Significance
We propose an innovative method to extract new knowledge from noisy and high-dimensional data. Our approach differs from previous methods in that it has an integrated procedure of validation of the results through maximization of cross-validated accuracy. In many cases, this method performs better than existing feature extraction methods and offers a general framework for analyzing any kind of complex data in a broad range of sciences. Examples ranging from genomics and metabolomics to astronomy and linguistics show the versatility of the method.
Keywords: dissimilarity matrix, mapping, multivariate statistics, clustering, data visualization
Abstract
Here we describe KODAMA (knowledge discovery by accuracy maximization), an unsupervised and semisupervised learning algorithm that performs feature extraction from noisy and high-dimensional data. Unlike other data mining methods, the peculiarity of KODAMA is that it is driven by an integrated procedure of cross-validation of the results. The discovery of a local manifold’s topology is led by a classifier through a Monte Carlo procedure of maximization of cross-validated predictive accuracy. Briefly, our approach differs from previous methods in that it has an integrated procedure of validation of the results. In this way, the method ensures the highest robustness of the obtained solution. This robustness is demonstrated on experimental datasets of gene expression and metabolomics, where KODAMA compares favorably with other existing feature extraction methods. KODAMA is then applied to an astronomical dataset, revealing unexpected features. Interesting and not easily predictable features are also found in the analysis of the State of the Union speeches by American presidents: KODAMA reveals an abrupt linguistic transition sharply separating all post-Reagan from all pre-Reagan speeches. The transition occurs during Reagan’s presidency and not from its beginning.
The last few decades have witnessed an explosion of data in almost all fields, from biology and the health sciences to economics and finance, to the extent that the bottleneck in research has shifted from data generation to data analysis (1).
As a result, there is an increasing need of unsupervised approaches to data mining especially, those geared toward the discovery of patterns in the data for exploratory tasks using clustering and feature extraction methods (2). A problem related to almost all such algorithms is that they do not provide estimates of the significance of the results returned (3). Therefore, the use of data mining methods is an intrinsically risky activity that can easily lead to the discovery of meaningless patterns. The reliability of a clustering solution can be verified a posteriori by evaluating the predictive accuracy of a supervised classifier by repeatedly leaving out one or a few randomly selected samples as a “test set,” whereas the remaining data objects are used as a “training set” (cross-validation) (4–6).
With this in mind we have devised an unsupervised feature extraction method, which we named KODAMA (knowledge discovery by accuracy maximization). KODAMA essentially integrates a validation procedure of the results in the method itself. The core idea is to derive an unsupervised measure of dissimilarity in multivariate data between pairs of samples by using, somewhat counter-intuitively, a supervised classifier followed by a cross-validation step.
An unsupervised classification with high cross-validated accuracy is more likely to contain meaningful information about the structure of data. However, inside a dataset, more than one possible classification may exhibit high cross-validation accuracy. For this reason, in KODAMA the identification of the best classifications is performed through a Monte Carlo (MC) procedure, in such a way as to maximize the cross-validated accuracy by randomly remodeling the classification itself. The cross-validated accuracy can be calculated using any supervised classifier. KODAMA can use several supervised classifiers such as k-nearest neighbors (kNN) (7), support vector machine (SVM) (8), and a combination of principal component analysis (PCA) and canonical analysis (CA) with kNN (PCA-CA-kNN) (9).
A large number of unsupervised feature extraction techniques have been designed, like KODAMA, to preserve the local structure of data. To better assess KODAMA’s performance against this metric, we compared KODAMA with several of these other unsupervised techniques: diffusion maps (DM) (10), isometric feature mapping (ISOMAP) (11), PCA (12), locally linear embedding (LLE) (13), random forest (RF) (14), Sammon’s nonlinear mapping (SAMMON) (15), stochastic proximity embedding (SPE) (16), and t-distributed stochastic neighbor embedding (t-SNE) (17). Despite the strong record of these methods, they are often not very successful when applied to noisy and/or high-dimensional data. Conversely, KODAMA demonstrates a high level of performance as an unsupervised method on datasets with these characteristics, ranging from simulated to a broad spectrum of scientific data. Finally, the greater flexibility of KODAMA is also apparent in a semisupervised context.
Methods
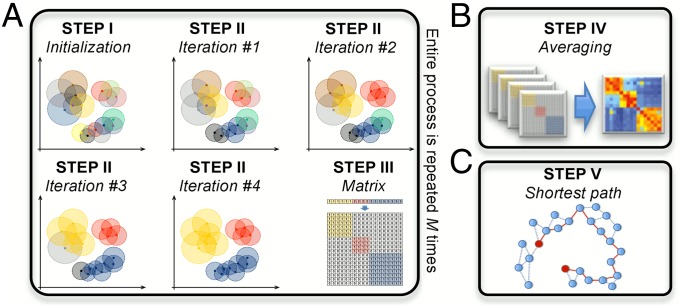
KODAMA consists of five steps, as illustrated in SI Appendix, Fig. S1. For a simple description of the method, we can divide KODAMA into two parts: (i) the maximization of cross-validated accuracy by an iterative process (steps I and II), resulting in the construction of a proximity matrix (step III), and (ii) the definition of a dissimilarity matrix (steps IV and V). The first part entails the core idea of KODAMA, that is, the partitioning of data guided by the maximization of the cross-validated accuracy, as shown in Fig. 1A and in the flowchart in SI Appendix, Fig. S1. At the beginning of this part, a fraction φ of the total samples (φ = 0.75 as default) are randomly selected from the original data. The whole iterative process (steps I–III) is repeated M times (M = 100 as default) to average the effects owing to the randomness of the iterative procedure. Each time that this part is repeated, a different fraction of samples is selected. The second part aims at collecting and processing these results by constructing a dissimilarity matrix to provide a holistic view of the data while maintaining their intrinsic structure (steps IV and V, Fig. 1 B and C). Although the method itself is a complex multistep procedure, to make things easy for the final user the source code of KODAMA written for the R statistical environment is freely available from www.kodama-project.com and a detailed user manual is being made available.
