Abstract
The 8;21 translocation is the most common chromosomal aberration occurring in acute myeloid leukemia (AML). This translocation causes expression of the RUNX1–ETO (AML1–ETO) fusion protein, which cooperates with additional mutations in leukemia development. We report here that interferons (IFNs) and IFN-stimulated genes are a group of genes consistently up-regulated by RUNX1–ETO in both human and murine models. RUNX1–ETO-induced up-regulation of IFN-stimulated genes occurs primarily via type I IFN signaling with a requirement for the IFNAR complex. Addition of exogenous IFN in vitro significantly reduces the increase in self-renewal potential induced by both RUNX1–ETO and its leukemogenic splicing isoform RUNX1–ETO9a. Finally, loss of type I IFN signaling via knockout of Ifnar1 significantly accelerates leukemogenesis in a t(8;21) murine model. This demonstrates the role of increased IFN signaling as an important factor inhibiting t(8;21) fusion protein function and leukemia development and supports the use of type I IFNs in the treatment of AML.
Keywords: RUNX1–ETO, acute myeloid leukemia, interferon, t(8;21)
Introduction
Acute myeloid leukemia (AML) is the most common form of adult leukemia and is currently treated primarily by induction and consolidation therapy, with a 5-year survival rate of approximately 50% [1–3]. Chromosomal translocations are common genetic events in AML, with the most common being t(8;21), present in 8–20% of all cases of AML and up to 40% of the French–American–British (FAB)-M2 AML subtype [4–8]. The 8;21 translocation causes expression of the fusion protein RUNX1–ETO (RE) and its splicing isoforms, including RUNX1–ETO9a (RE9a) [9] and RUNX1–ETO11a [10]. Both of these isoforms are present in t(8;21) + human AML patient samples and lack the C-terminal MYND/NHR4 domain. Interestingly, full-length RE alone will maintain the self-renewal potential of hematopoietic cells in vitro but requires additional cooperating mutations to induce leukemia in vivo [11–13]. Conversely, RE9a, which lacks the MYND domain, and RUNX1–ETO-W692A, a mutant with disrupted interaction between the MYND domain and the nuclear co-repressor NCOR1 [14,15], strongly promote leukemia development in a mouse model in the absence of additional mutagenic manipulations [9]. Although t(8;21) fusion proteins can activate and repress target genes via the RUNX1 binding motif, in general RE dysregulates genes to a greater extent and has a more negative effect on cell survival than RE9a and RUNX1–ETO-W692A [14,15]. Therefore, the attenuation of RE-induced gene dysregulation by RE9a and RUNX1–ETO-W692A may contribute to increased leukemogenic potential.
Interferons (IFNs) are a family of cytokines initially discovered in 1957 that are able to potently inhibit viral infection [16]. IFNs can be broken down into three classes, named type I to type III, based on the receptors to which they bind and activate to initiate signaling [17]. In humans, type I IFNs include 13 IFN-α subtypes and IFN-β, which bind to the receptor complex IFNAR, comprising the IFNAR1 and IFNAR2 subunits. Mice lacking the IFNAR1 subunit are completely unresponsive to type I IFNs and are unable to combat viral infections, although these mice are still resistant to some types of bacterial infection [18,19]. The type II class of IFNs in humans is composed solely of IFN-γ, which signals through the IFNGR complex. The type III IFNs (IFN-λ1, -λ2 and -λ3) are less well characterized, but are also involved in resistance to viral infections [20,21]. After binding to their respective receptors, IFNs activate the JAK/STAT (Janus kinase/signal transducer and activator of transcription) signaling pathway to exert transcriptional changes on the cell. All three types of IFNs induce STAT1 phosphorylation and consequent homo-dimerization, whereas only types I and III induce STAT1/STAT2 phosphorylation and hetero-dimerization [22]. Dimerized STAT1 proteins then bind to genes containing IFN-γ-activated site (GAS) elements in their promoters, leading to their activation [23]. Type I and type III IFNs differ from type II in their ability to induce formation of the ISGF3 complex, composed of IRF9 and the phosphorylated forms of STAT1 and STAT2, which then binds to IFN-stimulated response elements (ISREs) to activate gene expression [22]. Promoters of some interferon-stimulated genes contain both GAS and ISRE elements, whereas others contain only one or the other [23]. As a result, some interferon-stimulated genes can be activated by multiple types of IFNs, and others are responsive to only specific IFN types. Although type I and type III IFNs function through similar pathways, they act on overlapping but distinct groups of cells. Whereas virtually all nucleated cells respond to IFN-α and IFN-β, the majority of hematopoietic cells are unresponsive to IFN-λ, except for plasmacytoid dendritic cells, and the anti-viral effects of type III IFN signaling have been shown to be mediated mainly through the induction of an anti-viral state in epithelial cells [24].
