Significance
Recent studies have shown that actin-binding and -regulating proteins, originally characterized in the context of cytoskeletal events, can also modify gene expression through directly impacting actin-dependent transcription. This study shows α-catenin (α-cat), an actin-binding protein that is essential for cell–cell adhesion and contact-dependent growth inhibition, can antagonize Wnt/β-catenin–mediated transcription and impact nuclear actin properties, suggesting that these events may be related. These findings establish α-cat as one of a growing list of actin-binding proteins that can modulate transcription, possibly by controlling actin dynamics in the nucleus.
Keywords: alpha-catenin, beta-catenin
Abstract
α-Catenin (α-cat) is an actin-binding protein required for cell–cell cohesion. Although this adhesive function for α-cat is well appreciated, cells contain a substantial amount of nonjunctional α-cat that may be used for other functions. We show that α-cat is a nuclear protein that can interact with β-catenin (β-cat) and T-cell factor (TCF) and that the nuclear accumulation of α-cat depends on β-cat. Using overexpression, knockdown, and chromatin immunoprecipitation approaches, we show that α-cat attenuates Wnt/β-cat–responsive genes in a manner that is downstream of β-cat/TCF loading on promoters. Both β-cat– and actin-binding domains of α-cat are required to inhibit Wnt signaling. A nuclear-targeted form of α-cat induces the formation of nuclear filamentous actin, whereas cells lacking α-cat show altered nuclear actin properties. Formation of nuclear actin filaments correlates with reduced RNA synthesis and altered chromatin organization. Conversely, nuclear extracts made from cells lacking α-cat show enhanced general transcription in vitro, an activity that can be partially rescued by restoring the C-terminal actin-binding region of α-cat. These data demonstrate that α-cat may limit gene expression by affecting nuclear actin organization.
An emerging theme in adhesion signaling is that proteins integral to the structure of adherent-type junctions and focal adhesions are also found localized to the nucleus (1). β-Catenin (β-cat) is the prototypical example of such dual-function adhesion signaling proteins. At the cell surface, β-cat links the cytoplasmic domain of cadherin-type adhesion receptors to the actin-binding protein, α-catenin (α-cat), effectively coordinating the cortical actin networks of adjacent cells. In the nucleus, β-cat interacts with TCF-type transcription factors and promotes transcription at Wnt-regulated genes by recruiting chromatin-remodeling proteins (2). Similarly, p120 catenin binds and stabilizes cadherins at the cell surface, but also localizes to the nucleus to displace the repressor protein, Kaiso, resulting in gene activation (3). Although α-cat has been found to localize to the nucleus (4, 5), contain nuclear export sequences (6), and inhibit Wnt signaling (5, 7–9), the mechanism by which α-cat attenuates transcription has remained unclear.
Roles for α-cat are best understood at cell junctions, where it is essential for cell cohesion and tissue organization (10–12). As a homodimer, α-cat directly interacts with filamentous (F) actin (13) but α-cat can also indirectly associate with the cytoskeleton through other actin-binding proteins, such as epithelial protein lost in neoplasm (EPLIN) (14), vinculin (15), afadin (16), α-actinin (17), and zonula occludens-1 (ZO-1) (18). In addition, α-cat can impact F-actin remodeling by directly inhibiting Arp2/3-mediated actin polymerization in vitro (13), lamellipodial dynamics in cells (19), and by promoting F-actin bundling in vitro (20).
A substantial body of evidence indicates that nuclear actin coordinates gene expression. Nuclear β-actin incorporates into all three RNA polymerase complexes and is required for transcription (21–24). Although cytosolic actin exists in both G- and F-actin conformations, F actin is rarely detected in the nucleus. However, nuclear actin filaments (NAFs) are detected in various pathological conditions (25, 26), and fluorescence recovery after photobleaching (FRAP) studies of GFP actin have identified a slow recovering form of presumably polymeric actin as well as a dynamic pool of monomeric actin in the nucleus (27). Polymeric actin appears to mediate RNA synthesis because depolymerization of actin inhibits RNA polymerase (Pol) I-mediated transcription (28) and prevents reactivation of the pluripotency gene Oct4 in Xenopus oocytes (29). Based on the role of nuclear actin in coordinating gene expression, we predicted that the actin-binding function of α-cat might be conserved in the nucleus and used to modulate transcription.
Results
Nuclear Accumulation of α-Cat Depends on β-Cat.
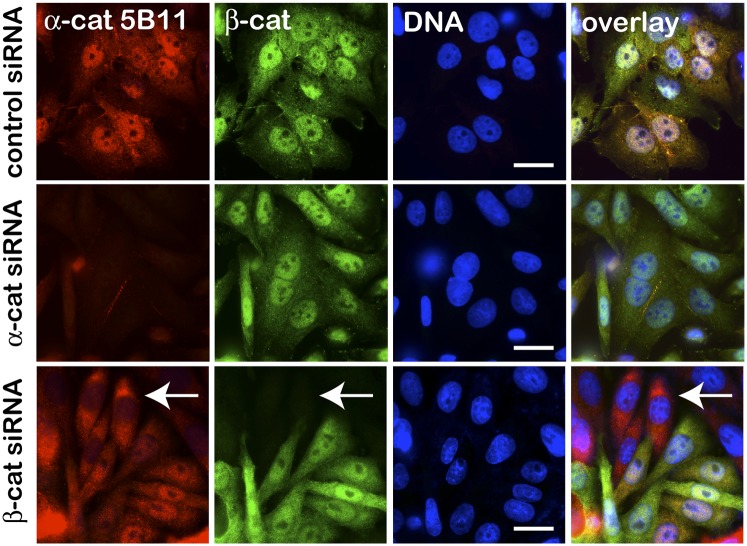
In normal epithelia, α-cat is enriched at sites of cell–cell contact, but roughly a third of total α-cat is cytosolic by fractionation methods (19), where it typically appears excluded from nuclei (SI Appendix, Fig. S1). In colon cancer and cell lines from patients with familial adenomatous polyposis, which manifest robust nuclear β-cat, α-cat has been reported to colocalize with nuclear β-cat (4, 5). We confirm this nuclear localization in two well-characterized colon cancer cell lines (SW480 and DLD1) by both immunofluorescence and nuclear fractionation methods (Fig. 1 and SI Appendix, Fig. S2). Nuclear α-cat staining is specific, as the signal is lost after siRNA-mediated knockdown of α-cat (Fig. 1). Nuclear α-cat also depends on β-cat, as siRNA-mediated knockdown of β-cat promotes α-cat redistribution to the cytoplasm. Conversely, forced expression of β-cat up-regulates and promotes the nuclear accumulation of α-cat (see Fig. 6C), and induction of cells with Wnt3a enhances α-cat nuclear accumulation via its β-cat–binding domain (SI Appendix, Fig. S3). Altogether, these data demonstrate that α-cat localizes to cell nuclei in a manner that depends on β-cat, raising that possibility that α-cat regulates an aspect of Wnt/β-cat signaling.