Fig. 1.
(A) The KODAMA accuracy maximization procedure is illustrated for 2D data points, where kNN (with k = 2) classifier was used. Each point is colored according to the cluster it belongs to; the circle represents its distance to the second-nearest neighbor. More details are shown in SI Appendix, Fig. S2. (B) In the fourth step the matrices are averaged. More details are shown in SI Appendix, Fig. S3. (C) In the fifth step the shortest path is calculated.
Maximization of Cross-Validated Accuracy (Steps I and II).
Taking a dataset constituted by N′ = φN samples and f variables, step I consists of the assignment of each sample to a class defined in the class-indicator vector W = {w1,w2,…,wi,…,wN′}, where wi is the class label of the ith sample. If W is not predefined, each sample is assigned to a different class. Therefore, in step I, N′ different classes are created. A 10-fold cross-validation procedure is performed on the basis of the classes defined in W. This procedure is performed using a supervised classifier such as kNN (7), SVM (8), or PCA-CA-kNN (9). Details on the use of these classifiers are given in SI Appendix. Because in this first step removal of samples for cross-validation implies removal of their classes, these samples are forced to join other classes. The global accuracy is calculated by summing up the number of correctly classified samples and dividing this number by the total number of samples. The obtained value is stored in the variable AW. A record of the predicted class labels for each sample is stored in the vector ZW = {z1,z2,…,zi,…,zN′}, where zi is the predicted class label of the ith sample. Obviously, after the first step AW = 0.
In step II, an iterative MC procedure optimizes the vector W by maximizing AW. At the end of these iterations, a classification with a high value of accuracy is stored in the vector W. This procedure includes the following points: (i) A new class-indicator vector V = {v1,v2,…,vi,…,vN′} is created by randomly swapping some class labels of the misclassified samples with the predicted class labels stored in ZW. (ii) A 10-fold cross-validation procedure is performed on the basis of the classes defined in V. The relative accuracy value is then stored in AV and the predicted class labels are stored in ZV = {z1,z2,…,zi,…,zN′}. (iii) If AV is equal to, or higher than, AW, the value of AW is changed to AV, the vector W is changed to V, and the vector ZW is changed to ZV.
This iterative procedure leads to a pruning of the classes because, in some cases, all samples belonging to one class may also be classified as belonging to a different class, and therefore the swapping of vector V will eliminate one or more classes (this is what always happens in step I). The loop is repeated until either AW becomes equal to 100% or the maximum number of iterations T is reached (the default value is T = 20). Fig. 1A illustrates how the classification gradually emerges during this iterative procedure. The evolution of the W and ZW vectors, as well as of AW and AV, is exemplified in SI Appendix, Fig. S2 for 2D data points.
Construction of the Proximity Matrix (Step III).
In step III, the classification obtained from the iterations described in step II is used to generate a proximity matrix P′ = {p′(i,j)} (N′ × N′) from the vector W, where p′(i,j) = 1 if i and j are assigned to the same class (i.e., wi = wj), p′(i,j) = 0 otherwise (i.e., wi ≠ wj).
Steps I–III are repeated M times, each time by randomly selected different N′ = φN subsets from the N samples, to generate M different P′ matrices. Each P′ (N′ × N′) matrix is then padded with zeroes to get a sparse P (N × N) matrix where the zeroes fill the cells of P corresponding to the unselected samples at the beginning of step I.
Definition of the Dissimilarity Matrix (Steps IV and V).
In step IV the M P matrices are then averaged to generate the average proximity matrix PM = {pM(i,j)} (N × N) (Fig. 1B). Each element of PM thus ranges from 0 to 1. More information on this process is provided in SI Appendix, Fig. S3.
High proximities are typical of intracluster relationships, whereas low proximities are expected for intercluster relationships. Very low proximities between samples are ignored by setting pM = 0 for pM < ε, where ε is a predefined cutoff. This ensures that occasional proximities between two otherwise unrelated samples are not taken as meaningful. We set ε = 0.05 as a default value. Then the Euclidean distances d(i,j) are calculated in the f-dimensional space between the N samples. By multiplying 1/pM(i,j) by d(i,j), a weighted dissimilarity matrix Dw = {dw(i,j)} (N × N) is obtained. For all PM elements pM(i,j) equaling 1, the corresponding dw(i,j) elements equal the Euclidean distance. For 0 < pM(i,j) < 1, dw(i,j) represents a distance weighted by the probability that i and j belong to the same class. If pM(i,j) = 0 then dK(i,j) = ∞.
In step V, the final KODAMA dissimilarity matrix DK is calculated by applying Floyd’s algorithm (18) to find the shortest path distances between all pairs of points (Fig. 1C): For each value of h = 1,2,…,N in turn, all entries dK(i,j) are defined as dK(i,j) = min[dw(i,j), dw(i,h) + dw(h,j)]. The final DK = {dK(i,j)} contains the shortest path distances between all pairs of points. This is a way to capture the global topology of the manifold embedded in the data, as reported in a previous study (11).
Initializing and Constraining.