Along with the inhibition of viral infection and replication, the downstream consequences of IFN signaling include anti-proliferative, pro-apoptotic and immunomodulatory effects, leading to additional clinical applications [23]. In particular, type I IFNs are used to treat various hematological malignancies, including chronic myeloid leukemia [25], hairy cell leukemia [26] and myeloproliferative neoplasms [27]. Furthermore, the anti-neoplastic effects of IFN-α have been taken advantage of in the treatment of AML with varying degrees of success depending upon the specific clinical application [28].
In this report we determine that type I IFNs and IFN-stimulated genes are specifically up-regulated by RUNX1–ETO and to a lesser degree by RE9a. We have further demonstrated that increased IFN signaling negatively impacts t(8;21)-induced self-renewal, and that loss of IFN signaling significantly increases the leukemogenic potential of RE9a. These findings provide an additional mechanism for RUNX1–ETO cellular toxicity and further support the use of type I IFNs in the treatment of AML.
Materials and methods
U937 cell culture and microarray
U937T-parental, RUNX1–ETO and RUNX1–ETO9a cell lines used in this study have been described previously [29]. Cells were cultured in RPMI 1640 (Corning Cellgro, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Omega Scientific, Tarzana, CA) either in the presence (uninduced) or absence (induced) of 1 μg/mL tetracycline (Sigma-Aldrich, St. Louis, MO). For microarray studies, cells were induced by washing three times in phosphate buffered saline (PBS) to remove residual tetracycline and then incubated in tetracycline-free medium for 24 h. RNA was harvested from both induced and uninduced cells (TRIzol; Life Technologies, Grand Island, NY) and analyzed using the Whole Human Genome Microarray 4 × 44K Chip (Agilent Technologies, Santa Clara, CA). For Western blot, cells were induced for 48 h and protein lysates from 106 cells were blotted with α-tubulin (Covance, San Diego, CA) or RUNX1 [9] antibodies. For quantitative real-time polymerase chain reaction (qRT-PCR), RNA from induced and uninduced cells was harvested using TRIzol, 1 μg of RNA was used to generate cDNA using oligo(dT) and random primers (qScript cDNA SuperMix; Quanta Biosciences, Gaithersburg, MD), and cDNA was subject to qRT-PCR on an iCycler (BioRad, Hercules, CA) using KAPA SYBR FAST Universal 2 × qRT-PCR Master Mix (KAPA Biosystems, Woburn, MA). Primers used are listed in Table I.
Table I.
List of primers used in qRT-PCR experiments.
| Gene symbol | Forward primer | Reverse primer |
|---|---|---|
| Human | 5′→3′ | 5′→3′ |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC |
| IFNA | TGGCAACCAGTTCCAGAAG | GACAACCTCCCAGGCACAA |
| HUMAN IFNB1 | TCAGAAGCTtCTGTGGCAAT | TCATGAGTTTTCCCCTGGTG |
| IRF7 | TATACCATCTACCTGGGCTTCGG | GCTCCAGCTTTCTGGAGTTCTCATT |
| OASL | AGGACTGTTGCTATGACAACAGGGA | TGCTGCTGAGAAGCTGCCTCTC |
| ISG15 | TTTGCCAGTACAGGAGCTTGTG | GCTCAGAGGTTCGTCGCATTT |
| IFI44 | TTCGATGCGAAGATTCACTG | CCCTTGGAAAACAGACCTCA |
| Mouse | 5′→3′ | 5′→3′ |
| Gapdh | GGTGCTGAGTATGTCGTGGAGTCTA | AAAGTTGTCATGGATGACCTTGG |
| Ifna | TGTCTGATGCAGCAGGTGG | AAGACAGGGCTCTCCAGAC |
| Ifnb1 | CACTTGAAGAGCTATTACTGGAGGG | CTCGGACCACCATCCAGG |
| Ly6a | GGAGGCAGCAGTTATTGTGGATTCT | GTGGGAACATTGCAGGACCCCA |
| OaSl2 | AGCCGTGATGGAGCTCCTCGT | GGATGATGGGCCGGTCTCCCT |
| Oasl1 | TGGAGGGTGAAGAGAGCACCCG | AGGCGAGCGTGCAATGGCTT |
| IRF7 | TCCGGTACCAGGGTCCAGCC | CGGGAGCGCACACGTGATGT |
| Ifng | TGCCAAGTTTGAGGTCAACAACCC | C C ACCCCG AATC AG C AGCG A |
qRT-PCR, quantitative real-time polymerase chain reaction.