Fig. 1.
α-Cat nucleoplasmic localization depends on β-cat. Epifluorescence images of SW480 cells transfected with control, α-cat, or β-cat siRNAs stained with antibodies to α-cat (5B11 hybridoma) and β-cat or DNA (Hoechst). The arrow denotes β-cat–silenced cells where α-cat is redistributed to the cytoplasm. (Scale bars: 10 μm.)
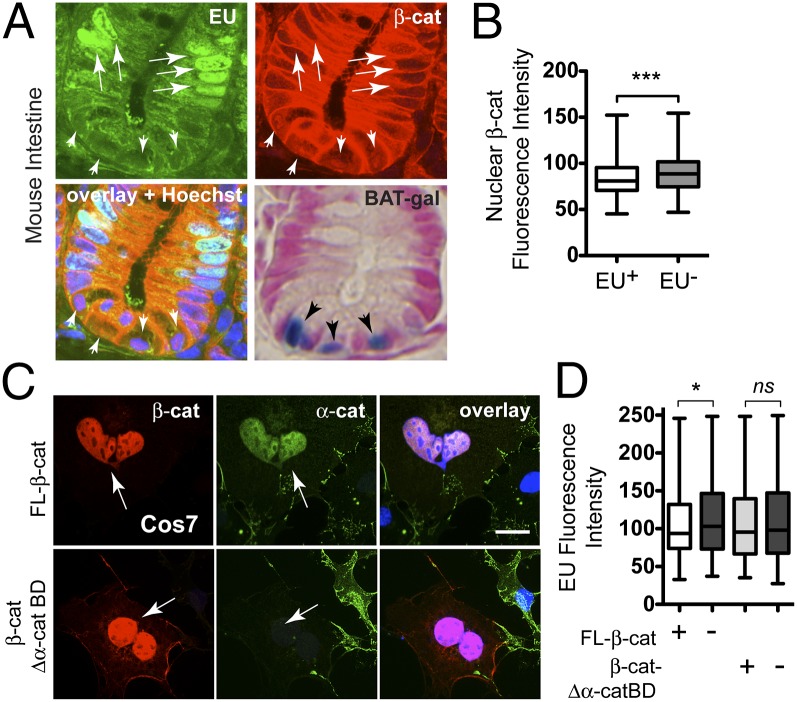
Fig. 6.
β-Cat attenuates transcription by its α-cat–binding domain. (A) Intestinal crypt image from a EU-labeled mouse (2 mg; 4 h) and RNA detection (green) by alkyne–azide click chemistry (SI Appendix, Materials and Methods) and double labeled with an antibody to β-cat (red). Nuclear β-cat+ stem cells at crypt base (arrowheads) incorporate less EU than nearby transit amplifying cells (arrows). X-gal staining (blue) of crypts from β-cat/TCF reporter mouse (BAT-gal; Jackson Labs) shows peak reporter activity in basal (stem) cells. Images taken with 63× objective. (B) Quantification of nuclear β-cat mean fluorescence intensity in EU+ and EU− cells (n = 276; ***P < 0.002). (C) Image of COS7 cells transfected with full-length β-cat– and β-cat-Δα-cat–binding domain and labeled with antibodies to β-cat (red), endogenous α-cat (green), and DNA (blue). (D) EU labeling of cells transfected with β-cat or β-cat lacking the α-cat–binding domain (β-cat-Δα-cat BD). Mean fluorescence intensities from 200 transfected (+) and adjacent untransfected (−) cells were quantified in MetaMorph (*P < 0.05 by t test; see also SI Appendix, Fig. S15). ns, not significant. Arrows point to transfected cells; error bars represent SEM. (Scale bar: 10 μm.)
α-Cat Inhibits Wnt/β-Cat–Dependent Transcription.
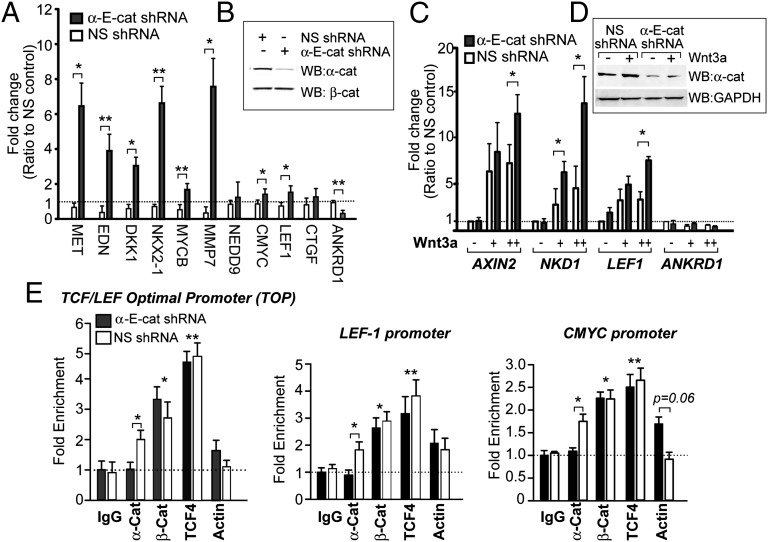
As previously observed (5, 7, 8), we find that overexpression of α-cat in HEK293T cells inhibits Wnt/β-cat signaling using the β-cat/TCF–optimal promoter luciferase reporter (TOPflash) (SI Appendix, Fig. S4). Quantitative real-time PCR (qPCR) also shows that α-cat reduces Wnt-mediated expression of C-MYC, a well-defined β-cat/TCF target gene. Conversely, stable knockdown of α-cat in SW480 cells reveals up-regulation of a number of established Wnt/β-cat responsive genes (Fig. 2 A and B). Importantly, α-cat knockdown in normal human skin fibroblasts activated with recombinant Wnt3a shows elevated expression of genes considered more selective to Wnt activation, such as AXIN2, NKD1, and LEF1 (Fig. 2 C and D). Despite evidence that α-cat inhibits the Hippo pathway effector, Yap (30, 31), we find no evidence for up-regulation of its targets, CTGF and ANKRD1 (Fig. 2 A and C) (32). Together these data show that α-cat limits the expression of a number of Wnt/β-cat–responsive genes across three different cell types.