The KODAMA procedure can be started by different initializations of the vector W. Without any a priori information the vector W can be initialized, as described above, with each wi being different from the others (i.e., each sample categorized in a one-element class). Alternatively, the vector W can be initialized by a clustering procedure, such as k-means, k-medoids, or hierarchical clustering. Finally, supervised constraints can be imposed by linking some samples in such a way that if one of them is changed the linked ones must change in the same way (i.e., they are forced to belong to the same class). This will produce solutions where linked samples are forced to have the lowest values in the KODAMA dissimilarity matrix.
Optimization of the Parameters.
As described above, KODAMA contains adjustable parameters (φ, T, M, and ε). These parameters do not show remarkable effects if changed within reasonable ranges. Their proposed default values are shown in SI Appendix, Table S1. We describe their optimization as default values in SI Appendix and SI Appendix, Fig. S4.
The result of KODAMA is also affected by the choice of the classification method to use in the cross-validation procedure. We tested kNN, SVM, and PCA-CA-kNN, the performance of which has been assessed several times in the analysis of multivariate data (9, 19, 20), but any other classifier can be used for the KODAMA analysis. Depending on the structure of the data, one classifier may perform better than others. After extensive tests, we found that the analysis of the distribution of proximity values pM(i,j) can be used to select the best classifier (and its relative parameters). To quantify the information contained in PM and to assess the significance of the KODAMA result on a high-dimensional dataset the Shannon entropy (H) (21) can be used. Details are provided in SI Appendix. The use of H to assess the significance of a KODAMA analysis was tested on three-clusters, Swiss-roll, and two multivariate Gaussian distributions datasets (SI Appendix, Fig. S5). The results are shown in SI Appendix, Table S2.
Visualization of the Data.
Visualization is an important aspect in the analysis of high-dimensional data (22). Feature extraction methods such as multidimensional scaling (MDS) can be used to provide a visual representation of the KODAMA dissimilarity matrix DK by a set of points in a low dimensional space where the distances between the points are approximately equal to the dissimilarities. Alternatively, t-SNE or tree preserving embedding (TPE) (23) can be used on the KODAMA dissimilarity matrix when the intrinsic dimensionality of the data largely exceeds the embedding dimensionality used to visualize the data—the so-called crowding problem (17). Other feature extraction methods applied to KODAMA are described in SI Appendix.
Time Complexity.
KODAMA has polynomial complexity, proportional to the product of the number of cross-validations performed by the time complexity of the classifier used. A 10-fold cross-validation performed with kNN classifier has thus a time complexity of O(0.9 × N′2 × f), where N′ is the overall number of data points (N′ = φN). KODAMA consequently has a time complexity at most of O(0.9 × M × T × N′2 × f), where M is the number of times that the maximization of the cross-validated accuracy is repeated, and T is the maximum number of MC iterations. Some optimizations can be used to improve the efficiency of the algorithm, for instance, using KD-Tree (24) to improve the storage efficiency or approximate nearest neighbor searching (25) to improve the speed at the cost of slightly lower accuracy. Efficient parallel formulation of the kNN search problem based on graphics processing units are proposed (26), observing speed-ups of 50–60 times compared with central processing unit implementation. Some tests of KODAMA performed with kNN, SVM, or PCA-CA-kNN are provided in SI Appendix both for synthetic (Tables S3 and S4) and experimental (Table S5) datasets. The running times are significantly longer than those of other unsupervised methods, but still acceptable. The performance of KODAMA is relatively insensitive to the number of variables but decreases substantially with increasing number of samples, although not as substantially as for other methods.
Results
Comparative Tests of KODAMA on Synthetic Datasets.
Manifolds.
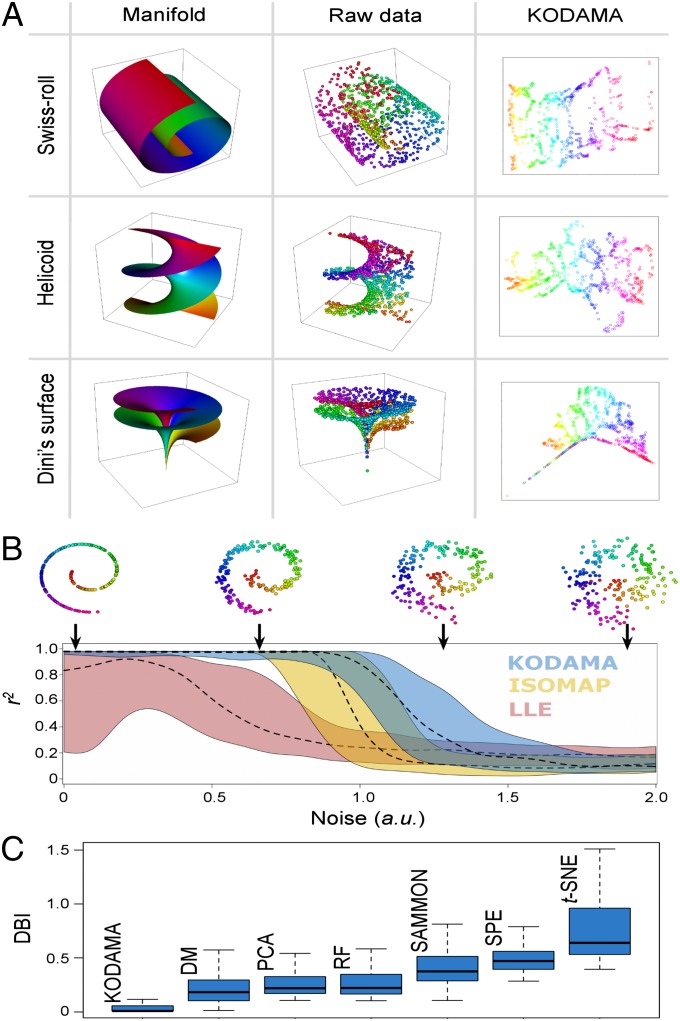
To visually and intuitively demonstrate the features of KODAMA, we tested its performance in representing 2D manifolds embedded in a 3D space. Fig. 2A illustrates three examples. All of them are intrinsically 2D datasets and can thus be projected onto a plane. The first is the well-known Swiss-roll, similar to the one used in ref. 11. The second is a minimal surface example discovered by Baptiste Meusnier in 1776, the so-called helicoid. Its name derives from its similarity to a helix: For every point on the helicoid there is a helix contained in the helicoid, which passes through that point. The third one is a surface (described by Ulisse Dini in 1866) with constant negative curvature that can be created by twisting a pseudosphere. H is calculated to test whether or not KODAMA is able to catch the internal structure of these highly nonlinear datasets. The calculated H values are 11.39, 10.94, and 10.92 for the Swiss-roll, the helicoid, and Dini’s surface, respectively. These values can be compared with those obtained on three sets of 100 random datasets (average H values of 13.80, 11.68, and 11.65, respectively). Statistically significant results (P < 0.01) were thus obtained for all three manifolds tested, demonstrating that the KODAMA proximities contain structured information (SI Appendix, Table S2).
Fig. 2.
The problem of nonlinear dimensionality reduction, as illustrated for 3D data sampled from 2D manifolds. The color coding reveals how the data are embedded in two dimensions. (A) From left to right, columns correspond to manifold structures, data points distribution, and the KODAMA map, respectively. KODAMA was performed using kNN as classifier and MDS was used to visualize the results. (B) Performance of achieving a low-dimensional representation from a manifold embedded in high-dimensional space as a function of the noise in the spiral datasets. KODAMA (in blue), ISOMAP (in yellow), and LLE (in red) were applied to spiral datasets. The coefficient of determination, r2, was used to evaluate the performance of each method. A higher r2 means that the low-dimensional embedding provides an accurate description of the original data. The dashed lines represent the median of r2 obtained with KODAMA, ISOMAP, and LLE from 100 datasets with different noise values. The solid lines indicate the lower hinge and the upper hinge. KODAMA was performed using kNN as classifier and MDS to process the KODAMA dissimilarity matrix. (C) Box-and-whiskers plot of DBI values obtained from the Gaussian datasets. For each dataset, the classifier of KODAMA was selected by minimizing the H value.
Among all tested methods, only KODAMA, ISOMAP, and LLE had the capacity to compute a 2D neighborhood preserving embeddings of the data as shown in Fig. 2A and SI Appendix, Fig. S6. LLE produced an incorrect solution when applied to the Dini’s surface.
Nonlinear datasets.
In a second series of experiments, KODAMA, LLE, and ISOMAP were applied to spiral datasets with 21 different degrees of noise. For each degree of noise, we created 100 different datasets. To assess the methods’ performance in achieving a low-dimensional representation from a noisy manifold embedded in high-dimensional space, we calculated the coefficient of determination, r2, between the first component of each method and the distribution of each point in the spiral. A high r2 means that the low-dimensional embedding provides an accurate description of the original data. LLE and ISOMAP clearly suffer from problems relative to the “short-circuits” in the neighborhood graph. Short-circuits can lead to low-dimensional embeddings that do not preserve a manifold’s true topology (27). Thus, LLE and ISOMAP performed poorly compared with KODAMA, as shown in Fig. 2B, whereas the other methods (i.e., DM, PCA, RF, SAMMON, SPE, and t-SNE) do not show the capability to compute a monodimensional neighborhood preserving embeddings of the data. Tests on noisy Swiss-roll and helicoid datasets were also performed and are described in SI Appendix, Fig. S7. KODAMA performs better than other methods on the Swiss-roll dataset, and it is comparable to ISOMAP on the helicoid dataset.
Gaussian datasets.
Most dimensionality reduction methods fail to preserve clusters (28). In multiclass data, ISOMAP and LLE cannot lead to successful embedding owing to unconnected subgraphs. These methods fail if data lie on disconnected manifolds. For further comparison, 100 datasets were generated with three clusters and dimensionalities ranging between 2 and 100. The number of data points for each cluster ranged between 50 and 200. Each cluster was created from a different multivariate normal distribution with a different covariance matrix of variables (29). Each covariance matrix was randomly generated with values that ranged between 0 and 1. The performance of each feature extraction method was analyzed by estimating the relative cluster overlap using the Davies–Bouldin index (DBI) (30), a function of the ratio of the sum of within-cluster scatter to between-cluster separation. Small values of DBI correspond to clusters that are compact and whose centers are far away from each other. The lowest value indicates the best solution. KODAMA achieved the best results compared with other methods, as shown in the box-and-whiskers plot in Fig. 2C.
The results of KODAMA, DM, PCA, RF, SAMMON, SPE, and t-SNE on datasets with different degrees of separation between clusters are also compared. DBI was used to quantify the degree of separation between the original clusters, and the performances of the methods were evaluated by the DBI of the outputs. KODAMA showed the lowest DBI independently of the DBI (i.e., cluster separation) of the original data. The obtained DBI values for the various methods are reported in SI Appendix, Fig. S8 as a function of the DBI values of the raw data. Moreover, we also show the results of KODAMA when applied to datasets with a continuous distribution of data points (i.e., single multivariate Gaussian distribution, test-1 and test-2 in SI Appendix, Fig. S5). In these tests, KODAMA correctly showed nonstatistically significant results.
Frequently, missing values occur in real-life experiments. KODAMA can handle missing data: A detailed procedure is provided in SI Appendix. SI Appendix, Fig. S9 shows that KODAMA has intermediate performance, behaving somewhat less well with respect to ISOMAP, PCA, and RF, comparably with SAMMON and SPE, and significantly better than LLE, DM, and t-SNE.
Comparative Tests of KODAMA on Experimental Datasets.
Lymphoma dataset.
KODAMA was tested on an experimental dataset (31) that is a popular benchmark for statistical analysis programs. This dataset consists of gene expression profiles of the three most prevalent adult lymphoid malignancies: diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), and B-cell chronic lymphocytic leukemia (B-CLL). The source study produced gene expression data for f = 4,682 genes in n = 62 mRNA samples: 42 samples of DLBCL, 9 samples of FL, and 11 samples of B-CLL. In the present work, it is assumed that the lymphoma data are unsupervised (i.e., the number of classes and the class of each sample are not given a priori). We imputed missing values and standardized the data as described in ref. 32.
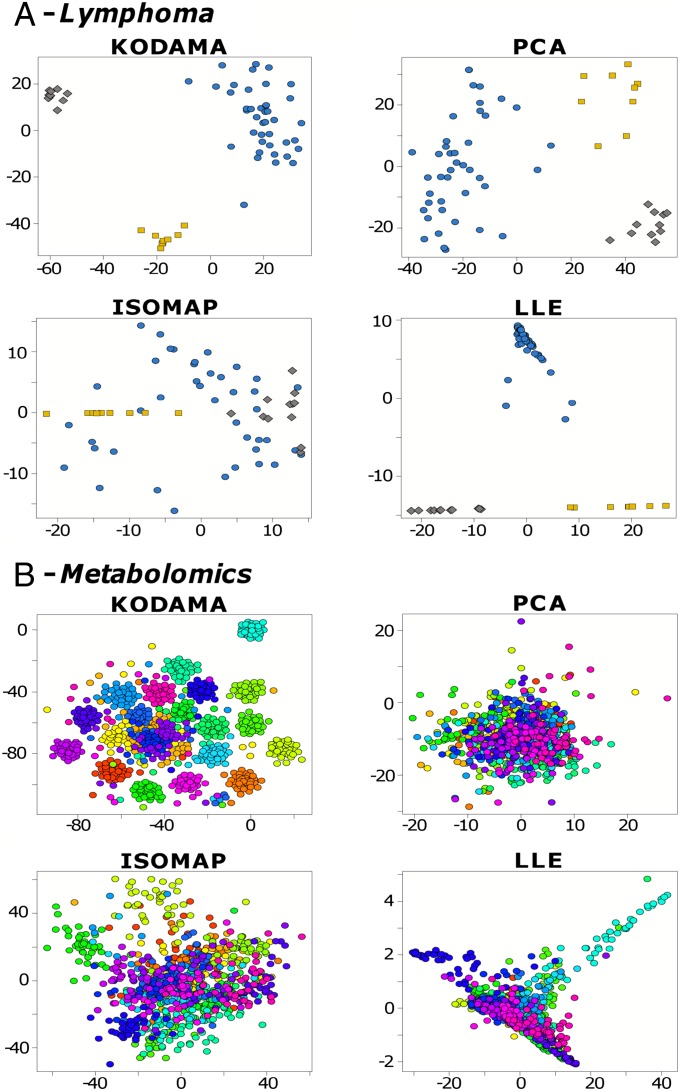
KODAMA performed with kNN identified three classes. A separation between DLBCL and FL/B-CLL is clearly apparent in the first two components, whereas FL and B-CLL can be distinguished in the third component. From this unsupervised KODAMA analysis, we may conclude that the lymphoma data consist primarily of two classes (DLBCL and FL/B-CLL) and that FL and B-CLL are secondary classes, confirming the results obtained in a previous study (5). KODAMA performed with SVM shows a clear separation of the three different malignancies, as does LLE and at variance with PCA and ISOMAP (Fig. 3A). The other methods are shown in SI Appendix, Fig. S10. The DBIs are reported in Fig. 4 and in SI Appendix, Table S6. Whereas KODAMA with either kNN or SVM achieved statistically significant results (P < 0.01) and comparable values of H (respectively 8.05 and 8.09), KODAMA with PCA-CA-kNN showed a higher and not statistically significant H value of 8.25 (P = 0.35), compared with an averaged H of 8.26 obtained on 100 random datasets. In terms of accuracy with respect to the biological classification, LLE and KODAMA with SVM yield no misclassifications, and KODAMA with kNN yields one misclassified sample. PCA, ISOMAP, and the other methods tested (SI Appendix, Fig. S10) perform less well.
Fig. 3.
(A) Lymphoma dataset. Blue circle, DLBCL; yellow square, FL; and gray diamond, B-CLL. Comparison between KODAMA, PCA, ISOMAP, and LLE. MDS was used to visualize the results of KODAMA dissimilarity matrix. The results of the other methods are shown in SI Appendix, Fig. S10. (B) Metabolomic dataset. Comparison between KODAMA, PCA, ISOMAP, and LLE. Color coding indicates samples from the same donor. The results of the other methods are shown in SI Appendix, Fig. S10. PCA-CA-kNN classifier for KODAMA was selected by minimizing the H value. TPE was used to visualize the results of KODAMA dissimilarity matrix.
Fig. 4.

Bar plot of DBI values obtained with KODAMA, PCA, ISOMAP, and LLE on the lymphoma, metabolomic, and ETGs datasets.
Metabolomic dataset.
The global analysis of metabolites in biological fluids, tissues, or related biological samples is a promising area of research, owing to its potential relevance for human health. To examine KODAMA in this context, we address the task of clustering a dataset of NMR spectra of urines (9). The data belong to a cohort of 22 healthy donors (11 male and 11 female) where each provided about 40 urine samples over the time course of approximately 2 mo, for a total of n = 873 samples and f = 416 variables (9). KODAMA was performed with kNN, SVM, and PCA-CA-kNN. Moreover, KODAMA was initialized with different class-indicator vectors W. We obtained the lowest value of H in KODAMA with PCA-CA-kNN and W initialized by k-means. KODAMA was then compared with classical and state-of-the-art methods. Comparisons between KODAMA, PCA, ISOMAP, and LLE are shown in Fig. 3B and the solutions of the other methods are shown in SI Appendix, Fig. S10. The results are summarized in SI Appendix, Table S6. Moreover, a comparison of the different visualization methods is shown in SI Appendix, Fig. S11. Clearly, KODAMA performs better than all other methods also in this case.
The ability of feature extraction methods to highlight local structures permits their use for clustering purposes. When we applied k-medoids (with k = 22) to the KODAMA dissimilarity matrix DK, we obtained only 10.7% of misclusterized samples in the urine dataset. The performances of the different unsupervised clustering methods were compared with the adjusted Rand index (ARI) (SI Appendix) (33), a function that measures similarity between two classifications. ARI spans from −1 to 1; perfect agreement is scored 1, whereas 0 corresponds to a random partition. Negative values indicate less agreement than expected by chance. In all cases the presence of 22 clusters was imposed. The ARI value for KODAMA was 0.769. The other methods tested (34–39) (SI Appendix) provided ARI values ranging from 0.439 to 0.212 (SI Appendix, Table S7).
We also applied KODAMA in a semisupervised context, by providing the information regarding sample groupings from each individual in such a way that the spectra belonging to each individual were forced to maintain the same classification. No information about sex was provided. We performed a PCA-CA analysis as described in ref. 9: The data were projected into their PCA subspace representing 90.0% of the variance, and the resulting PCA scores were projected into the 2D CA subspace. SI Appendix, Fig. S12 shows the result obtained by PCA-CA and KODAMA. We observed that in terms of showing a clear sex separation the semisupervised KODAMA performed better than PCA-CA.
Knowledge Discovery by KODAMA.
Early-type galaxies datasets.
We next explored two early-type galaxies (ETGs) datasets, available from the ATLAS3D project (40). The ATLAS3D project combines a multiwavelength survey of a complete set of n = 260 ETGs. Various parameters are collected, such as the largest equivalent aperture radius (Rmax), the moment ellipticity measured within one effective radius Re and one-half effective radius Re/2 (respectively εe and εe/2), and other photometric and integral-field spectroscopic parameter (i.e., V/σe, V/σe/2, λRe, and λRe/2 indexes). A dataset contains the Rmax, εe, V/σe, and λRe parameters; the other dataset contains Rmax, εe/2, V/σe/2, and λRe/2. Recent work by Emsellem et al. (41) shows how these parameters can be used to define a refined and optimized criterion for disentangling the so-called fast rotators (FRs) and slow rotators (SRs). SI Appendix, Figs. S10 and S13 show the comparison between KODAMA and other unsupervised feature extraction methods. In both ETGs datasets, KODAMA achieved excellent results, comparable only to those of ISOMAP (Fig. 4 and SI Appendix, Table S6). KODAMA correctly highlighted the differences between FRs and SRs, further suggesting that the SRs may be part of a larger well-defined cluster, with some exceptions falling outside the boundaries.
State of the Union dataset.
Sparse data, in which each individual record contains values only for a small fraction of attributes, present a challenge for data mining methods. An interesting case study is offered by the annual addresses presented by the presidents of the United States to the Congress (the “State of the Union” speech), which are available from The American Presidency Project repository. The State of the Union speeches have been the subject of numerous linguistic analyses (ref. 42 and references therein). We selected only the spoken, not written, addresses from 1900 until the sixth address by Barack Obama in 2014. Punctuation characters, numbers, words shorter than three characters, and stop-words (e.g., “that,” “and,” and “which”) were removed from the dataset. This resulted in a dataset of n = 86 speeches containing f = 834 different meaningful words each. Term frequency-inverse document frequency (TF-IDF) (43) was used to get the feature vectors for the unsupervised analysis. It is often used as a weighting factor in information retrieval and text mining. The TF-IDF value increases proportionally to the number of times a word appears in the document, but is offset by the frequency of the word in the corpus, which helps to control for the fact that some words are generally more common than others.
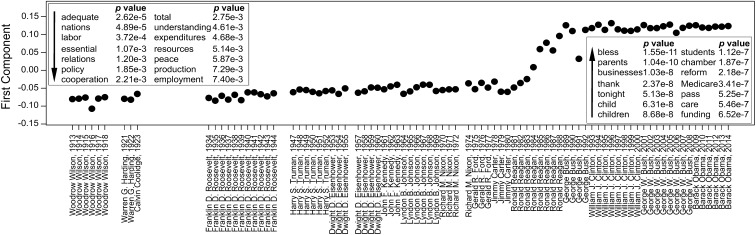
KODAMA was performed with kNN, SVM, and PCA-CA-kNN. We found the lowest value of H in KODAMA with kNN. The first component of MDS applied to the KODAMA dissimilarity matrix (Fig. 5) shows a single, clear, and abrupt transition over a period of more than 100 y, which occurred during the presidency of Ronald Reagan. The pre-Reagan and post-Reagan speeches are recognized with 100% cross-validated accuracy using the kNN classifier. Words such as “labor,” “expenditures,” “employment,” “relations,” “resources,” and “production” suddenly decrease in frequency, whereas words such as (parents,” “students,” “pass,” “children,” “Medicare,” and “reform” suddenly increase in frequency (Fig. 5, Insets). It can be noted that the third address of G. Bush (January 29, 1991) is somewhat different from the other Bush’s addresses and marks a partial reversal to the pre-Reagan style. Interestingly, this speech was held in the middle of Operation Desert Storm (January 17, 1991–February 28, 1991) and probably reflects the emotional atmosphere of the nation.
Fig. 5.
First component of MDS applied to the KODAMA dissimilarity matrix obtained from the selected addresses of American presidents, in chronological order. The Insets show the words that have most significantly decreased (left) or increased (right) their frequencies from before to after Reagan’s presidency, and the associated Bonferroni corrected P values. kNN classifier for KODAMA was selected by minimizing the H value.
It is noteworthy that if KODAMA is performed in a semisupervised way, by providing information grouping together the speeches of each president, Reagan seems to represent a “hinge” between past and present rhetorical modes. The performances of the other feature extraction methods are lower when compared on the basis of the pre- and post-Reagan discrimination (SI Appendix, Table S6). The results of each method are shown in SI Appendix, Fig. S14. Interestingly, no distinction between Republicans and Democrats is evident. Although it is widely accepted that Reagan’s rhetoric was unique and that it influenced all subsequent speeches—in the words of Obama, Ronald Reagan “changed the trajectory of America in a way that, you know, Richard Nixon did not and in a way that Bill Clinton did not” (44)—our analysis clearly points to a sharp change of Reagan’s rhetoric during his presidency, and more precisely toward the end of his first mandate.
Discussion
Some limitations of conventional algorithms, such as PCA (12), stem from the fact that they use imposed distance measures defined in a globally linear space or with limited degrees of freedom. Linear methods are usually not appropriate to modeling curved manifolds, because they focus on preserving the distances between widely separated data points rather than on preserving the distances between nearby data points. Nonlinear dimensionality reduction methods (11, 13) are capable of discovering nonlinear degrees of freedom (11) but are negatively affected by increasing dimensionality of the embedded manifold (the so-called curse of dimensionality) (45) and by the problem of short-circuit edges in the presence of noisy or sparse data (27).
To illustrate another type of drawback, t-SNE reduces the dimensionality of data in a manner dependent on the local properties of that data. This makes t-SNE likewise sensitive to the curse of dimensionality of the data (17). Manifold learners such as ISOMAP and LLE suffer from precisely the same problem (17). Even a single short-circuit error (27) can alter many entries into the neighborhood graph, which in turn can lead to a drastically different and incorrect low-dimensional embedding.
We thus propose KODAMA as a method of performing feature extraction on noisy and high-dimensional data. Thus, we have demonstrated its performance on real datasets chosen for their different structures and properties. Our approach differs from previous methods in that it is based on an integrated procedure of validation of the results through an embedded MC procedure that maximizes cross-validated accuracy. Overall, KODAMA outperformed the existing feature extraction methods that we tested. KODAMA offers a general framework for analyzing any kind of complex data in a broad range of sciences. It also makes it possible to perform analyses in unsupervised or semisupervised contexts.
Finally, its ability to resolve meaningful clusters within the data makes the KODAMA dissimilarity matrix useful in conjunction with classical clustering algorithms (e.g., k-medoids), because it strongly improves their performances.
Supplementary Material
Acknowledgments
We thank S. Tyekucheva, A. K. Smilde, E. Saccenti, C. Sander, and M. A. Andrade-Navarro for support and several critical discussions. We also thank E. Emsellem for expressing interest in our analysis of the early-type galaxies datasets, and the anonymous reviewer of the earlier versions of this manuscript for the many constructive criticisms and suggestions that allowed us to significantly improve the paper. This work was partly supported by the European Commission-funded FP7 projects COSMOS (312941), WeNMR (261572), CHANCE (266331), BioMedBridges (284209), and by the Programmi di Ricerca Nazionale (2009FAKHZT_001). S.C. was supported by a fellowship from Fondazione Italiana per la Ricerca sul Cancro.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220873111/-/DCSupplemental.
References
- 1.Schadt EE, Linderman MD, Sorenson J, Lee L, Nolan GP. Computational solutions to large-scale data management and analysis. Nat Rev Genet. 2010;11(9):647–657. doi: 10.1038/nrg2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 3.Handl J, Knowles J, Kell DB. Computational cluster validation in post-genomic data analysis. Bioinformatics. 2005;21(15):3201–3212. doi: 10.1093/bioinformatics/bti517. [DOI] [PubMed] [Google Scholar]
- 4.Jiang D, Tang C, Zhang A. Cluster analysis for gene expression data: A survey. IEEE Trans Knowl Data Eng. 2004;16(11):1370–1386. [Google Scholar]
- 5.Kim C, Cheon M, Kang M, Chang I. A simple and exact Laplacian clustering of complex networking phenomena: Application to gene expression profiles. Proc Natl Acad Sci USA. 2008;105(11):4083–4087. doi: 10.1073/pnas.0708598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golub TR, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 7.Cover TM, Hart PE. Nearest neighbor pattern classification. IEEE Trans Inf Theory. 1967;13(1):21–27. [Google Scholar]
- 8.Vapnik VN. The Nature of Statistical Learning Theory. New York: Springer; 1995. p. 188. [Google Scholar]
- 9.Assfalg M, et al. Evidence of different metabolic phenotypes in humans. Proc Natl Acad Sci USA. 2008;105(5):1420–1424. doi: 10.1073/pnas.0705685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coifman RR, et al. Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc Natl Acad Sci USA. 2005;102(21):7426–7431. doi: 10.1073/pnas.0500334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenenbaum JB, de Silva V, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290(5500):2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 12.Ringnér M. What is principal component analysis? Nat Biotechnol. 2008;26(3):303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 13.Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290(5500):2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 14.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 15.Sammon JWJ. A nonlinear mapping for data structure analysis. IEEE Trans Comput. 1969;C-18(5):401–409. [Google Scholar]
- 16.Agrafiotis DK. Stochastic proximity embedding. J Comput Chem. 2003;24(10):1215–1221. doi: 10.1002/jcc.10234. [DOI] [PubMed] [Google Scholar]
- 17.van der Maaten LJP, Hinton GE. Visualizing high-dimensional data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 18.Floyd RW. Algorithm 97: Shortest path. Commun ACM. 1962;5(6):345. [Google Scholar]
- 19.Bertini I, et al. Metabolomic NMR fingerprinting to identify and predict survival of patients with metastatic colorectal cancer. Cancer Res. 2012;72(1):356–364. doi: 10.1158/0008-5472.CAN-11-1543. [DOI] [PubMed] [Google Scholar]
- 20.Wei X, Li KC. Exploring the within- and between-class correlation distributions for tumor classification. Proc Natl Acad Sci USA. 2010;107(15):6737–6742. doi: 10.1073/pnas.0910140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27(3):379–423. [Google Scholar]
- 22.Shiffrin RM, Börner K. Mapping knowledge domains. Proc Natl Acad Sci USA. 2004;101(Suppl 1):5183–5185. doi: 10.1073/pnas.0307852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shieh AD, Hashimoto TB, Airoldi EM. Tree preserving embedding. Proc Natl Acad Sci USA. 2011;108(41):16916–16921. doi: 10.1073/pnas.1018393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman JH, Bentley JL, Raphael AF. An algorithm for finding best matches in logarithmic expected time. ACM Trans Math Softw. 1977;3(3):209–226. [Google Scholar]
- 25.Arya S, Mount DM, Netanyahu NS, Silverman R, Wu A. An optimal algorithm for approximate nearest neighbor searching. J ACM. 1998;45(6):891–923. [Google Scholar]
- 26.Arefin AS, Riveros C, Berretta R, Moscato P. GPU-FS-kNN: A software tool for fast and scalable kNN computation using GPUs. PLoS ONE. 2012;7(8):e44000. doi: 10.1371/journal.pone.0044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balasubramanian M, Schwartz EL. The isomap algorithm and topological stability. Science. 2002;295(5552):7. doi: 10.1126/science.295.5552.7a. [DOI] [PubMed] [Google Scholar]
- 28.Venna J, Peltronen J, Nybo K, Aidos H, Samuel K. Information retrieval perspective to nonlinear dimensionality reduction for data visualization. J Mach Learn Res. 2010;11:451–490. [Google Scholar]
- 29.Ripley BD. Stochastic Simulation. New York: Wiley; 1987. p. 98. [Google Scholar]
- 30.Davies DL, Bouldin DW. A cluster separation measure. IEEE Trans Pattern Anal Mach Intell. 1979;1(2):224–227. [PubMed] [Google Scholar]
- 31.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 32.Dudoit S, Fridlyand J, Speed TP. Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc. 2002;97(417):77–87. [Google Scholar]
- 33.Hurbet L, Arabie P. Comparing partitions. J Classif. 1985;2(1):193–218. [Google Scholar]
- 34.Bouveyron C, Girard S, Schmid C. High-dimensional data clustering. Comput Stat Data Anal. 2006;52(1):502–519. [Google Scholar]
- 35.Ng AY, Jordan MI, Weiss Y. On spectral clustering: Analysis and an algorithm. Adv Neural Inf Process Syst. 2001;14:849–856. [Google Scholar]
- 36.Shy J, Malik J. Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell. 2000;22(8):888–905. [Google Scholar]
- 37.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley; 1990. p. xiv, 342. [Google Scholar]
- 39.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315(5814):972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 40.Cappellari M, et al. The Atlas3D project – I. A volume-limited sample of 260 nearby early-type galaxies: Science goals and selection criteria. Mon Not R Astron Soc. 2011;413(2):813–836. [Google Scholar]
- 41.Emsellem E, et al. The ATLAS3D project – III. A census of the stellar angular momentum within the effective radius of early-type galaxies: Unveiling the distribution of Fast and Slow Rotators. Mon Not R Astron Soc. 2011;414(2):888–912. [Google Scholar]
- 42.Schonhardt-Bailey C, Yager E, Lahlou S. Yes, Ronald Reagan’s rhetoric was unique–but statistically, how unique? Pres Stud Q. 2012;42(3):482–513. [Google Scholar]
- 43.Wu HC, Luk RWP, Wong KF, Kwok KL. Interpreting TF-IDF term weights as making relevance decisions. ACM Trans Inf Syst. 2008;26(3):1–37. [Google Scholar]
- 44.Halperin M. 2010. What Obama can learn from Reagan. Time Magazine. Available at http://content.time.com/time/magazine/article/0,9171,1957469,00.html. Accessed March 7, 2014. [Google Scholar]
- 45.van der Maaten L, Postma E, van den Herik J. Dimensionality reduction: A comparative review. J Mach Learn Res. 2009;10:1–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.