Murine bone marrow retroviral transduction and cell culture
C57 background IFN-α/β receptor R1 knock-out mice (Ifnar1−/−) were kindly provided by Jonathan Sprent (The Scripps Research Institute) [18]. Retroviral transduction and replating assays were performed as previously described [30]. Briefly, total bone marrow cells from wild-type or Ifnar1−/− mice were transduced twice with retrovirus encoding MSCV-IRES-Puro (MIP) [31] vector control, MIP-HA-RUNX1–ETO or MIP-HA-RUNX1–ETO9a, as indicated. Infected cells were selected 1 week in 1 μg/mL puromycin in M3434 (STEMCELL Technologies, Vancouver, BC, Canada). Twenty thousand cells from each transduction were replated in duplicate every 7 days after colony and cell counting in the presence or absence of 500 units/mL of universal human type I interferon (PBL Biomedical Laboratories, Piscataway, NJ). For Western blot, protein lysates from 106 cells were blotted with α-tubulin or HA (Roche, Basel, Switzerland) antibodies. For qRT-PCR studies, transduced cells were selected 48 h in 2 μg/mL puromycin and then RNA extraction, cDNA synthesis and qRT-PCR studies were performed as above, with primers listed in Table I.
Fetal liver cell isolation, transduction and transplantation
C57BL/6J wild-type and Ifnar1−/− mice used in this study were housed in a pathogen-free facility and all procedures were approved by the Institutional Animal Care and Use Committee of the University of California, San Diego, CA. Transplant experiments were performed as described previously [9]. Briefly, fetal liver cells were harvested from embryonic day 13.5–16.5 wild-type or Ifnar1−/− mouse embryos and transduced twice with MSCV-IRES-GFP (MIGR1) or MIG-RUNX1–ETO9a retrovirus. Wild-type recipient mice were lethally irradiated at 900 rad and intravenously transplanted with the transduced fetal liver cells. Kaplan–Meier survival curves and statistical analyses were performed using GraphPad Prism4 (GraphPad Software, San Diego, CA). Peripheral blood smears and cytocentrifugation of bone marrow and spleen cells from leukemic mice were stained with Wright–Giemsa solutions (Sigma-Aldrich).
Results
RUNX1–ETO induces a stronger type I IFN response than RUNX1–ETO9a in human leukemia cells
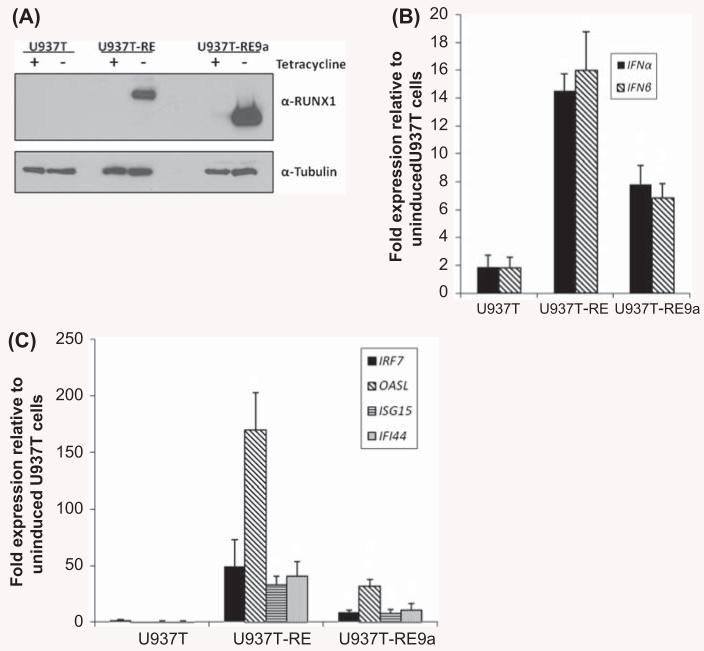
In order to examine which classes of genes are regulated by RE and RE9a, we performed gene expression analysis by microarray using human U937T leukemia cells with inducible RE and RE9a expression [29]. One group of genes consistently up-regulated by RE was IFN-stimulated genes, the top 10 most up-regulated of which are shown in Table II. Nine of these genes were also up-regulated by RE9a, with the lone exception being ISG15. However, the fold induction for these genes was lower in RE9a as compared to RE samples (Table II, ratios in rightmost column). We next sought to confirm these microarray results by qRT-PCR. Following induction of RE or RE9a expression in U937T cells [Figure 1(A)], we found that both type I IFNs α and β as well as multiple IFN-stimulated genes were strongly and consistently up-regulated by both RE and RE9a [Figures 1(B) and 1(C)]. In addition, the majority of these genes were significantly more up-regulated by RE than RE9a, corresponding well with the microarray data (Table II).
Table II.
List of top 10 interferon-stimulated genes up-regulated by RUNX1-ETO.
| Gene symbol |
Fold change in U937T-RUNX1-ETO |
Fold change in U937T-RUNX1-ETO9a |
Ratio RE/RE9a |
|---|---|---|---|
| IFI44 | 11.65 | 2.72 | 4.28 |
| IFIT1 | 10.36 | 2.97 | 3.49 |
| OASL | 8.8 | 2.96 | 2.97 |
| IFIT2 | 5.56 | 2.32 | 2.40 |
| CXCL10 | 4.59 | 1.66 | 2.77 |
| OAS1 | 4.43 | 1.53 | 2.90 |
| SRF7 | 4.35 | 1.86 | 2.34 |
| ISG15 | 4.16 | 0.84 | 4.95 |
| DDX58 | 3.94 | 1.4 | 2.81 |
| IRF9 | 3.1 | 1.64 | 1.89 |
Figure 1.
Expression of RUNX1–ETO and RUNX1–ETO9a induces IFN response in U937 cells. (A) RE and RE9a expression in U937T cell lines. 48 h after tetracycline withdrawal, U937T cells were analyzed by Western blot to confirm RE and RE9a induction. α-Tubulin serves as a loading control. (B) Expression of IFN-α and IFN-β in induced U937T cells was examined by qRT-PCR. Expression levels were normalized to GAPDH and each respective uninduced cell line. Data show averages and standard deviations of three independent experiments. *p < 0.05 relative to U937T. #p < 0.05 relative to U937T-RE. (C) Expression of IFN-stimulated genes in induced U937T cells. Expression levels were normalized to GAPDH and each respective uninduced cell line. Data show averages and standard deviations of three independent experiments. *p < 0.05 relative to U937T. #p < 0.05 relative to U937T-RE.
Interestingly, nine of the top 10 IFN-stimuated genes up-regulated by RE in our U937T system were also shown by another group to be up-regulated by RE in primary murine lineage-negative Sca-1 + c-Kit+ (LSK) bone marrow cells [14] (Supplementary Table I to be found online at http://informahealthcare.com/doi/abs/10.3109/10428194.2013.815351). This group compared gene expression changes in LSK cells induced by RUNX1–ETO and RUNX1–ETO-W692A and found that W692A, like RE9a, showed reduced dysregulation of many genes compared to full-length RE, including those listed in Supplementary Table I to be found online at http://informahealthcare.com/doi/abs/10.3109/10428194.2013.815351. Of note, when compared to full-length RE, both RUNX1–ETO-W692A and RE9a display attenuated cellular dysregulation and strongly induce leukemia in mice [14,15]. The close correlation between these two datasets demonstrates a high level of conservation of IFN-related RE target gene regulation between the mouse and human systems. Furthermore, there is a good correlation between gene regulation and leukemogenic potential of the fusion gene, as RE strongly up-regulates this gene set but is not leukemogenic on its own, but both W692A and RE9a more weakly regulate these genes and are leukemogenic. This led us to hypothesize that the IFN response induced by t(8;21) fusion proteins may functionally inhibit their leukemic potential.
RE-induced IFN response is dependent upon type I IFN receptor IFNAR
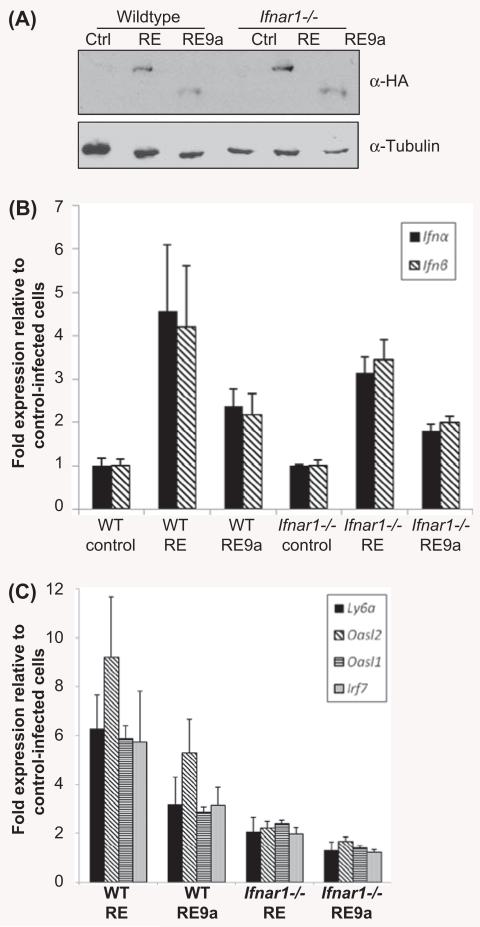
To examine a functional role for type I IFN signaling in inhibition of RE function, we made use of a mouse model lacking Ifnar1, which is required for signaling via IFNs α and β [18]. First, to confirm induction of IFNs and IFN-stimulated genes in this system, we retrovirally transduced wild-type and Ifnar1−/− murine bone marrow cells with control, RE or RE9a virus and confirmed transgene expression [Figure 2(A)]. We next examined type I IFN expression by qRT-PCR and found that IFNs α and β are up-regulated by both RE and RE9a and this up-regulation is independent of Ifnar1 expression [Figure 2(B)]. Finally, although multiple IFN-stimulated genes were up-regulated in wild-type bone marrow cells, these same genes were only weakly up-regulated when Ifnar1 was lacking, demonstrating a requirement for the type I IFN receptor in IFN-stimulated gene regulation by RE and RE9a [Figure 2(C)]. A possible explanation as to why these IFN-stimulated genes are still weakly up-regulated in Ifnar1−/− cells is that there is significant overlap between type I and type II IFN signaling and target genes [23,32,33], and that the type II IFN-γ is also up-regulated by RE and RE9a in this system (Supplementary Figure 1 to be found online at http://informahealthcare.com/doi/abs/10.3109/10428194.2013.815351), which could lead to some induction of these response genes. In conclusion, although RE and RE9a are still able to up-regulate type I IFNs in the Ifnar1−/− background, the lack of a functional type I IFN receptor blocks their downstream signaling, clearly indicating a requirement of Ifnar1 expression for the RE-induced IFN response.
Figure 2.
RUNX1–ETO- and RUNX1–ETO9a-induced IFN response requires IFNAR. (A) RE and RE9a expression in wild-type and Ifnar1−/− in total murine bone marrow cells. Following retroviral transduction and selection, bone marrow cells were analyzed by Western blot to confirm RE and RE9a expression. α-Tubulin serves as a loading control. (B) Expression of Ifn-α and Ifn-β in transduced bone marrow cells. Expression levels were normalized to Gapdh with control-transduced cells of each genotype set to 1. Data show averages and standard deviations of three independent experiments. (C) Expression of IFN-stimulated genes in transduced bone marrow cells. Expression levels were normalized to Gapdh and respective wild-type or Ifnar1−/− control-transduced cells. Data show averages and standard deviations of three independent experiments.
Type I IFN inhibits increased self-renewal induced by t(8;21) fusion proteins
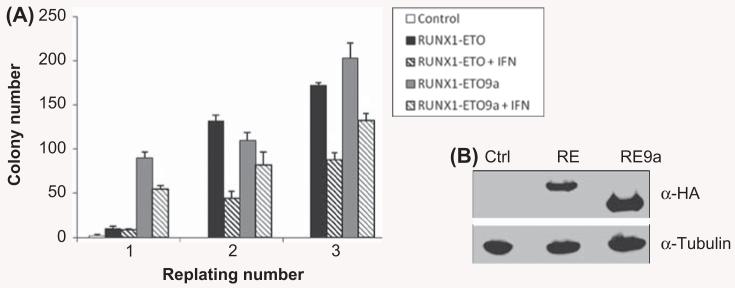
Having demonstrated that the t(8;21)-induced IFN response requires Ifnar1, we next sought to determine the functional consequences of increased IFN signaling using the colony replating assay. In this assay, wild-type cells are transduced by control, RE or RE9a retrovirus and cells are replated weekly in methylcellulose culture. As shown in Figure 3, both RE and RE9a increase cellular self-renewal capacity over control-transduced cells, leading to a corresponding increase in the number of colonies formed. However, when these cells were treated with 500 units/mL of universal type I IFN, colony numbers were significantly reduced in both RE and RE9a samples between 26 and 66% as compared to controls [Figure 3(A)]. This dramatic reduction in colony number clearly demonstrates the negative effect of type I IFN signaling on the increased self-renewal capacity induced by RE and RE9a, and indicates that the increased interferon response caused by these fusion proteins may be inhibitory for the development of leukemia.
Figure 3.
IFN inhibits RUNX1–ETO- and RUNX1–ETO9a-induced increase in self-renewal. (A) Colony numbers from wild-type bone marrow cells transduced with control (MIP), MIP-RE or MIP-RE9a retrovirus and serially replated in methylcellulose ± treatment with 500 units/mL universal human type I IFN. Data shown are averages and standard deviations of a representative experiment. Three independent assays were performed, each in duplicate. *p < 0.01 relative to respective non-IFN treated sample. (B) Confirmation of expression of RE and RE9a in 293T cells used to produce retrovirus for bone marrow cell transduction. α-Tubulin serves as a loading control.
Lack of Ifnar1 significantly increases RE9a leukemogenecity
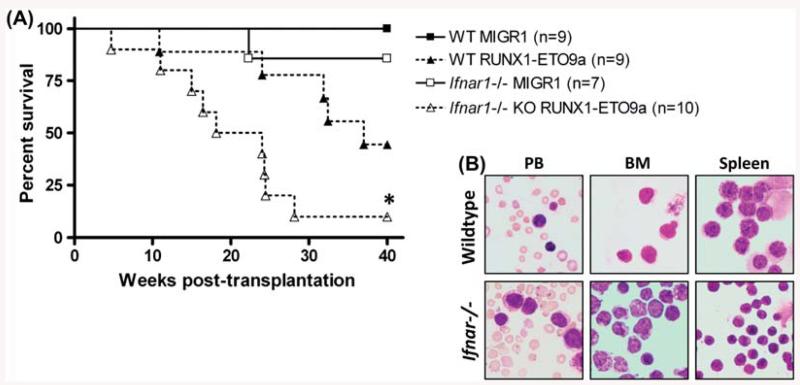
Since type I IFN inhibits t(8;21) fusion protein function in vitro, we next examined the consequences of loss of this signaling on RE9a-induced leukemia development using an in vivo mouse model. Wild-type and Ifnar1−/− fetal liver cells were transduced with retrovirus expressing GFP alone (MIGR1) or GFP with RE9a, and infected cells were transplanted intravenously into lethally irradiated wild-type recipient mice. As shown in Figure 4(A), mice receiving MIGR1-infected cells of either genotype did not develop leukemia. One Ifnar1−/− MIGR1 recipient mouse died approximately 22 weeks post-transplant; however, this mouse did not display splenomegaly, increased percent of GFP + cells in the peripheral blood, bone marrow or spleen or presence of blast cells in peripheral blood. We therefore concluded that this mouse died of non-leukemic causes. As expected, mice receiving RE9a-expressing wild-type cells developed leukemia and displayed the presence of blast cells in the peripheral blood, bone marrow and spleen (Figure 4). Interestingly, mice receiving RE9a Ifnar1−/− cells also developed leukemia, but significantly faster than mice developing leukemia from wild-type cells (median survival: wild-type 37.00 weeks vs. Ifnar1−/− 21.07 weeks; p = 0.02. This clearly demonstrates that disruption of type I IFN signaling significantly increases the leukemogenic potential of RE9a and indicates that the IFN response induced by t(8;21) fusion proteins is an inhibitory factor that must be overcome for leukemia development.
Figure 4.
Loss of type I IFN signaling significantly increases RUNX1-ETO9a leukemogenic potential. (A) Survival curves of mice transplanted with wildtype or Ifnar1−/− fetal liver cells transduced by control (MIGR1) or MIG-RE9a retrovirus. Number of mice in each cohort shown at right. Wild-type median survival: 37.00 weeks; Ifnar1−/− median survival: 21.07 weeks; *p = 0.02. Data shown are combined from two independent transductions and transplants. (B) Presence of hematopoietic blast cells in indicated tissues of mice transplanted with RE9a-transduced wildtype or Ifnar1−/− cells as analyzed by Wright–Giemsa staining.
Discussion
Although full-length RUNX1–ETO requires cooperating mutations to induce leukemia development, it has previously been reported that loss or mutation of RE C-terminus allows the protein to become leukemogenic [9,34]. Additionally, specific mutation or loss of the MYND domain impaired the ability of RE to block granulocytic differentiation and proliferation [14], decreased rates of RE-induced apoptosis and allowed the protein to induce leukemia in mice [15]. All of these phenotypic changes observed in the leukemic RE forms correlated with attenuated gene dysregulation by the fusion protein [14,15].
In this study, we confirm that the leukemogenic RE9a, which lacks the C-terminus of RE, shows attenuated dysregulation of IFNs and IFN-stimulated genes as compared to full-length RE (Figures 1 and 2 and Table II). This appears to occur mainly via type I IFN signaling, as both IFN-α and IFN-β are consistently up-regulated by RE and RE9a, and the up-regulation of IFN-stimulated genes is largely dependent upon the presence of Ifnar1, which is required for type I IFN signaling. However, there is still some up-regulation of IFN-stimulated genes in the absence of Ifnar1. This is not surprising, as knockout of Ifnar1 will only block type I IFN signaling. Since RE and RE9a both also up-regulate IFN-γ (Supplementary Figure 1 to be found online at http://informahealthcare.com/doi/abs/10.3109/10428194.2013.815351) and type I and type II IFNs have overlapping target genes [23], it is likely that these genes still show some up-regulation due to this increase in IFN-γ and consequent type II IFN signaling. Additionally, interferon response factors (IRFs) can be activated independently of IFN signaling. For instance, it is known that cellular stress such as that caused by DNA-damaging agents both induces nuclear localization and increases transcriptional activity of IRF3 [35]. Similarly, T-lymphocytes treated with DNA-damaging agents show induction of IRF1 and IRF1-dependent sensitization to apoptosis [36]. Given that RE expression causes repression of DNA repair genes and the accumulation of DNA damage [37], these findings provide a further mechanism by which RE-induced increases in DNA damage could lead to induction of IRFs and consequent IFN-stimulated gene expression. However, in light of our data, we believe that the majority of RE-induced IFN-stimulated gene expression occurs via type I IFN signaling and the IFNAR receptor complex. It will be interesting to study the mechanism behind t(8;21) up-regulation of type I IFNs. The promoters of both IFNB1 and IFNA contain elements bound and activated by IRF1, IRF3, IRF5 and IRF7 homodimers [38]. It is possible that RE and RE9a up-regulate type I IFNs through one or multiple of these proteins, and this will be an interesting avenue for future studies.
We further demonstrate that the increase in IFN signaling has negative effects on t(8;21) fusion protein function. Both RE- and RE9a-transduced bone marrow cells treated with universal type I IFN show a significant reduction in colony forming ability in vitro, demonstrating the deleterious impact of excess IFN signaling on self-renewal in t(8;21) + progenitor cells (Figure 3). More importantly, increased IFN signaling induced by RE9a clearly negatively affects leukemia development, as loss of type I IFN signaling via knockout of Ifnar1 significantly decreased AML latency in vivo (Figure 4). This strongly indicates that increased type I IFN signaling through IFNAR is one factor that needs to be overcome for t(8;21) fusion proteins to induce leukemia. Since RE shows even greater increases in IFN signaling, it stands to reason that the negative effects on leukemia development are stronger for RE than RE9a, and this may be part of the reason why RE requires additional mutations to induce leukemia, whereas RE9a is leukemogenic on its own. In this vein, it is interesting to note that none of the top 10 IFN-stimulated genes listed in Table II are significantly up-regulated in human t(8;21) AML patient samples [39]. This demonstrates that although expression of RE and RE9a clearly up-regulates these genes, this up-regulation is lost after leukemia develops. The lack of IFN-stimulated genes up-regulated in leukemia may be due to a transient up-regulation by RE and RE9a or to further t(8;21) cooperating mutations that reduce the IFN signaling effect. In either case, this change in gene expression further indicates that the negative effects of IFN signaling is one factor that needs to be overcome for the induction of leukemia by t(8;21) fusion proteins. However, increased IFN signaling by RE is likely not the sole factor preventing leukemia development, as recipient mice transplanted with RE-transduced Ifnar1−/− fetal liver cells failed to develop leukemia (data not shown), indicating the important effects of additional differences between RE and its leukemogenic isoforms.
The finding that an increase in type I IFN signaling negatively affects RE function and leukemia development fits well with the downstream functions of IFN-α and -β, which include increased apoptotic rate, decreased proliferation and increased AML cell immunogenicity [28]. In fact, type I IFN has been used clinically to treat AML, but has fallen out of favor due to inconsistent patient responses and adverse side effects [28]. These side effects include fever, nausea, myalgia and hematologic toxicity, and increase with increasing dose of IFN treatment [40]. Inconsistent clinical response may be partly explained by the short half-life of unmodified IFNs (4–16 h for IFN-α; 1-2 h for IFN-β) [41]. This is supported by the finding that continuous administration of IFN-β was a critical factor for treatment response in a murine AML xenograft model [42]. In this study, the authors found that continuous low dose IFN treatment showed significantly greater reductions in tumor load than intermittent IFN-β doses. This held true despite the fact that the intermittent doses resulted in peak plasma levels of IFN that were up to 20 times higher than those generated by the continuous IFN dose strategy, indicating the importance of duration of signaling as opposed to simply strength of signaling. This is a very important finding, as lower IFN doses would also result in decreased adverse side effects [40,41]. Formulations of IFNs fused to human serum albumin or modified by polyethylene glycol (pegylation) have recently been created, both of which demonstrate significantly increased half-lives over unmodified IFN, facilitating a continuous low dose IFN treatment strategy [43,44].
Our report elucidates a strong, RE-induced type I IFN response as an additional mechanism of RE cellular toxicity and one factor that is overcome in leukemia development. Given the lesser IFN response induced by RE9a, it also helps to explain why RE induces greater adverse cellular effects than does RE9a. More importantly, the clear role that loss of IFN signaling plays in accelerating RE9a-induced leukemia development further supports the use of these new long half-life IFNs in the treatment of AML.
Supplementary Material
Acknowledgements
We thank all members of the D.-E.Z. laboratory for valuable discussions.
This work was supported by National Institutes of Health grants R01 CA104509, R01 CA177305, 5T32GM007240, and 5T32CA67754-17
Footnotes
Supplementary material available online
Supplementary Table I & Figure 1.
Potential conflict of interest: Disclosure forms provided by the authors are available with the full text of this article at www.informahealthcare.com/lal.
References
- [1].Rai KR, Holland JF, Glidewell OJ, et al. Treatment of acute myelocytic leukemia: a study by cancer and leukemia group B. Blood. 1981;58:1203–1212. [PubMed] [Google Scholar]
- [2].Roboz GJ. Current treatment of acute myeloid leukemia. Curr Opin Oncol. 2012;24:711–719. doi: 10.1097/CCO.0b013e328358f62d. [DOI] [PubMed] [Google Scholar]
- [3].Byrd JC, Mrozek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- [4].Groupe Francais de Cytogenetique Hematologique Acute myelogenous leukemia with an 8;21 translocation. A report on 148 cases from the Groupe Francais de Cytogenetique Hematologique. Cancer Genet Cytogenet. 1990;44:169–179. [PubMed] [Google Scholar]
- [5].Nucifora G, Rowley JD. AML1 and the 8;21 and 3;21 translocations in acute and chronic myeloid leukemia. Blood. 1995;86:1–14. [PubMed] [Google Scholar]
- [6].Langabeer SE, Walker H, Rogers JR, et al. Incidence of AML1/ETO fusion transcripts in patients entered into the MRC AML trials. MRC Adult Leukaemia Working Party. Br J Haematol. 1997;99:925–928. doi: 10.1046/j.1365-2141.1997.4663270.x. [DOI] [PubMed] [Google Scholar]
- [7].Rowe D, Cotterill SJ, Ross FM, et al. Cytogenetically cryptic AML1-ETO and CBF beta-MYH11 gene rearrangements:incidence in 412 cases of acute myeloid leukaemia. Br J Haematol. 2000;111:1051–1056. doi: 10.1046/j.1365-2141.2000.02474.x. [DOI] [PubMed] [Google Scholar]
- [8].Rege K, Swansbury GJ, Atra AA, et al. Disease features in acute myeloid leukemia with t(8;21)(q22;q22). Influence of age, secondary karyotype abnormalities, CD19 status, and extramedullary leukemia on survival. Leuk Lymphoma. 2000;40:67–77. doi: 10.3109/10428190009054882. [DOI] [PubMed] [Google Scholar]
- [9].Yan M, Kanbe E, Peterson LF, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- [10].Kozu T, Fukuyama T, Yamami T, et al. MYND-less splice variants of AML1-MTG8 (RUNX1-CBFA2T1) are expressed in leukemia with t(8;21) Genes Chromosomes Cancer. 2005;43:45–53. doi: 10.1002/gcc.20165. [DOI] [PubMed] [Google Scholar]
- [11].Mulloy JC, Cammenga J, Berguido FJ, et al. Maintaining the self-renewal and differentiation potential of human CD34 + hematopoietic cells using a single genetic element. