1 cm–1. Sodium selenate (Sigma) was chosen as an internal standard44 and added at a final concentration of 100 mM to 500 μM samples of Y75A and H83A holo-HasAp, to determine their relative resonance enhancement based on the ν4 intensity at 406 nm excitation and 105 K. EPR spectra of holo-HasAp variants were recorded on a Bruker E500 X-band EPR spectrometer equipped with a superX microwave bridge and a dual-mode cavity and a helium-flow cryostat (ESR 900, Oxford Instruments, Inc.). Quantitation of the EPR signals was performed at 8 K under nonsaturating conditions by double integration and comparison with Cu(II)-EDTA standards.
Stopped-Flow UV–Vis Spectroscopy
Stopped-flow experiments were performed with an SX20 stopped-flow UV–vis spectrometer (Applied Photophysics) with a 1 cm path length cell equilibrated at 4 °C. Solutions of apo-HasAp variants in 200 mM HEPES buffer (pH 7.0) were prepared, and their concentrations were calculated as previously described on the basis of a 280 nm molar extinction coefficient (ε280) of 27.13 mM–1 cm–1 for both variants. The apoproteins were diluted to yield concentrations ranging from ∼40 to 400 μM in the same buffer. Hemin was dissolved in 1 mM NaOH and diluted to a final concentration of 10 μM just before the stopped-flow experiments using an ε385 of 58.4 mM–1 cm–1. After each measurement, remaining premixed solutions were recovered from the stopped-flow apparatus to confirm the protein and hemin concentrations. Control experiments using 1:1 mixtures of a hemin solution and buffer were run to confirm that the UV–vis spectrum of hemin remains unchanged during the course of stopped-flow experiments. Complete sets of time-resolved spectra were examined by global analysis using a Marquardt–Levenberg algorithm (Pro-K software, Applied Photophysics), which results in pseudo-first-order rate constants k1obs and k2obs. The reported rate constants are from global analyses and are the average of at least three different rapid mixing experiments.
Rapid-Freeze-Quench Experiments
Our protocol for the preparation of RFQ samples was described in detail recently.45 Glass syringes (2 mL) were loaded with 0.6 mM apo-HasAp variants in 200 mM HEPES (pH 7.0) and a 0.6 mM hemin solution in 10 mM NaOH before being mounted on a System 1000 Chemical/Freeze Quench Apparatus (Update Instruments) equipped with a water bath maintained at 4 °C. Reaction times were controlled by varying the syringe displacement rate from 2 to 8 cm/s or by varying the length of the reactor hose after the mixer. Five milliseconds was added to the calculated reaction times to account for the time of flight and the freezing time in liquid ethane. Mixed samples (250 μL) were trapped in liquid ethane at or below −120 °C in glass funnels attached to NMR tubes. The frozen samples were packed into the tube as the assembly sat within a Teflon block cooled to −120 °C with liquid nitrogen. Liquid ethane was subsequently removed by incubating samples at −80 °C for 2 h. RR analyses before and after cryosolvent removal showed no spectral changes except for the loss of ethane bands. End point samples were generated by collecting reaction mixtures in NMR tubes at room temperature before they were frozen in liquid ethane.
Results
Y75A and H83A Holo-HasAp Mutants Adopt a Monomeric Structure in Solution
As is the case for wt HasAp, the bacterial expression of Y75A and H83A HasAp results in accumulation of a much larger proportion of apo hemophore relative to its holo counterpart in the E. coli host cells. Hence, the two mutants were purified in their apo form using a previously reported protocol, which utilizes hydrophobic affinity chromatography to separate apo and holo hemophores.14 The holo hemophore was prepared from its corresponding apo form by heme reconstitution in vitro, as described in Experimental Procedures. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis shows that both mutant proteins migrate with apparent masses of ∼19 kDa. Mass values measured with the aid of ESI-MS are 18667 Da (Y75A) and 18693 Da (H83A), which are in good agreement with the molecular masses calculated from amino acid sequences, 18668 Da (Y75A) and 18694 Da (H83A). The apparent molecular masses of Y75A and H83A determined by size exclusion chromatography are 16.9 and 17.6 kDa, respectively, indicating that both mutant proteins are monomeric in solution (Figure S1 of the Supporting Information).
Like wt Holo-HasAp, the Y75A and H83A Variants Exist as Six-Coordinate High-Spin–Low-Spin Equilibrium Mixtures
Figure 2 shows the electronic absorption spectra of wt (black), Y75A (red), and H83A (blue) HasAp. The UV–vis absorption spectrum of Y75A exhibits a Soret peak at 407 nm and a characteristic high-spin marker charge transfer band at 624 nm. Intense Q bands at 535 and 568 nm also support the presence of a low-spin contribution. This high-spin–low-spin equilibrium is consistent with axial coordination that includes a His side chain and an exogenous ligand as revealed by the X-ray crystal structure of Y75A holo-HasAp (see below). The UV–vis spectrum of H83A is primarily indicative of a high-spin conformation at room temperature with a broader Soret band at 405 nm, Q bands at 489 and 550 nm, and a high-spin marker band at 608 nm (Figure 2). UV–vis spectra recorded at low temperatures confirm that the low-spin conformation becomes dominant below 100 K (Figure S2 of the Supporting Information).
Figure 2.
Room-temperature UV–vis spectra of wt, Y75A, and H83A holo-HasAp.
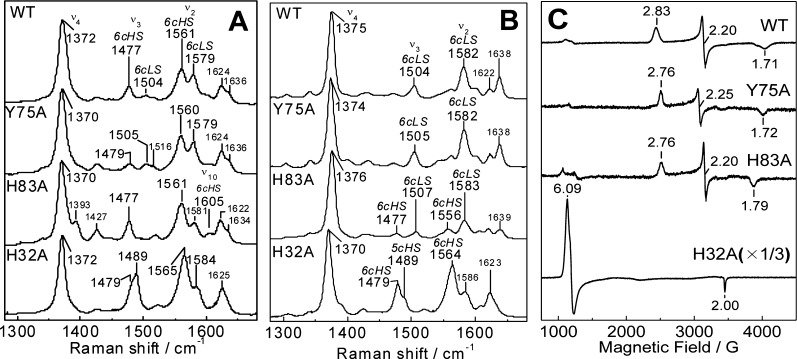
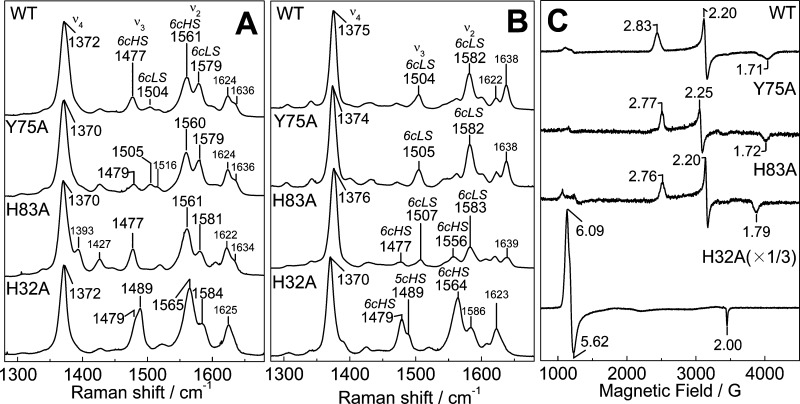
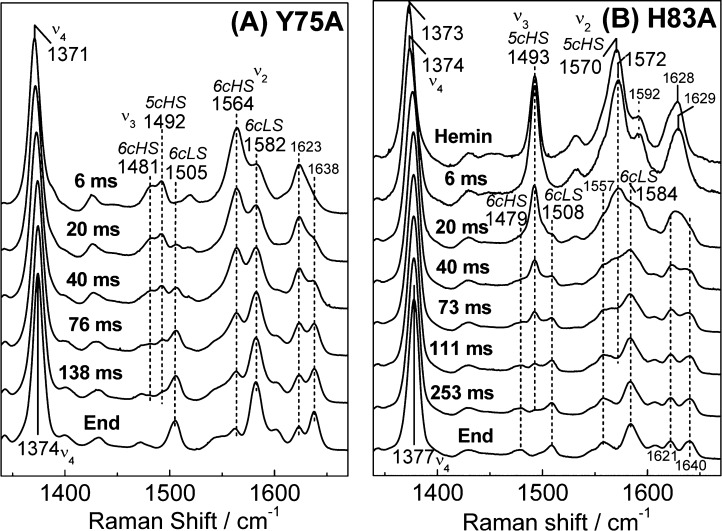
The room-temperature RR spectrum of Y75A is characteristic of a mixture of six-coordinate high-spin (6cHS) and six-coordinate low-spin (6cLS) hemes with ν3 and ν2 at 1479 and 1560 cm–1, and 1505 and 1579 cm–1, respectively (Figure 3A). Observed frequencies and intensity ratios between 6cHS and 6cLS bands in the RR spectrum of Y75A differ from those of the wt protein, but qualitatively, the RR spectra of Y75A are comparable to those of the wt protein. The RR spectra of H83A confirm the presence of a 6cHS–6cLS temperature-dependent equilibrium, with the 6cHS configuration, ν3, ν2, and ν10 at 1477, 1561, and 1605 cm–1, respectively, dominating at room temperature and 6cLS contributions beginning to prevail at 110 K (Figure 3B). Accordingly, the EPR spectra of Y75A and H83A holo-HasAp recorded at 8 K show rhombic signatures with resonances centered around g = 2 as expected for S = 1/2 low-spin ferric heme species (Figure 3C).
Figure 3.
Room-temperature RR (A), low-temperature RR (B), and 8 K EPR (C) spectra of wt, Y75A, H83A, and H32A holo-HasAp.
The Crystal Structures of Y75A, H83A, and wt Holo-HasAp Are Highly Homologous
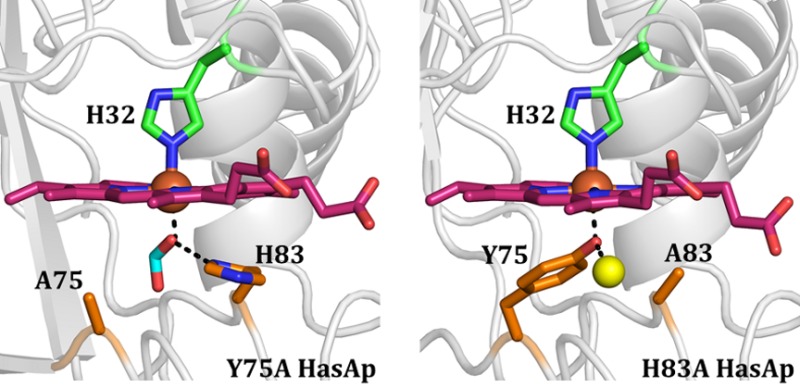
The structure of Y75A holo-HasAp was refined using data diffracting to 0.95 Å resolution (Table 1). Crystals of H83A HasAp were obtained from three different conditions (see Experimental Procedures), and the corresponding structures were refined using data diffracting to 0.89 Å (H83Aortho), 1.32 Å (H83Amono), and 1.25 Å (H83ApH5.4) resolution. The overall structure of the mutants is nearly identical to that of wt holo-HasAp (Figure 4 and Figure S3 of the Supporting Information). It is noteworthy that in the H83A mutant, Tyr75 remains coordinated to the heme iron in both structures obtained at pH 6.5 (Figure 4C and Figure S3A of the Supporting Information) as well as in the structure obtained at pH 5.4 (Figure S3C of the Supporting Information). In the Y75A HasAp structure (Figure 4B), His83 remains in the position it occupies in the wt protein and therefore does not coordinate the heme iron. In the H83Amono structure, the hairpin loop adjoining β6 and β7 is disordered, as suggested by the absence of electron density, which is demarcated by asterisks in Figure 4C. In the H83Aortho structure, strand β8 is shorter than in the wt protein (Figure S3A of the Supporting Information). Surprisingly, although both mutations are on the Y75 loop, the conformation of the H32 loop is significantly more affected than the conformation of the Y75 loop. This is illustrated in Figure 5, which depicts the conformational changes affecting the backbone and side chains of residues in the H32 loop of H83A HasAp (cyan) and Y75A HasAp (green) relative to their conformations in wt HasAp (magenta). In contrast, the structure of the Y75 loop remains nearly unchanged relative to the Y75 loop in wt HasAp. Note, however, that despite the conformational changes in the H32 loop, the coordinated imidazole rings of His32 in the wt and mutant proteins are identical, as is the seating of the heme.
Figure 4.
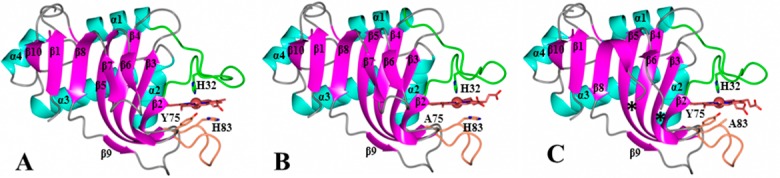
Overall structure of the Y75A and H83A mutants that is very similar to that of wt HasAp (PDB entry 3ELL). The Cα root-mean-square deviations (rmsds) from comparing the wild-type structure (A) to the structure of Y75A HasAp (B) is 0.571 Å, and that from comparing the wild-type structure to the structure of H83Amono HasAp (C) is 0.448 Å. Secondary structures are colored cyan (α-helices), magenta (β-strands), and gray (loops). Residues Ser2–Ala183 were used to calculate the rmsds using the secondary structure matching algorithm in Superpose58 via the CCP4 interface.59
Figure 5.
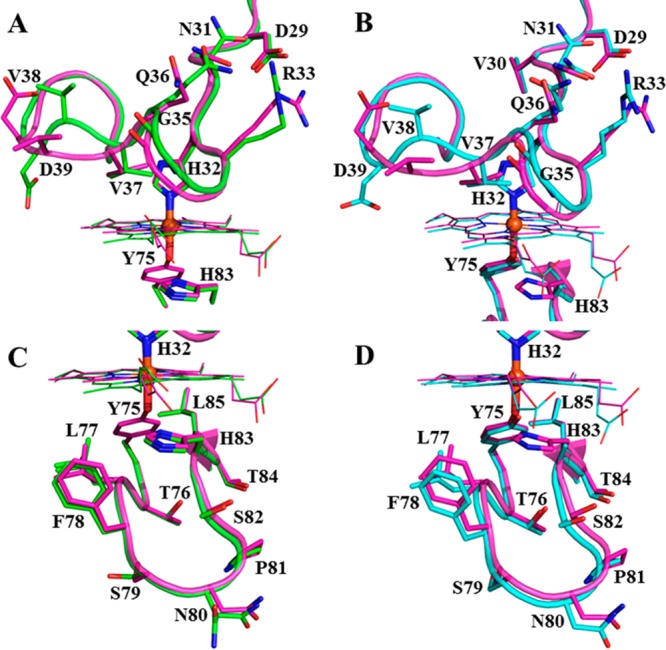
Replacing Tyr75 or His83 in the Y75 loop of HasAp with a noncoordinating Ala causes conformational changes in the His32 loop. Close-up views of the H32 (top) and Y75 (bottom) loops of H83A HasAp (cyan) and Y75A HasAp (green) superposed with the structure of wt HasAp (magenta).
Heme binds to the Y75A HasAp mutant in a single orientation, which is evident from the Fobs – Fcalc electron density map in Figure 6A. Given that the heme seating is also identical to that observed in wt HasAp, it allows similar interactions between the heme propionates and Gly35 and Arg129 (Figure S4 of the Supporting Information). Prominent difference electron density (Fobs – Fcalc) was observed near the heme on the side that would normally be occupied by Tyr75 in the wt protein. Structure refinement with an ethanol molecule at this site resulted in positive Fobs – Fcalc electron density at the C2 atom of ethanol, indicating an underestimation of electrons. Placement of a formate ion at this site did not yield residual electron density upon refinement; thus, a formate ion was modeled at this site. It is noteworthy that the same electron density was observed in four distinct diffraction data sets ranging in resolution from 0.95 to 1.3 Å, which were obtained from four different crystallization conditions (see Experimental Procedures). Herein, we report only the structure obtained from crystals diffracting to 0.95 Å resolution. Given that the protein is initially purified in its apo form and then reconstituted with heme in vitro, we surmise that the most likely source of formate stems from the heme or the dimethyl sulfoxide used to dissolve heme for the heme reconstitution experiments. The plane of the modeled formate ion is aligned with the heme α–γ-meso axis, which allows one of the formate oxygen atoms to coordinate the heme and also form a hydrogen bond with Nδ of His83 (2.6 Å). This hydrogen bonding interaction is similar to the hydrogen bond formed in the wt protein between the coordinated phenolic oxygen of Tyr75 and Nδ of His83. The second oxygen in the formate molecule interacts with the backbone carbonyl group of Thr76 (3.8 Å). The side chain of His83, which also displays very well-defined electron density, is placed in a position identical to the position it occupies in the wt protein, where it does not coordinate the heme iron.
Figure 6.
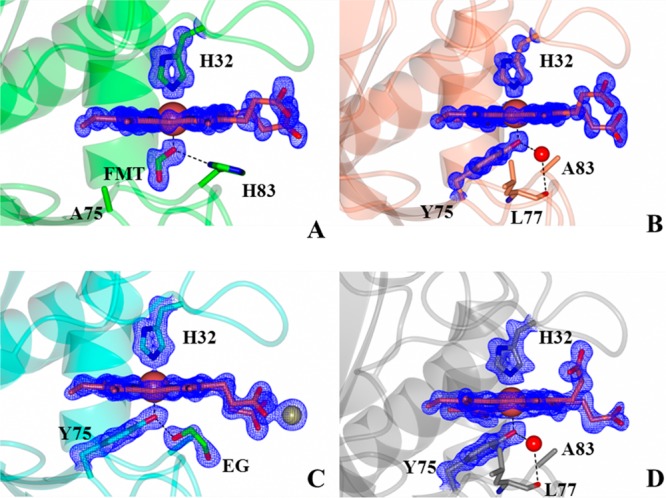
Close-up views of the heme binding sites in (A) Y75A HasAp, (B) H83Aortho HasAp, (C) H83Amono HasAp, and (D) H83ApH5.4 HasAp. Fobs – Fcalc electron density omit maps contoured at 3σ are shown as blue mesh. FMT denotes the formate ion and EG ethylene glycol. The Zn2+ ion in panel C is shown as a sphere.
Tyr75 and His32 axially coordinate the heme iron in the H83Aortho (Figure 6B), H83Amono (Figure 6C), and H83ApH5.4 (Figure 6D) structures. The Fobs – Fcalc electron density maps show that both axial ligands exhibit very well-defined electron density in all structures, as does the heme, which is bound to the protein in a single orientation, thus clearly demonstrating that Tyr75 remains coordinated to the heme iron, even at pH 5.4. The seating of the heme is the same in all three structures and identical to that observed in wt HasAp, although the heme propionates adopt different conformations in the three structures (also see Figure S4 of the Supporting Information). In all structures, Tyr75 coordinates the heme iron in a conformation identical to that seen in wt HasAp. In addition to coordinating the heme iron, the phenolic oxygen of Tyr75 forms a hydrogen bond with a water molecule in the H83Aortho (Figure 6B) and H83ApH5.4 structures (Figure 6D), or with the oxygen atom of an adventitious ethylene glycol (used as a cryoprotectant) in the H83Amono structure (Figure 6C). These hydrogen bonding interactions experienced by Tyr75 in the H83A mutant effectively replace the Tyr75 Oη···His83 Nδ hydrogen bond seen in wt HasAp. These observations are in agreement with the significance of the hydrogen bond between Tyr75 and His83 in allowing coordination of Tyr75 to the heme iron.28,31 Importantly, however, our finding that Tyr75 coordinates the heme in all available structures is in contrast with conclusions derived from spectroscopic studies suggesting that the Tyr–Fe bond is severed in the H83A mutant of HasAsm.27,30
During refinement of the H83Amono structure, large peaks greater than 10σ were observed in the Fobs – Fcalc maps, which were tentatively attributed to zinc ions from the crystallization solution. No anomalous signal was observed at these sites using diffraction data from Cu Kα X-rays. An X-ray fluorescence scan at the synchrotron indicated that zinc was indeed present in the crystal (Figure S5A of the Supporting Information). Hence, data for structural refinement that were collected at a wavelength of 1.0000 Å yielded an appreciable anomalous signal for Zn. In contrast, diffraction data collected at a low-energy remote wavelength (1.28414 Å), which resulted in the disappearance of the anomalous difference density at the Zn sites and increased the magnitude of the anomalous signal at the heme iron site, confirmed the presence of Zn (Figure S5B of the Supporting Information). One of the zinc ions interacts with the heme propionates, which induces a conformation distinct from that observed for the heme propionates in wt HasAp. In the H83ApH5.4 structure, heme propionate 6 interacts with the side chain of Arg129 and thus adopts a conformation similar to that in the wt protein. In contrast, heme propionate 7 adopts a different conformation, which places the carboxylate group within hydrogen bonding distance of the backbone NH and CO groups of Arg33, Gly35, and Val37 (Figure S4E of the Supporting Information).
Y75A and H83A Apo-HasAp Have Disordered Y75 Loops
The X-ray crystal structures of Y75A and H83A holo-HasAp indicate minimal changes in the structure of the Y75 loop and small changes in the H32 loop relative to those of the wt protein. Despite considerable effort, however, we were unable to grow crystals of Y75A or H83A apo-HasAp. We hypothesized that conformational disorder in the apoproteins may prevent crystal growth; thus, we conducted NMR spectroscopic studies in solution aimed at probing disorder in the apoproteins. A suite of five experiments (see Experimental Procedures) allowed sequential backbone assignments for 93% of non-proline residues in wt apo-HasAp (Table S1 of the Supporting Information). Chemical shift indexing46 of Cα chemical shifts was used to identify elements of secondary structure in solution, which were found to be consistent with elements of secondary structure observed in the crystal structure of wt apo-HasAp (Figure S6A of the Supporting Information). The 1H–15N HSQC spectrum of each of the mutant apoproteins, with the exception of a few cross-peaks, can be nearly superimposed with the HSQC spectrum of wt apo-HasAp (Figure S7 of the Supporting Information), which indicates that overall, the structures of the mutant apo hemophores are similar to the structure of wt apo-HasAp. Given the high degree of similarity in the HSQC spectra of wt and mutant proteins, the sequential backbone assignments of Y75A and H83A apo-HasAp were obtained relatively readily (Tables S2 and S3, respectively, of the Supporting Information). Plots comparing Cα chemical shift indexes of wt and mutant proteins (Figure S6 of the Supporting Information) also demonstrate that neither the Y75A mutation nor the H83A mutation caused significant changes in the structure of the apoproteins.
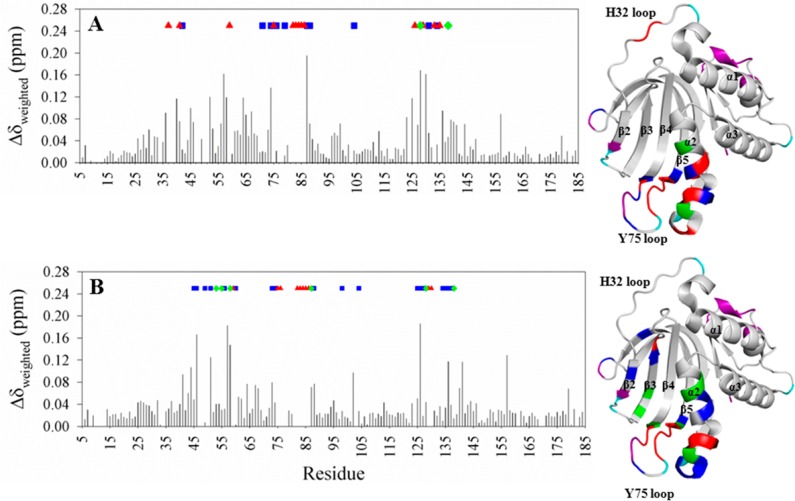
Closer inspection of the HSQC spectra shows mostly small differences in the weighed chemical shifts (Δδweighed) obtained from comparing the chemical shifts of cross-peaks in Y75A apo-HasAp (Figure 7A) or H83A apo-HasAp (Figure 7B) with corresponding cross-peaks in wt apo-HasAp. Importantly, the comparison also reveals that several cross-peaks in the spectra of the mutants (i) are missing (red triangles in Figure 7), (ii) are broadened and have intensities significantly lower than those of their counterparts in the spectrum of wt HasAp (blue squares), or (iii) are split (green diamonds). In addition, some of the affected resonances exhibit pronounced changes in intensity, split farther apart, or disappear with changes in temperature. An example is shown in Figure S8 of the Supporting Information, where the two cross-peaks originating from G128 (α2) in Y75A apo-HasAp become progressively more separated as the temperature is lowered from 32 to 15 °C. In comparison, G128 in the wt protein gives rise to a unique cross-peak at all temperatures. The observations described above indicate that the corresponding residues in the mutant apoproteins are indeed affected by conformational disorder. Mapping the missing, broadened, and split cross-peaks in the spectra of the mutants onto the X-ray crystal structure of wt apo-HasAp using the same color scheme as in the plots (Figure 7) reveals that the Y75 loop, the neighboring α2 helix, some of the β-strands near the loop, and a portion of the distal loop emerging from β2 are affected by conformational disorder in both mutants.
Figure 7.
Per-residue differences obtained from comparing cross-peaks in the HSQC spectrum of wt apo-HasAp with cross-peaks in the HSQC spectra of (A) Y75A apo-HasAp and (B) H83A apo-HasAp. Residues in the mutants whose cross-peaks disappear are denoted with red triangles; residues whose cross-peaks split are denoted with green diamonds, and residues whose cross-peaks are broadened and have decreased intensity are denoted with blue squares. These residues have been mapped on the wt apo-HasAp structure (PDB entry 3MOK) using the same color code; purple denotes residues with no assignment, and cyan identifies prolines. Δδweighed = {[(ΔδN/5)2 + (ΔδH)2]/2}1/2.
The Y75A and H83A Apo-HasAp Variants Conserve the Characteristic Biphasic Kinetics of Hemin Loading Observed in the wt Protein
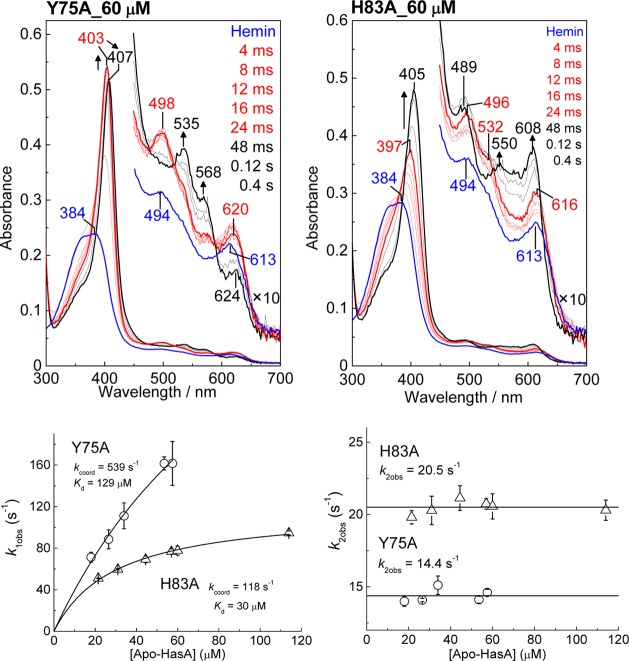
As previously described for wt apo-HasAp,26 the stopped-flow absorption data of the acquisition of hemin by the Y75A and H83A variants correspond to a biphasic process. The stopped-flow traces obtained with Y75A are very similar to those observed with wt HasAp as they reveal a millisecond intermediate species with Soret, Q, and high-spin CT bands at 403, 498, and 620 nm, respectively (Figure 8 and Figure S9 of the Supporting Information). Low-spin Q bands at 535 and 568 nm begin to grow ∼20 ms after mixing at 4 °C. The H83A variant also reveals the formation of a high-spin intermediate with Soret, Q, and high-spin CT bands at 397, 496, 532, and 616 nm (Figure S9 of the Supporting Information). After completion of this first millisecond phase, the UV–vis spectra further evolve to new predominantly high-spin components at 405, 489, 550, and 608 nm (Figure 8).
Figure 8.
Stopped-flow absorption spectra of the association of 4.7 μM hemin with (A) Y75A and (B) H83A apo-HasAp at 60 μM. Red and black traces are representative of early and late phases of the reaction, respectively; the blue traces are hemin/buffer controls. Also shown are plots of observed rate constants vs apo-HasAp concentration.
In previous work,26 we analyzed the biphasic behavior of loading of hemin to wt apo-HasAp with a multistep model, in which formation of a reversible hemin–apoprotein complex proceeds quickly before slower iron(III) coordination steps begin to take place:
Assuming that k1[apo-HasAp] and k–1 ≫ kcoord1 and kcoord1 ≫ kcoord2 produces eq 1:
| 1 |
This model explains the hyperbolic dependence of k1obs on the apo-HasAp concentration. In contrast, k2obs shows no dependence on the apo-HasAp concentration, as expected for an intramolecular reaction that corresponds to the closure of the H32 loop and coordination of the iron by His32.
This same analysis was used to extract Kd, kcoord1, and kcoord2 values from the stopped-flow data obtained with the variant proteins (Table 2). The Y75A substitution results in a 4-fold higher Kd value without a significant change in the rate of the first coordination step, kcoord1. The second kinetic phase reflects kcoord2 and is ∼3 times faster in Y75A than in wt HasAp, but it remains low, i.e., in the millisecond time scale expected for a significant protein loop rearrangement (Table 2).
Table 2. Kinetic Parameters for the Binding of Hemin to wt and Mutant HasAp Proteins at 4 °C.
| wt | H32A | Y75A | H83A | |
|---|---|---|---|---|
| Kd (μM) | 35 | 61 | 129 | 30 |
| kcoord1 (s–1) | 552 | 830 | 539 | 118 |
| kcoord2 (s–1) | 5.2 | n.o. | 14.4 | 20.5 |
The k1obs values for H83A apo-HasAp also follow a hyperbolic dependence on apoprotein concentration as seen with the Y75A variant, but the curve for H83A rises more quickly and achieves a lower plateau, indicating lower values of Kd and kcoord1 (Figure 8). Compared to those of wt HasAp, the H83A substitution does not appear to change the Kd value but does decrease kcoord1 5-fold. In contrast, the kcoord2 value is increased ∼4-fold in the H83A variant compared to that of wt HasAp (Table 2). In addition to kinetic rates, the global fitting analyses provide electronic absorption spectra of the millisecond intermediates in individual proteins (Figure S9 of the Supporting Information). These computed spectra suggest that distinct high-spin heme–protein intermediate complexes form in Y75A, H83A, and wt HasAp.
The Millisecond Intermediate in Y75A Is Likely Coordinated by His83, while in H83A, Y75 Is the Likely Axial Ligand to the Hemin Iron
Rapid-freeze-quench resonance Raman (RFQ-RR) experiments were performed in an effort to characterize the millisecond intermediate heme–protein complexes in Y75A and H83A HasAp. The high-frequency RR spectrum of the 6 ms RFQ sample of Y75A shows porphyrin modes indicative of a mixture of 5cHS and 6cHS species with ν3 modes at 1492 and 1481 cm–1, respectively (Figure 9A). High-spin heme iron(III) species with O atom axial coordination typically exhibit ν3 modes that are intense relative to the ν4 and ν2 modes,47−54 and the high ν3/ν4, ν2 intensity ratio observed in the RFQ-RR spectra of the millisecond intermediate formed in wt HasAp supported the notion that Y75 is the first coordinating ligand to the iron(III).26 In contrast, the RR spectra of the 6 ms samples of Y75A show ν3 modes that are weak relative to the ν2 and ν4 modes, which suggest that the initial coordination to the iron(III) involves an endogenous ligand, presumably His83, rather than a solvent molecule. Over time, these high-spin contributions are replaced by 6cLS modes with ν3 and ν2 at 1505 and 1582 cm–1, respectively. On the basis of the crystal structure of Y75A holo-HasAp, we assign this transition to the closure of the H32 loop on the heme pocket and intercalation of a solvent molecule between the heme iron(III) and His83.
Figure 9.
High-frequency RR spectra of RFQ samples of the reaction of 1 equiv of hemin with (A) Y75A apo-HasAp and (B) H83A apo-HasAp (protein concentration after mixing of 300 μM and excitation wavelength of 406 nm; all spectra are normalized to the intensity of the ν4 feature observed between 1371 and 1377 cm–1).
The high-frequency RR spectra of the 6 ms RFQ sample of H83A exhibit ν4, ν3, and ν2 modes at 1374, 1493, and 1572 cm–1, respectively, that are characteristic of a pure 5cHS species with an O atom axial ligand; they are analogous to but spectrally distinguishable from free hemin (Figure 9B). These 5cHS porphyrin skeletal modes are also different from the 5cHS signals observed in the 6 ms RFQ sample of wt HasAp (Figure S10 of the Supporting Information). Longer reaction times allow for the conversion of this five-coordinate complex to a mixture of 6cHS and 6cLS heme conformers. Attempts to detect vibrational modes involving iron(III) axial ligands such as tyrosinate modes or ν(Fe–OH) modes using off-Soret excitation and 18OH2–D2O solvent exchange were unsuccessful for both the Y75A and H83A variants (data not shown).
Discussion
Y75A and H83A Substitutions Do Not Affect the Structure of Holo-HasAp
Previous investigations of the process of loading of hemin onto HasAp showed that within 20 ms hemin binds first to the Y75 loop. This rapid process, which is believed to include the coordination of Tyr75 to the heme iron, is followed by a slower reorganization of the H32 loop and coordination of the heme by His32, which occurs within ∼1 s at 4 °C.14,26 In contrast to the large rearrangement of the His32-bearing loop, the Y75 loop retains its structure in the apo and holo forms of the hemophore. The structural conservation includes the Tyr75 Oη···His83 Nδ hydrogen bond, which is thought to play a pivotal role in hemin binding by increasing the phenolate character of the Tyr75 axial ligand and allowing it to coordinate the ferric ion in hemin, yet our high-resolution crystal structures of the Y75A and H83A holo-HasAp mutants are remarkably similar to that of wt holo-HasAp. In particular, in all three H83A holo-HasAp structures, the heme iron is coordinated by both His32 and Tyr75 and the structure of the Y75 loop is unaltered relative to that of wt holo-HasAp (see Figures 4 and 6). Interestingly, the void left by the His83 to Ala substitution is filled by a water molecule in the H83Aortho structure at pH 6.5 and in the structure at pH 5.4, and this crystallographically defined water molecule is hydrogen bonded to the phenol oxygen of the coordinated Tyr75. In the H83Amono structure, the same void is filled with an ethylene glycol molecule that acts as a hydrogen bond partner to the coordinated Tyr75. It is also noteworthy that the side chain of Tyr75 remains coordinated to heme, even at pH 5.4, despite the absence of the His83 side chain. Clearly, these observations demonstrate that His83 and the Tyr75 Oη···His83 Nδ hydrogen bond are not essential for maintaining the integrity of the heme-binding pocket in holo-HasAp or the coordination of hemin iron by Tyr75.
In the Y75A holo-HasAp structure, we observed well-defined elongated electron density below the hemin iron, which was modeled as a formate ion (Figure 6A); as indicated above, the most likely source of this formate ion is our hemin stock. Nevertheless, the structure of the loop, including the position of the His83 side chain, can be superimposed with that observed in the Y75 loop of wt holo-HasAp, suggesting that in the absence of formate, the heme iron is coordinated to a water molecule and His32, as has been suggested on the basis of spectroscopic studies of Y75A holo-HasAsm.27
The Y75 Loop Is Partially Disordered in the Variant Apoproteins
In contrast to the success we experienced growing single crystals of wt, Y75A, and H83A holo-HasAp, we were unable to grow crystals of the apo form of these variant proteins despite extensive screening of conditions. Because we suspected conformational disorder was hindering crystal growth, we turned to solution NMR and obtained the sequential backbone assignments of wt, Y75A, and H83A apo-HasAp. As expected, the NMR spectra of wt apo-HasAp are consistent with a well-ordered structure, which is in agreement with the well-defined electron density defining its X-ray crystal structure.14 The HSQC spectra of Y75A or H83A apo-HasAp are nearly identical to the HSQC spectrum of their wt counterpart. Detailed inspection, however, reveals that several cross-peaks corresponding to residues in the Y75 loop, helix α2, and several β-strands in Y75A and H83A apo-HasAp are affected by conformational disorder (see Figure 7). The crystal structure of wt apo-HasAp (Figure 10) shows that Tyr75 and His83 are involved in several hydrogen bonding and hydrophobic interactions within the loop. Figure 10A shows the hydrogen bond between the phenol oxygen of Tyr75 and Nδ of His83, as well as the hydrogen bonds between the carbonyl oxygen of His83 and the side chains of Arg129 and His134 in helix α2, and the Leu85 carbonyl···His134 NH hydrogen bond, which mediate interactions between the Y75 loop and helix α2. Also apparent is a group of hydrophobic interactions (Figure 10B) that include the hydrophobic portions of the Tyr75 and His83 side chains, helix α2 (Met141), strands β5 (Leu85), β4 (Leu73), β3 (Tyr56), and β2 (Phe46), and the hairpin loop connecting strands β2 and β3 (Phe51). This extensive and intricate network of H-bonding and hydrophobic interactions that propagates into a significant portion of the structure stabilizes the structure of the Y75 loop in the apoprotein, but replacing Tyr75 or His83 with Ala results in conformational disorder in the Y75 loop in apo-HasAp.
Figure 10.
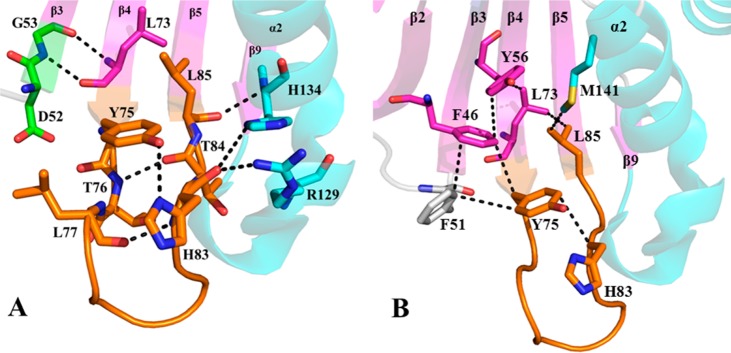
Close-up view of (A) hydrophilic and (B) hydrophobic interactions in wt apo-HasAp (PDB entry 3MOK) emanating from the Tyr75 loop into helix α2 and nearby β-strands. β-Strands are colored magenta and α-helices cyan. The Tyr75 loop is colored orange, with corresponding residues shown as sticks.
The Kinetics of Hemin Binding in the Two Variants Are Only Marginally Affected Relative to Those of the wt, but the Two Variants Form Distinct Millisecond High-Spin Intermediates
The relatively severe changes introduced into the Y75 loop and the concomitant conformational disorder introduced into neighboring structural domains in Y75A and H83A apo-HasAp are likely to result in the initial distribution of [apo-HasAp·hemin] substates during the process of hemin loading. However, rapid interconversion between these [apo-HasAp·hemin] substates is likely to take place, and the conformational disorder observed in Y75A and H83A apo-HasAp does not dramatically impact the rates of hemin loading in the variants compared to that in the wt protein (Table 2). Both variants show Kd values for the initial [apo-HasAp·hemin] complex in the same micromolar range as that of the wt protein, and the first coordination event defined by kcoord1 occurs in similar time frames for all three proteins. The variants show kcoord2 values slightly higher than that of the wt, but here again the differences are small.
In contrast, the RFQ-RR analyses of the two variant proteins clearly reveal that they form distinct millisecond intermediates. In Y75A, the intermediate shows an intensity ratio between ν4 and ν3 porphyrin skeletal modes consistent with coordination of His83 to the heme iron before closure of the H32 loop, coordination of H32, and displacement of the H83 ligand by a water molecule; coordination of His32 and rearrangement at the Fe–His83 site are not kinetically resolved and are possibly coupled events. In comparison, the RFQ-RR spectrum of the intermediate in H83A supports iron coordination by an O atom, either from solvent or from Tyr75, before coordination of the second axial ligand, His32. Importantly, despite these differences in coordination geometry in the [HasAp-hemin] millisecond intermediates of the variant and wt proteins, all three proteins follow equivalent kinetic profiles.
We have previously proposed that π–π stacking interactions between residues in the Y75 loop and hemin contribute significantly to a “sticky” hydrophobic platform that facilitates hemin capture.14,16 Within this framework, the Tyr75 Oη···His83 Nδ hydrogen bond contributes to the hemin loading process by positioning the Oη of Tyr to coordinate the heme iron (Figure 11). In absence of a Tyr75 Oη···His83 Nδ hydrogen bond in the apo Y75A mutant, the conformational disorder of residues in the Y75 loop may permit coordination of the incoming hemin by H83. Subsequent scission of the His83–Fe coordination bond and movement of the His83 side chain away from the iron and toward its position in the structure of wt holo-HasAp are likely a consequence of loop reorganization driven by the establishment of productive π–π stacking and hydrophobic interactions between hemin and side chains in the Y75 loop. Isothermal titration calorimetry measurements with HasAsm revealed ≈250- and ≈400-fold decreases in heme affinity in the H83A and Y75A variants, respectively, relative to that of the wt protein.31 Our pre-steady state measurements show that these mutations have little impact on the rates of formation of holoproteins and therefore suggest that changes in binding affinity primarily reflect increases in dissociation rates. Consequently, the data suggest that axial ligation plays a prominent role in slowing the loss of hemin from the hemophore, but the on rate of loading of hemin onto the apo hemophore is governed by noncovalent hydrophobic interactions. In this context, the changes in Kd and kcoord1 observed with the Y75A and H83A variants relative to those of wt HasAp likely reflect changes in hydrophobic and π–π stacking interactions between the porphyrin macrocycle and the Y75 loop. Hence, the data at hand suggest that HasA hemophores utilize hydrophobic interactions to capture hemin while controlling its release via the heme iron axial coordination. Future studies aimed at dissecting the thermodynamics and kinetics of hemin capture and release with complementary variants, such as Y75F and H83F, Y75F/H83F, and Y75A/H83A, will contribute to the establishment of this mechanistic model.
Figure 11.
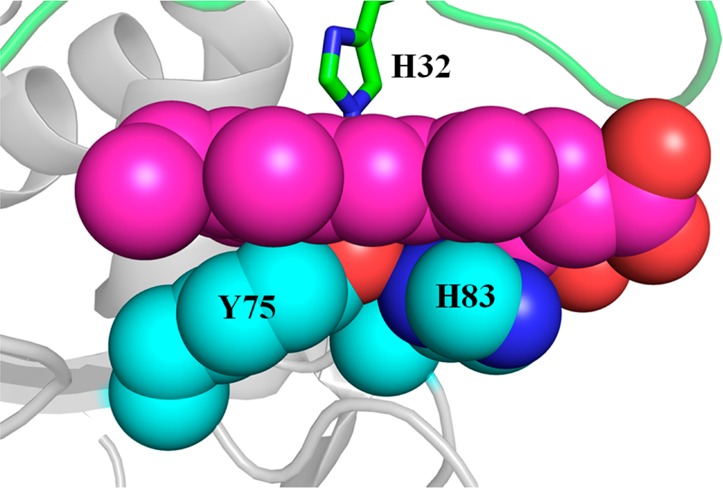
Close-up view of the Y75 loop in wt holo-HasAp (PDB entry 3ELL) illustrating the hydrophobic and π-stacking interactions between the Y75 and H83 side chains with the heme.
Acknowledgments
The assistance of Dr. Grace Jepkorir in the preparation of site-directed mutants and in protein crystallization is acknowledged. Use of IMCA-CAT beamlines 17-BM and 17ID at the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. Use of the KU COBRE-PSF Protein Structure Laboratory was supported by grants from the National Center for Research Resources (5P20RR017708-10) and the National Institute of General Medical Sciences (8 P20 GM103420-10) from the National Institutes of Health.
Supporting Information Available
Elution volumes of H83A and Y75A from a calibrated size exclusion column (Figure S1), temperature dependence of UV–vis spectra of H83A holo-HasAp (Figure S2), comparison of H83A HasAp mutants (Figure S3), interaction of heme propionates in HasAp mutants (Figure S4), X-ray fluorescence scan of the H83Amono HasAp crystal and phased anomalous difference map using data collected at λ values of 1.0000 and 1.28414 Å with the H83Amono HasAp crystal (Figure S5A,B), elements of secondary structure in wt apo-HasAp (Figure S6), 1H–15N HSQC spectra of apo-HasAp (Figure S7), effect of temperature on selected portions of the HSQC spectra of wt, Y75A, and H83A apo-HasAp (Figure S8), global analysis of stopped-flow data (Figure S9), RR spectra of 6 ms RFQ samples (Figure S10), and backbone resonance assignments for wt apo-HasAp (Table S1), Y75A apo-HasAp (Table S2), and H83A apo-HasAp (Table S3). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This study was supported by National Science Foundation Grants MCB0818488 and MCB1158469 to M.R. and MCB0811888 to P.M.-L. and by a fellowship from the Japan Society for the Promotion of Science to H.M.
Accession Codes
Coordinates and crystallographic structure factors for the distinct HasAp structures have been deposited in the Protein Data Bank as entries 4O6Q (Y75A HasAp), 4O6S (H83Amono HasAp), 4O6U (H83Aortho HasAp), and 4O6T (H83pH5.4). Sequential backbone chemical shift assignments (H, N, and Cα) have been deposited in the BMRB as entries 19720 (wt apo-HasAp), 19721 (Y75A apo-HasAp), and 19722 (H83A apo-HasAp).
Supplementary Material
References
- Nobles C. L.; Maresso A. W. (2011) The theft of host heme by Gram-positive pathogenic bacteria. Metallomics 3, 788–796. [DOI] [PubMed] [Google Scholar]
- Grigg J. C.; Ukpabi G.; Gaudin C. F.; Murphy M. E. (2010) Structural biology of heme binding in the Staphylococcus aureus Isd system. J. Inorg. Biochem. 104, 341–348. [DOI] [PubMed] [Google Scholar]
- Weinberg E. D. (2009) Iron Availability and Infection. Biochim. Biophys. Acta 1790, 600–605. [DOI] [PubMed] [Google Scholar]
- Bullen J. J.; Rogers H. J.; Spalding P. B.; Ward C. G. (2005) Iron and Infection: The Heart of the Matter. FEMS Immunol. Med. Microbiol. 43, 325–330. [DOI] [PubMed] [Google Scholar]
- Otto B. R.; Verweij-van Vught A. M.; MacLaren D. M. (1992) Transferrins and Heme-Compounds as Iron Sources of Pathogenic Bacteria. Crit. Rev. Microbiol. 18, 217–233. [DOI] [PubMed] [Google Scholar]
- Ascenzi P.; Bocedi A.; Visca P.; Altruda F.; Tolosano E.; Beringhelli T.; Fasano M. (2005) Hemoglobin and heme scavenging. IUBMB Life 57, 749–759. [DOI] [PubMed] [Google Scholar]
- Benson D. R.; Rivera M. (2013) Heme Uptake and Metabolism in Bacteria. Met. Ions Life Sci. 12, 279–332. [DOI] [PubMed] [Google Scholar]
- Andrews S. C. (1998) Iron Storage in Bacteria. Adv. Microb. Physiol. 40, 281–351. [DOI] [PubMed] [Google Scholar]
- Tullius M. V.; Harmston C. A.; Owens C. P.; Chim N.; Morse R. P.; McMath L. M.; Iniguez A.; Kimmey J. M.; Sawaya M. R.; Whitelegge J. P.; Horwitz M. A.; Goulding C. W. (2011) Discovery and characterization of a unique mycobacterial heme acquisition system. Proc. Natl. Acad. Sci. U.S.A. 108, 5051–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo J. M.; Letoffe S.; Wandersman C. (1997) A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J. Bacteriol. 179, 3572–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letoffe S.; Ghigo J. M.; Wandersman C. (1994) Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc. Natl. Acad. Sci. U.S.A. 91, 9876–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoux P.; Haser R.; Izadi N.; Lecroisey A.; Delepierre M.; Wandersman C.; Czjzek M. (1999) The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat. Struct. Biol. 6, 516–520. [DOI] [PubMed] [Google Scholar]
- Wolff N.; Izadi-Pruneyre N.; Couprie J.; Habeck M.; Linge J.; Rieping W.; Wandersman C.; Nilges M.; Delepelairre P.; Lecroisey A. (2008) Comparative Analysis of Structural and Dynamic Properties of the Loaded and Unloaded Hemophore HasA: Functional Implications. J. Mol. Biol. 376, 517–525. [DOI] [PubMed] [Google Scholar]
- Jepkorir G.; Rodríguez J. C.; Rui H.; Im W.; Lovell S.; Battaille K. P.; Alontaga A. Y.; Yukl E. T.; Moënne-Loccoz P.; Rivera M. (2010) Structural, NMR Spectroscopic and Computational Investigation of Hemin Loading in the Hemophore HasAp from Pseudomonas aeruginosa. J. Am. Chem. Soc. 132, 9857–9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alontaga A. Y.; Rodríguez J. C.; Schönbrunn E.; Becker A.; Funke T.; Yukl E. T.; Hayashi T.; Stobaugh J.; Moënne-Loccoz P.; Rivera M. (2009) Structural Characterization of the Hemophore HasAp from Pseudomonas aeruginosa: NMR Spectroscopy Reveals Protein-Protein Interactions Between Holo-HasAp and Hemoglobin. Biochemistry 48, 96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R.; Lovell S.; Matsumura H.; Battaile K. P.; Moenne-Loccoz P.; Rivera M. (2013) The Hemophore HasA from Yersinia pestis (HasAyp) Coordinates Hemin with a Single Residue, Tyr75, and with Minimal Conformational Change. Biochemistry 52, 2705–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aki Y.; Nagai M.; Nagai Y.; Imai K.; Aki M.; Sato A.; Kubo M.; Nagatomo S.; Kitagawa T. (2010) Differences in coordination states of substituted tyrosine residues and quaternary structures among hemoglobin M probed by resonance Raman spectroscopy. JBIC, J. Biol. Inorg. Chem. 15, 147–158. [DOI] [PubMed] [Google Scholar]
- Das T. K.; Couture M.; Lee H. C.; Peisach J.; Rousseau D. L.; Wittenberg B. A.; Wittenberg J. B.; Guertin M. (1999) Identification of the ligands to the ferric heme of Chlamydomonas chloroplast hemoglobin: Evidence for ligation of tyrosine-63 (B10) to the heme. Biochemistry 38, 15360–15368. [DOI] [PubMed] [Google Scholar]
- Lecomte J. T.; Smit J. D.; Winterhalter K. H.; La Mar G. N. (1989) Structural and electronic properties of the liver fluke heme cavity by nuclear magnetic resonance and optical spectroscopy. Evidence for a distal tyrosine residue in a normally functioning hemoglobin. J. Mol. Biol. 209, 235–247. [DOI] [PubMed] [Google Scholar]
- Williams P. A.; Fulop V.; Garman E. F.; Saunders N. F.; Ferguson S. J.; Hajdu J. (1997) Haem-ligand switching during catalysis in crystals of a nitrogen-cycle enzyme. Nature 389, 406–412. [DOI] [PubMed] [Google Scholar]
- Jin Y.; Nagai M.; Nagai Y.; Nagatomo S.; Kitagawa T. (2004) Heme structures of five variants of hemoglobin M probed by resonance Raman spectroscopy. Biochemistry 43, 8517–8527. [DOI] [PubMed] [Google Scholar]
- Wardell M.; Wang Z.; Ho J. X.; Robert J.; Ruker F.; Ruble J.; Carter D. C. (2002) The atomic structure of human methemalbumin at 1.9 Å. Biochem. Biophys. Res. Commun. 291, 813–819. [DOI] [PubMed] [Google Scholar]
- Reid T. J. III; Murthy M. R.; Sicignano A.; Tanaka N.; Musick W. D.; Rossmann M. G. (1981) Structure and heme environment of beef liver catalase at 2.5 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 78, 4767–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho W. W.; Li H.; Eakanunkul S.; Tong Y.; Wilks A.; Guo M.; Poulos T. L. (2007) Holo- and apo-bound structures of bacterial periplasmic heme-binding proteins. J. Biol. Chem. 282, 35796–35802. [DOI] [PubMed] [Google Scholar]
- Mattle D.; Zeltina A.; Woo J. S.; Goetz B. A.; Locher K. P. (2010) Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis. J. Mol. Biol. 404, 220–231. [DOI] [PubMed] [Google Scholar]
- Yukl E. T.; Jepkorir G.; Alontaga A.; Pautsch L.; Rodriguez J. C.; Rivera M.; Moënne-Loccoz P. (2010) Kinetic and Spectroscopic Studies of Hemin Acquisition in the Hemophore HasAp from Pseudomonas aeruginosa. Biochemistry 49, 6646–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillet-Saguy C.; Piccioli M.; Turano P.; Lukat-Rodgers G.; Wolff N.; Rodgers K. R.; Izadi-Pruneyre N.; Delepierre M.; Lecroisey A. (2012) Role of the iron axial ligands of heme carrier HasA in heme uptake and release. J. Biol. Chem. 287, 26932–26943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff N.; Deniau C.; Letoffe S.; Simenel C.; Kumar V.; Stojiljkovic I.; Wandersman C.; Delepierre M.; Lecroisey A. (2002) Histidine pKa shifts and changes of tautomeric states induced by the binding of gallium-protoporphyrin IX in the hemophore HasA(SM). Protein Sci. 11, 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillet-Saguy C.; Delepierre M.; Lecroisey A.; Bertini I.; Piccioli M.; Turano P. (2006) Direct-Detected 13C NMR to Investigate the Iron(III) Hemophore HasA. J. Am. Chem. Soc. 128, 150–158. [DOI] [PubMed] [Google Scholar]
- Caillet-Saguy C.; Turano P.; Piccioli M.; Lukat-Rodgers G. S.; Czjzek M.; Guigliarelli B.; Izadi-Pruneyre N.; Rodgers K. R.; Delepierre M.; Lecroisey A. (2008) Deciphering the structural role of histidine 83 for heme binding in hemophore HasA. J. Biol. Chem. 283, 5960–5970. [DOI] [PubMed] [Google Scholar]
- Deniau C.; Gilli R.; Izadi-Pruneyre N.; Lettofe S.; Delepierre M.; Wandersman C.; Briand C.; Lecroisey A. (2003) Thermodynamics of Heme Binding to the HasASM Hemophore: Effect of Mutations at the Three Key Residues for Heme Uptake. Biochemistry 42, 10627–10633. [DOI] [PubMed] [Google Scholar]
- Letoffe S.; Deniau C.; Wolff N.; Dassa E.; Delepelaire P.; Lecroisey A.; Wandersman C. (2001) Haemophore-mediated bacterial haem transport: Evidence for a common or overlapping site for haem-free and haem-loaded haemophore on its specific outer membrane receptor. Mol. Microbiol. 41, 439–450. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (2010) Xds. Acta Crystallogr. D66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D62, 72–82. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1988) Automatic-Indexing of Rotation Diffraction Patterns. J. Appl. Crystallogr. 21, 67–71. [Google Scholar]
- Vagin A.; Teplyakov A. (1997) MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025. [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Lovell S. C.; Davis I. W.; Arendall W. B. III; de Bakker P. I.; Word J. M.; Prisant M. G.; Richardson J. S.; Richardson D. C. (2003) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins 50, 437–450. [DOI] [PubMed] [Google Scholar]
- Potterton L.; McNicholas S.; Krissinel E.; Gruber J.; Cowtan K.; Emsley P.; Murshudov G. N.; Cohen S.; Perrakis A.; Noble M. (2004) Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D60, 2288–2294. [DOI] [PubMed] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu W.; Pfeifer J.; Bax A. (1995) NMRPipe: A Multidimensional Spectral Processing System Based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Goddard T. D., and Kneller D. G. (2008) Sparky 3, University of California, San Francisco. [Google Scholar]
- Wishart D. S.; Bigam C. G.; Yao J.; Abildgaard F.; Dyson H. J.; Oldfield E.; Markley J. L.; Sykes B. D. (1995) 1H, 13C and 15N Chemical Shift Referencing in Biomolecular NMR. J. Biomol. NMR 6, 135–140. [DOI] [PubMed] [Google Scholar]
- Song S. H.; Asher S. A. (1991) Internal intensity standards for heme protein UV resonance Raman studies: Excitation profiles of cacodylic acid and sodium selenate. Biochemistry 30, 1199–1205. [DOI] [PubMed] [Google Scholar]
- Matsumura H.; Moënne-Loccoz P. (2014) Characterizing Millisecond Intermediates in Hemoproteins Using Rapid-Freeze-Quenched Resonance Raman Spectroscopy. Methods Mol. Biol. 1122, 107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Sykes B. D. (1994) The 13C Chemical-Shift Index: A Simple Method for the Identification of Protein Secondary Structure Using 13C Chemical Shift Data. J. Biomol. NMR 4, 171–180. [DOI] [PubMed] [Google Scholar]
- Chuang W. J.; Johnson S.; Van Wart H. E. (1988) Resonance Raman spectra of bovine liver catalase: Enhancement of proximal tyrosinate vibrations. J. Inorg. Biochem. 34, 201–219. [DOI] [PubMed] [Google Scholar]
- Sharma K. D.; Andersson L. A.; Loehr T. M.; Terner J.; Goff H. M. (1989) Comparative spectral analysis of mammalian, fungal, and bacterial catalases. Resonance Raman evidence for iron-tyrosinate coordination. J. Biol. Chem. 264, 12772–12779. [PubMed] [Google Scholar]
- Egeberg K. D.; Springer B. A.; Martinis S. A.; Sligar S. G.; Morikis D.; Champion P. M. (1990) Alteration of sperm whale myoglobin heme axial ligation by site-directed mutagenesis. Biochemistry 29, 9783–9791. [DOI] [PubMed] [Google Scholar]
- Adachi S.; Nagano S.; Ishimori K.; Watanabe Y.; Morishima I.; Egawa T.; Kitagawa T.; Makino R. (1993) Roles of proximal ligand in heme proteins: Replacement of proximal histidine of human myoglobin with cysteine and tyrosine by site-directed mutagenesis as models for P-450, chloroperoxidase, and catalase. Biochemistry 32, 241–252. [DOI] [PubMed] [Google Scholar]
- Das T. K.; Franzen S.; Pond A.; Dawson J. H.; Rousseau D. L. (1999) Formation of a Five-Coordinate Hydroxide-Bound Heme in the His93Gly Mutant of Sperm Whale Myoglobin. Inorg. Chem. 38, 1952–1953. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Moenne-Loccoz P.; Hildebrand D. P.; Wilks A.; Loehr T. M.; Mauk A. G.; Ortiz de Montellano P. R. (1999) Replacement of the proximal histidine iron ligand by a cysteine or tyrosine converts heme oxygenase to an oxidase. Biochemistry 38, 3733–3743. [DOI] [PubMed] [Google Scholar]
- Uchida T.; Stevens J. M.; Daltrop O.; Harvat E. M.; Hong L.; Ferguson S. J.; Kitagawa T. (2004) The interaction of covalently bound heme with the cytochrome c maturation protein CcmE. J. Biol. Chem. 279, 51981–51988. [DOI] [PubMed] [Google Scholar]
- Eakanunkul S.; Lukat-Rodgers G. S.; Sumithran S.; Ghosh A.; Rodgers K. R.; Dawson J. H.; Wilks A. (2005) Characterization of the periplasmic heme-binding protein shut from the heme uptake system of Shigella dysenteriae. Biochemistry 44, 13179–13191. [DOI] [PubMed] [Google Scholar]
- Evans P. R. (2011) An Introduction to Data Reduction: Space-Group Determination, Scaling and Intentisy Statistics. Acta Crystallogr. D67, 282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs K.; Karplus P. A. (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4, 269–275. [DOI] [PubMed] [Google Scholar]
- Weiss M. S. (2001) Global indicators of X-ray data quality. J. Appl. Crystallogr. 34, 130–135. [Google Scholar]
- Krissinel E.; Henrick K. (2004) Secondary Structure Matching (SSM), a New Tool for Fast Protein Structure Alignment in Three Dimensions. Acta Crystallogr. D60, 2256–2268. [DOI] [PubMed] [Google Scholar]
- Winn M. D.; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.