Fig. 2.
α-Cat inhibits β-cat signaling. (A) qPCR of β-cat target genes from SW480 cells stably transfected with nonspecific (NS) and α-E-cat shRNAs and (B) the corresponding immunoblot. (C) qPCR of β-cat target genes from human dermal fibroblasts stably transfected with NS and α-E-cat shRNAs and incubated ±Wnt3a. Genes in A and C are normalized to 18S and fold induction was calculated with the ΔΔCt method from three independent experiments. (D) Corresponding immunoblot. (E) ChIP analysis of α-cat, β-cat, TCF4, and actin enrichment at three established β-cat/TCF–responsive promoters in nonsilenced and α-cat–silenced SW480 cells: integrated 4xTOP promoter, LEF1, and C-MYC. Asterisks denote significance by standard t test (*P < 0.05 and **P < 0.01). Those without brackets correspond to both silenced and nonsilenced cells compared with the IgG control. Error bars represent SEM for 2–3 independent experiments.
α-Cat Interacts with β-Cat/TCF but Does Not Affect Promoter Loading.
A previous study found that α-cat can coprecipitate with β-cat and TCF, but could not interact with a TCF promoter probe by mobility shift assay (5), suggesting that α-cat may be unable to exist in a complex with β-cat/TCF on DNA. We revisited this question using chromatin immunoprecipitation (ChIP). We find that α-cat, β-cat, and TCF can be significantly enriched at both endogenous (C-MYC and LEF-1) and integrated 4xTOP promoters (Fig. 2E). α-Cat also coprecipitates β-cat and TCF from un–cross-linked nuclear extracts (SI Appendix, Fig. S5). Of interest, α-cat silencing does not impact β-cat/TCF loading to these three promoters. Together with evidence that α-cat binds β-cat in a region that would not compete with β-cat/TCF binding (33), these data suggest that α-cat can localize to β-cat/TCF–occupied promoters and attenuate transcription in a manner that is downstream of β-cat/TCF–promoter binding.
Inhibition of Wnt-Dependent Transcription by α-Cat Requires Binding to β-Cat.
To determine if Wnt pathway regulation by α-cat depends on binding to β-cat, we generated dimerization mutants of both α- and β-cat and expressed them in HEK293T cells. Deletion of the first 81 aa of α-cat (Δβ BD–α-cat) is sufficient to reduce binding to β-cat, and an internal deletion of residues 118–146 from β-cat (Δα BD-β-cat) effectively reduces binding to α-cat (SI Appendix, Fig. S6). Whereas full-length α-cat inhibits Wnt-mediated induction of TOPflash, the α-cat mutant that cannot bind to β-cat has no effect. Likewise, β-cat stimulated TOPflash activity can be inhibited by full-length α-cat, but not the α-cat mutant that cannot bind to β-cat.
Wnt Inhibition by α-Cat Requires the C-Terminal Actin-Binding Domains.
To understand how α-cat inhibits β-cat signaling, we designed a panel of truncation mutants that progressively removes sequences from the C-terminal, F actin-binding region of α-cat (residues 697–906) (15, 34, 35), as well as the middle (M) domain (residues 377–633) (36), which interacts with a number of actin-binding proteins (17, 34, 37, 38). When analyzed in the TOPflash assay, constructs containing residues 279–500 and 864–906 showed the strongest inhibitory activity of α-cat (SI Appendix, Fig. S7), suggesting that the actin-binding functions of α-cat may be critical for transcription inhibition. These mutants localize to nuclei similarly by fractionation (SI Appendix, Fig. S8). A previous study indicated that an N-terminal fragment of α-cat containing the β-cat–binding domain (residues 46–149) was sufficient to inhibit Wnt signaling (8). However, we find that N-terminal fragments of α-cat encoding either β-cat heterodimerization (residues 1–163) or α-cat homodimerization domains (residues 82–279) enhance Wnt signaling, and their coexpression activates the reporter additively (SI Appendix, Fig. S7 C–E). Moreover, coexpression of the N-terminal fragments with full-length α-cat also prevents inhibition mediated by the full-length construct (SI Appendix, Fig. S7F), suggesting that α-cat amino acids 1–279 dominantly displace endogenous α-cat. Altogether, these data show that the entire full-length α-cat protein participates in Wnt-signaling inhibition, as both N-terminal dimerization and C-terminal actin-binding regions are required.
α-Cat Impacts Nuclear Actin.
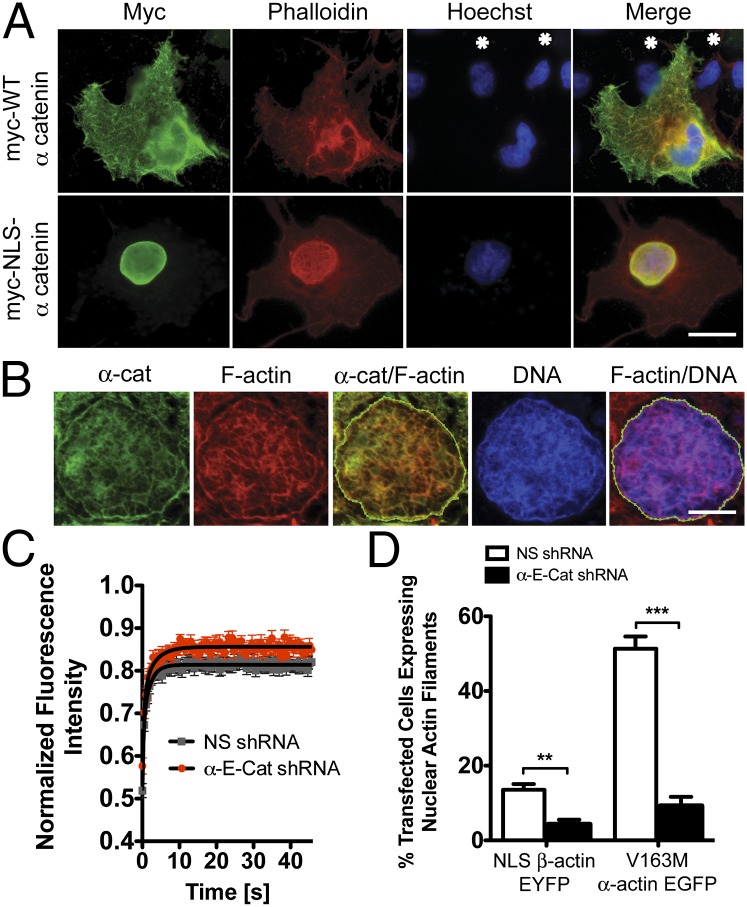
Because nuclear localized actin and actin-binding proteins such as myosin I (39), N-WASP (40), Arp2/3 (41), cofilin (42), profilin (43), afadin (44), and α-actinin (25) have been documented, it is possible that the actin-binding/-bundling function of α-cat is conserved in the nucleus. To test this prediction, COS7 cells were transfected with either wild-type (WT) or nuclear localization signal (NLS)-tagged α-cat and subsequently stained with phalloidin to identify F-actin structures. Overexpression of WT α-cat enhances cytoplasmic F-actin staining compared with adjacent untransfected cells (Fig. 3A). Remarkably, expression of NLS-α-cat leads to the formation of nuclear F-actin filaments in ∼25% of expressing cells. The F actin-binding region of α-cat is required and sufficient for nuclear F-actin staining (SI Appendix, Figs. S9 and S10). Confocal imaging and correlation analysis reveal that NLS-α-cat partially colocalizes with nuclear F actin (Fig. 3B and Movie S1). Importantly, no difference in actin abundance was observed in nuclear fractions of WT and NLS-α-cat–transfected cells (SI Appendix, Fig. S11), suggesting that α-cat may promote NAFs from a preexisting polymeric pool. Confocal microscopy showed that formation of nuclear actin bundles appears to affect DNA organization, as revealed by Hoechst staining intensity (Fig. 3B and Movie S1), suggesting that induced nuclear actin structures can impact chromatin organization. Importantly, SW480 cells lacking α-cat show small but significant differences in nuclear actin. α-Cat knockdown cells have a higher mobile fraction of NLS-actin-YFP assessed by FRAP (Fig. 3C) and the pool of nuclear actin is more soluble by ultracentrifugation (SI Appendix, Fig. S12). α-Cat knockdown cells are also less prone to forming NAFs using both a pathogenic V163M skeletal muscle α-actin mutation that causes intrauclear rod myopathy (25) or by overexpression of NLS β-actin YFP (Fig. 3D and SI Appendix, Fig. S12). Thus, α-cat has consequences for nuclear actin using both gain- and loss-of-function approaches.
Fig. 3.
α-Cat affects nuclear actin. (A) Epifluorescence images of COS7 cells transfected with myc-WT or NLS-tagged α-cat and stained using an myc antibody (green), phalloidin (red), and Hoescht (blue). (Scale bar: 20 μm.) Asterisks denote untransfected cells. (B) Confocal microscopy of the nucleus from the cell shown in E and stained as in A. (Scale bar: 5 μm.) Correlation analysis for myc and phalloidin (R = 0.73, overlap coefficient = 1.0) and phalloidin and Hoechst (R = 0.43, overlap coefficient of 0.9). (C) FRAP analysis of control (NS) and α-cat–silenced cells transiently transfected with YFP-NLS-β-actin. Mobile fraction = 81.41% ± 0.15 for control versus 85.62% ± 0.24 in α-cat knockdown cells (P < 0.001 by t test). (D) Graph of the percentage of control (NS) and α-cat knockdown cells exhibiting NAFs upon transfection with NLS-β-actin-EYFP or V163M α-actin-EGFP. ***P < 0.001, **P < 0.01. Error bars represent SEM of duplicate transfections.
α-Cat Inhibits RNA Synthesis in Vivo and in Vitro.
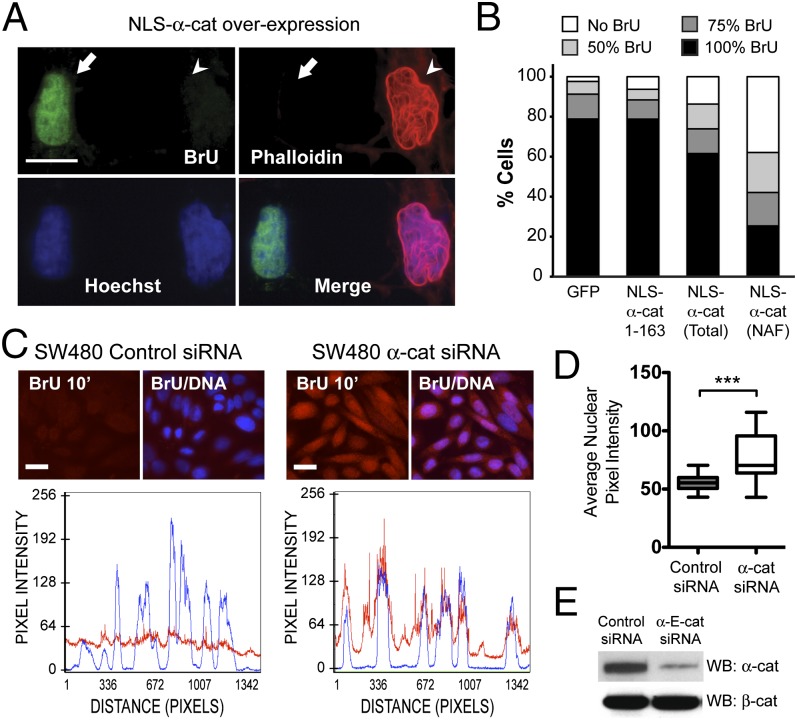
Nuclear actin is required for RNA Pol I-, II-, and III-mediated transcription (21–24). To determine whether α-cat–induced changes in nuclear actin impact transcription, we investigated the effect of expressing NLS-tagged α-cat in COS7 cells on bromouridine (BrU) incorporation into newly synthesized RNA. Cells expressing phalloidin-positive NAFs exhibited a pronounced reduction in BrU signal intensity compared with cells expressing GFP or a myc-tagged NLS-α-cat fragment (Fig. 4 A and B and SI Appendix, Fig. S13). To test the contribution of α-cat to RNA synthesis using a loss-of-function approach, we quantified BrU incorporation in SW480 cells where α-cat protein levels were silenced using siRNAs. We found that the fluorescence intensity of α-cat knockdown cells was ∼40% greater than that of control knockdown cells (Fig. 4 C–E). Together, these data indicate that α-cat can also limit RNA expression generally.
Fig. 4.
α-Cat inhibits RNA synthesis. (A) Epifluorescence image of COS7 cells transfected with mycNLS-WT-α-cat, incubated with BrU, and stained as indicated. Arrow indicates a cell without nuclear F actin that is BrU+; arrowhead indicates a cell with nuclear F actin that is BrU−. (Scale bar: 20 μm.) (B) Average BrU fluorescence intensities were quantified using Zeiss Axioplan software and binned according to percent staining intensity (N = 300 cells). Intensities were quantified from all cells expressing mycNLS-α-cat 1–163 and mycNLS-WT-α-cat (total) and cells containing NAFs. The percentage of cells with no BrU labeling is significantly different between NLS-α-cat 1–163 and NLS-WTα-cat (total; P < 0.05) and NAFs (P < 0.001) by t test. (C) Image of SW480 cells transfected with control or α-cat siRNAs, incubated with BrU for 10 min, and stained for RNA (red) and DNA (blue). Line scans of BrU and DNA intensities are shown below. (Scale bar: 10 μm.) (D) BrU fluorescence intensities were quantified in MetaMorph (Molecular Devices). Error bars represent SEM of duplicate transfections. N = 250 cells. ***P < 0.001 by t test. (E) Immunoblot of cells in C.
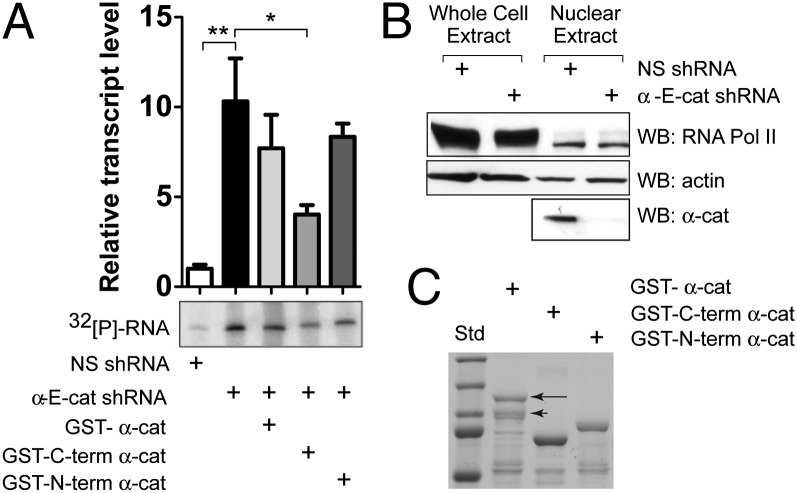
To ask whether α-cat is a proximal inhibitor of transcription, we examined the effects of α-cat on RNA Pol II activity in a standard in vitro transcription assay that uses the adenoviral major late promoter. As this promoter lacks classic TCF-recognition sites, this template allows one to quantify transcription generally. Using nuclear extracts made from SW480 cells where α-cat is stably knocked down by two independent shRNAs, we find that α-cat silencing enhances transcript production by ∼10-fold (Fig. 5 and SI Appendix, Fig. S14). Evidence that WT levels of transcription activity can be partially restored by adding back the C-terminal actin-binding region of α-cat suggests that α-cat inhibits transcription directly, rather than indirectly through its known effects on cell shape and cohesion. Given evidence that actin is important for RNA Pol II activities in vitro (21–24), and can be recruited to activated promoters under stimulated conditions (21, 45), we asked whether α-cat can impact nuclear actin in the vicinity of β-cat/TCF promoters. Of interest, α-cat silencing leads to a nearly significant increase in actin recruitment to the C-MYC promoter (Fig. 2E), suggesting that α-cat binding to β-cat/TCF may limit the recruitment of actin to some promoters.
Fig. 5.
α-Cat C-terminal actin-binding domain is sufficient to inhibit transcription in an in vitro assay. (A) Graph of 32[P]-labeled RNA transcripts from an adenovirus major late promoter cassette incubated with nuclear extracts from NS hairpin and α-cat–silenced SW480 cells. Exposure from one experiment is shown. Error bars represent SD of three independent experiments. (**P < 0.01, *P < 0.05.) Identical findings are observed in cells silenced with a second α-cat hairpin (SI Appendix, Fig. S14). (B) Immunoblot of SW480 whole-cell (45 μg) and nuclear (15 μg) extracts. (C) Coomassie-stained gel of 100 pmoles of recombinant GST-α-cat proteins added to the assay. Arrow indicates full-length α-cat. Arrowhead shows N-terminally truncated α-cat.
β-Cat Attenuates Transcription via Its α-Cat–Binding Domain.
To reconcile how α-cat impacts both β-cat/TCF–dependent and general transcription, we asked whether transcription rates are regulated in a tissue with established Wnt signaling relationships. We injected mice with a single bolus of a modified nucleotide 5′ ethnyl uridine (EU) and followed bulk RNA synthesis by immunofluorescence detection. We confirm that the rates of general transcription are quite different along the intestinal crypt (Fig. 6A) (46). Specifically, the transit-amplifying cells incorporate the most EU, whereas the stem cell compartment (marked by nuclear β-cat, or X-gal staining in the β-cat/TCF reporter mouse, BAT-gal) shows substantially less incorporation. Of interest, fluorescence intensity of nuclear β-cat is significantly reduced in EU+ versus EU− cells (Fig. 6B), suggesting that general transcription may be antagonized by β-cat signaling. To test this, we investigated EU uptake by transfected COS7 cells. Although transcription rates of these cells are quite variable, mean EU fluorescence intensity is significantly reduced upon full-length β-cat transfection (Fig. 6 C and D). Importantly, when we express a form of β-cat that lacks the α-cat–binding domain (and which fails to up-regulate and recruit α-cat to the nucleus; Fig. 6C), no significant difference in EU incorporation is observed between transfected and adjacent untransfected cells (Fig. 6D). Together, these data suggest that β-cat signaling may also attenuate general transcription, and that α-cat binding to β-cat is required for this inhibition.
Discussion
α-Cat is an F actin-binding protein and an essential component of cadherin-based cell–cell adhesions (47). Curiously, cells contain a substantial amount of nonjunctional α-cat, whose functions are just emerging. Studies show that this pool of α-cat can limit the actin-polymerizing activity of the Arp2/3 complex in vitro (13), and lamellipodial dynamics in vivo (19). Given that a cadherin-free form of β-cat is the major stoichiometric binding partner of cytosolic α-cat (48), we sought to understand whether α-cat is a regulator of nuclear β-cat. We show that α-cat can indeed localize to the nucleus and to β-cat/TCF–occupied promoters, where its nucleoplasmic localization depends on β-cat. α-Cat overexpression, silencing, and ChIP experiments show that α-cat attenuates Wnt/β-cat–responsive genes in a manner that is downstream of β-cat/TCF loading on promoters. These data indicate that β-cat, in addition to recruiting coactivators of transcription (2), recruits α-cat as a negative regulator of gene expression. We provide evidence that inhibition of Wnt/β-cat–mediated transcription by α-cat requires not only its β-cat dimerization domain but also its actin-binding domains, and that α-cat impacts the organization of nuclear actin. Evidence that α-cat silencing is associated with actin recruitment to the C-MYC promoter supports the possibility that the actin-binding function of α-cat may be conserved in the nucleus to regulate transcription.
Our study builds on previous studies showing that α-cat overexpression can attenuate β-cat/TCF transcriptional activity in vivo (9) and in vitro (5, 7, 8). Using a α-cat knockdown approach, we show that a number of established Wnt/β-cat–responsive genes are elevated in cells with reduced α-cat protein, indicating that α-cat normally serves to limit the expression of Wnt targets. Although Lien et al. (10) found that targeted loss of α-E-cat in the developing brain did not impact β-cat signaling in the TOP-gal reporter mouse or by following canonical targets AXIN2, C-MYC, and CYCLIND1, factors may contribute to this discrepancy. For instance, presence of the neural isoform of α-N-cat could compensate for the loss of α-E-cat; and AXIN2, C-MYC, and CYCLIND1 can receive Wnt/β-cat–independent inputs (49, 50). Alternatively, expression differences may be underrepresented in a tissue where only a subset of cells receive Wnt signals. In contrast, our study assessed the contribution of α-cat to β-cat target gene expression in a cancer cell line where β-cat signaling is uniformly activated (and α-cat is robustly nuclear), thus favoring detection of changes in gene expression.
Our domain mapping analysis reveals that each of the known functional domains in α-cat contributes to β-cat/TCF reporter inhibition, from the N-terminal β-cat–binding and α-cat homodimerization domains to the M- and C-terminal F actin-binding domains. Of interest, the M domain is known to bind to a number of actin-binding proteins, including vinculin (51), α-actinin (38), afadin (34), and formin-1 (17), which cooperate with α-cat to bind actin at the cell membrane (52, 53). Most of these proteins can localize to the nucleus or its substructures (25, 54–56) and one variant can promote nuclear receptor transcription through the coactivator GRIP1 (57). Therefore, it is possible that these actin-binding proteins cooperate with α-cat to attenuate β-cat–dependent transcription.
Whereas our data show that α-cat inhibits Wnt/β-cat–dependent gene expression with apparent specificity (i.e., α-cat reduces gene/reporter expression relative to an abundant housekeeping gene/reporter control), BrU-labeling studies reveal that α-cat also inhibits transcription more generally, which we reason may be mediated through an ability of α-cat to affect nuclear actin. First, α-cat knockdown significantly increases the rate of transcription in vivo and in vitro, and this effect can be partially reversed by adding back the actin-binding domain of α-cat. Second, NLS-tagged α-cat can promote formation of NAFs that are strongly associated with cells that cannot incorporate BrU. Third, the C-MYC promoter in α-cat–silenced cells shows enhanced recruitment of actin compared with nonsilenced cells. Lastly, we find an increase in the mobile fraction of nuclear actin in α-cat knockdown cells, as well as greater solubility in nuclear actin and a reduced capacity to form NAFs upon overexpression of a pathogenic actin mutant. Together, these data are consistent with a model where α-cat binding to β-cat/TCF–occupied promoters antagonizes transcriptional activity by limiting the recruitment of actin (SI Appendix, Fig. S16).
It was also interesting to find that bulk transcription rates along intestinal crypts appear highest outside of the nuclear β-cat+ stem cell region, and that β-cat expression itself can limit general transcription in a manner that requires its α-cat–binding domain. Together, these data support a model where α-cat binding to β-cat is likely required for both Wnt-specific and general effects on transcription. How can we reconcile a role for α-cat in both Wnt/β-cat–dependent and general transcription? One way is to view the role for α-cat in transcription through the lens of α-cat functions at cell contacts, which can be divided into cadherin/β-cat–associated and –free pools. Evidence indicates that the cadherin/β-cat/α-cat complex can directly link to the F-actin cytoskeleton (58), whereas a cadherin-free pool of α-cat limits the activity of Arp2/3 (19). Together, these complementary activities promote cell cohesion. Perhaps analogously, nuclear α-cat is present in both β-cat/TCF–bound and –unbound forms (e.g., monomer, homodimer, and α-/β-cat heterodimer), which contribute distinct but complementary activities to attenuate transcription. For example, promoter-localized α-cat may inhibit Wnt target genes, whereas nucleoplasmic α-cat may antagonize transcription more generally via nuclear actin (SI Appendix, Fig. S16). Evidence that Xenopus organizer genes are poised by β-cat/TCF, but not transcriptionally active until the midblastula transition (59), may present a system in which to address such distinct nuclear functions of α-cat.
Evidence that the C-terminal actin-binding domain of α-cat is required and sufficient to promote nuclear staining by phalloidin may allow inferences as to the normal structure of nuclear actin. Although the nucleus contains all components necessary to drive actin polymerization in vitro (27), nuclear actin appears to be largely monomeric, where an association with the actin-depolymerizing protein, cofilin, likely prevents the formation of NAFs (42). Because α-cat promotes the formation of F-actin bundles in vitro (20) but has not been shown to promote actin polymerization on its own, it is likely that NLS-α-cat promotes phalloidin-stainable F actin through direct association with preexisting polymeric actin.
How might α-cat limit transcription via nuclear actin? Substantial evidence indicates that nuclear actin participates in most aspects of gene expression from chromatin remodeling to basic RNA polymerase activity to mRNA processing (60–62). RNA Pol II has recently been shown to associate with nuclear-localized actin-polymerizing and -branching proteins, Arp2/3 and N-WASP (40, 41), an association required for RNA synthesis. Others have shown that α-cat reduces Arp2/3-mediated actin polymerization in vitro by directly competing Arp3 binding to actin (13). In the cell, this inhibition correlates with a transition from actin polymerization in lamellipodia to actin bundling during cell contact maturation (19). It is possible that nuclear α-cat inhibits RNA Pol II-associated Arp2/3 by competing for access to nuclear actin directly. Alternatively, α-cat–induced nuclear actin filamentation might deplete actin from RNA Pol II, and thereby attenuate transcription (SI Appendix, Fig. S16).
In summary, we demonstrate that α-cat limits expression of both β-cat/TCF–dependent and –independent promoters in a manner that depends on both dimerization and actin-binding regions of α-cat. Future studies will be needed to understand how endogenous levels of α-cat affect nuclear actin organization, and how this organization impacts transcription.
Materials and Methods
Cell lines were obtained from American Type Culture Collection. Lentiviral knock-down (shRNA) plasmids are provided by Open Biosystems (Waltham, MA). All other reagents, transcription assays, fractionation and imaging methods are detailed in SI Appendix.
Note Added in Proof.
While this paper was under review, Choi et al. (63) also reported that α-cat inhibits Wnt/β-cat signaling, apparently through a distinct mechanism that involves binding to the adenomatous polyposis coli tumor suppressor protein.
Supplementary Material
Acknowledgments
We thank D. Rimm (Yale University) for the original human α-cat cDNA; Arnie Levine (Institute for Advanced Study, Princeton University) for hybridomas to α-cat; Stacy Hyunji Ryu and Terrance Barrett (Northwestern Division of Gastroenterology) for the BAT-gal mouse image; and the Cell Imaging, Recombinant Protein Production and DNA/RNA Delivery Core of the Skin Diseases Research Center and Skin Diseases Research Cores (Northwestern University). This work was supported by National Institutes of Health GM076561 (to C.J.G.), GM80587 (to P.d.), the Northwestern University Physical Sciences Oncology Center associated with National Cancer Institute U54CA143869 (to C.J.G, P.d., and S.T.K.), American Heart Association (AHA) 13PRE17050060 (to L.S.), and AHA 09PRE2060342 and Public Health Service T32-HL076139 (to R.L.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308663111/-/DCSupplemental.
References
- 1.Benmerah A, Scott M, Poupon V, Marullo S. Nuclear functions for plasma membrane-associated proteins? Traffic. 2003;4(8):503–511. doi: 10.1034/j.1600-0854.2003.00102.x. [DOI] [PubMed] [Google Scholar]
- 2.Willert K, Jones KA. Wnt signaling: Is the party in the nucleus? Genes Dev. 2006;20(11):1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 3.Park JI, et al. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell. 2005;8(6):843–854. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 4.El-Bahrawy M, Talbot I, Poulsom R, Alison M. Variable nuclear localization of alpha-catenin in colorectal carcinoma. Lab Invest. 2002;82(9):1167–1174. doi: 10.1097/01.lab.0000028821.41246.6a. [DOI] [PubMed] [Google Scholar]
- 5.Giannini AL, Vivanco Md, Kypta RM. Alpha-catenin inhibits beta-catenin signaling by preventing formation of a beta-catenin*T-cell factor*DNA complex. J Biol Chem. 2000;275(29):21883–21888. doi: 10.1074/jbc.M001929200. [DOI] [PubMed] [Google Scholar]
- 6.Giannini A, et al. Nuclear export of alpha-catenin: Overlap between nuclear export signal sequences and the beta-catenin binding site. Exp Cell Res. 2004;295(1):150–160. doi: 10.1016/j.yexcr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Hwang SG, et al. Regulation of beta-catenin signaling and maintenance of chondrocyte differentiation by ubiquitin-independent proteasomal degradation of alpha-catenin. J Biol Chem. 2005;280(13):12758–12765. doi: 10.1074/jbc.M413367200. [DOI] [PubMed] [Google Scholar]
- 8.Merdek KD, Nguyen NT, Toksoz D. Distinct activities of the alpha-catenin family, alpha-catulin and alpha-catenin, on beta-catenin-mediated signaling. Mol Cell Biol. 2004;24(6):2410–2422. doi: 10.1128/MCB.24.6.2410-2422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sehgal RN, Gumbiner BM, Reichardt LF. Antagonism of cell adhesion by an alpha-catenin mutant, and of the Wnt-signaling pathway by alpha-catenin in Xenopus embryos. J Cell Biol. 1997;139(4):1033–1046. doi: 10.1083/jcb.139.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lien WH, Klezovitch O, Fernandez TE, Delrow J, Vasioukhin V. alphaE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science. 2006;311(5767):1609–1612. doi: 10.1126/science.1121449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stocker AM, Chenn A. Focal reduction of alphaE-catenin causes premature differentiation and reduction of beta-catenin signaling during cortical development. Dev Biol. 2009;328(1):66–77. doi: 10.1016/j.ydbio.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 2001;104(4):605–617. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- 13.Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123(5):903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abe K, Takeichi M. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc Natl Acad Sci USA. 2008;105(1):13–19. doi: 10.1073/pnas.0710504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss EE, Kroemker M, Rüdiger AH, Jockusch BM, Rüdiger M. Vinculin is part of the cadherin-catenin junctional complex: Complex formation between alpha-catenin and vinculin. J Cell Biol. 1998;141(3):755–764. doi: 10.1083/jcb.141.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tachibana K, et al. Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J Cell Biol. 2000;150(5):1161–1176. doi: 10.1083/jcb.150.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobielak A, Pasolli HA, Fuchs E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat Cell Biol. 2004;6(1):21–30. doi: 10.1038/ncb1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh M, Nagafuchi A, Moroi S, Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to alpha catenin and actin filaments. J Cell Biol. 1997;138(1):181–192. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamin JM, et al. AlphaE-catenin regulates actin dynamics independently of cadherin-mediated cell-cell adhesion. J Cell Biol. 2010;189(2):339–352. doi: 10.1083/jcb.200910041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci USA. 1995;92(19):8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann WA, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6(11):1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 22.Hu P, Wu S, Hernandez N. A role for beta-actin in RNA polymerase III transcription. Genes Dev. 2004;18(24):3010–3015. doi: 10.1101/gad.1250804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol. 2005;12(3):238–244. doi: 10.1038/nsmb904. [DOI] [PubMed] [Google Scholar]
- 24.Philimonenko VV, et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol. 2004;6(12):1165–1172. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 25.Domazetovska A, et al. Mechanisms underlying intranuclear rod formation. Brain. 2007;130(Pt 12):3275–3284. doi: 10.1093/brain/awm247. [DOI] [PubMed] [Google Scholar]
- 26.Munsie L, et al. Mutant huntingtin causes defective actin remodeling during stress: Defining a new role for transglutaminase 2 in neurodegenerative disease. Hum Mol Genet. 2011;20(10):1937–1951. doi: 10.1093/hmg/ddr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172(4):541–552. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye J, Zhao J, Hoffmann-Rohrer U, Grummt I. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 2008;22(3):322–330. doi: 10.1101/gad.455908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto K, Pasque V, Jullien J, Gurdon JB. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes Dev. 2011;25(9):946–958. doi: 10.1101/gad.615211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlegelmilch K, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 2011;144(5):782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silvis MR, et al. α-Catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal. 2011;4(174):ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 33.Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105(3):391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 34.Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. Biochemical and structural definition of the l-afadin- and actin-binding sites of alpha-catenin. J Biol Chem. 2002;277(21):18868–18874. doi: 10.1074/jbc.M201463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pappas DJ, Rimm DL. Direct interaction of the C-terminal domain of alpha-catenin and F-actin is necessary for stabilized cell-cell adhesion. Cell Commun Adhes. 2006;13(3):151–170. doi: 10.1080/15419060600726142. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Dokurno P, Tonks NK, Barford D. Crystal structure of the M-fragment of alpha-catenin: Implications for modulation of cell adhesion. EMBO J. 2001;20(14):3645–3656. doi: 10.1093/emboj/20.14.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi HJ, et al. αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc Natl Acad Sci USA. 2012;109(22):8576–8581. doi: 10.1073/pnas.1203906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nieset JE, et al. Characterization of the interactions of alpha-catenin with alpha-actinin and beta-catenin/plakoglobin. J Cell Sci. 1997;110(Pt 8):1013–1022. doi: 10.1242/jcs.110.8.1013. [DOI] [PubMed] [Google Scholar]
- 39.Pestic-Dragovich L, et al. A myosin I isoform in the nucleus. Science. 2000;290(5490):337–341. doi: 10.1126/science.290.5490.337. [DOI] [PubMed] [Google Scholar]
- 40.Wu X, et al. Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat Cell Biol. 2006;8(7):756–763. doi: 10.1038/ncb1433. [DOI] [PubMed] [Google Scholar]
- 41.Yoo Y, Wu X, Guan JL. A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J Biol Chem. 2007;282(10):7616–7623. doi: 10.1074/jbc.M607596200. [DOI] [PubMed] [Google Scholar]
- 42.Chhabra D, dos Remedios CG. Cofilin, actin and their complex observed in vivo using fluorescence resonance energy transfer. Biophys J. 2005;89(3):1902–1908. doi: 10.1529/biophysj.105.062083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stüven T, Hartmann E, Görlich D. Exportin 6: A novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 2003;22(21):5928–5940. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchert M, et al. AF6/s-afadin is a dual residency protein and localizes to a novel subnuclear compartment. J Cell Physiol. 2007;210(1):212–223. doi: 10.1002/jcp.20853. [DOI] [PubMed] [Google Scholar]
- 45.Xu YZ, Thuraisingam T, Morais DA, Rola-Pleszczynski M, Radzioch D. Nuclear translocation of beta-actin is involved in transcriptional regulation during macrophage differentiation of HL-60 cells. Mol Biol Cell. 2010;21(5):811–820. doi: 10.1091/mbc.E09-06-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci USA. 2008;105(41):15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maiden SL, Hardin J. The secret life of α-catenin: Moonlighting in morphogenesis. J Cell Biol. 2011;195(4):543–552. doi: 10.1083/jcb.201103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gottardi CJ, Gumbiner BM. Distinct molecular forms of beta-catenin are targeted to adhesive or transcriptional complexes. J Cell Biol. 2004;167(2):339–349. doi: 10.1083/jcb.200402153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang IC, et al. Foxm1 mediates cross talk between Kras/mitogen-activated protein kinase and canonical Wnt pathways during development of respiratory epithelium. Mol Cell Biol. 2012;32(19):3838–3850. doi: 10.1128/MCB.00355-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein EA, Assoian RK. Transcriptional regulation of the cyclin D1 gene at a glance. J Cell Sci. 2008;121(Pt 23):3853–3857. doi: 10.1242/jcs.039131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imamura Y, Itoh M, Maeno Y, Tsukita S, Nagafuchi A. Functional domains of alpha-catenin required for the strong state of cadherin-based cell adhesion. J Cell Biol. 1999;144(6):1311–1322. doi: 10.1083/jcb.144.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watabe-Uchida M, et al. Alpha-Catenin-vinculin interaction functions to organize the apical junctional complex in epithelial cells. J Cell Biol. 1998;142(3):847–857. doi: 10.1083/jcb.142.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. Alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12(6):533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 54.Chan DC, Leder P. Genetic evidence that formins function within the nucleus. J Biol Chem. 1996;271(38):23472–23477. doi: 10.1074/jbc.271.38.23472. [DOI] [PubMed] [Google Scholar]
- 55.Dingová H, Fukalová J, Maninová M, Philimonenko VV, Hozák P. Ultrastructural localization of actin and actin-binding proteins in the nucleus. Histochem Cell Biol. 2009;131(3):425–434. doi: 10.1007/s00418-008-0539-z. [DOI] [PubMed] [Google Scholar]
- 56.Simcha I, et al. Differential nuclear translocation and transactivation potential of beta-catenin and plakoglobin. J Cell Biol. 1998;141(6):1433–1448. doi: 10.1083/jcb.141.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang SM, Cheng YS. Analysis of two CBP (cAMP-response-element-binding protein-binding protein) interacting sites in GRIP1 (glucocorticoid-receptor-interacting protein), and their importance for the function of GRIP1. Biochem J. 2004;382(Pt 1):111–119. doi: 10.1042/BJ20040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Desai R, et al. Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat Cell Biol. 2013;15(3):261–273. doi: 10.1038/ncb2685. [DOI] [PubMed] [Google Scholar]
- 59.Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS. Beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev Cell. 2010;19(2):220–231. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castano E, et al. Actin complexes in the cell nucleus: New stones in an old field. Histochem Cell Biol. 2010;133(6):607–626. doi: 10.1007/s00418-010-0701-2. [DOI] [PubMed] [Google Scholar]
- 61.Zheng B, Han M, Bernier M, Wen JK. Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 2009;276(10):2669–2685. doi: 10.1111/j.1742-4658.2009.06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Lanerolle P, Serebryannyy L. Nuclear actin and myosins: Life without filaments. Nat Cell Biol. 2011;13(11):1282–1288. doi: 10.1038/ncb2364. [DOI] [PubMed] [Google Scholar]
- 63.Choi SH, Estarás C, Moresco JJ, Yates JR, 3rd, Jones KA. α-Catenin interacts with APC to regulate β-catenin proteolysis and transcriptional repression of Wnt target genes. Genes Dev. 2013;27(22):2473–2488. doi: 10.1101/gad.229062.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.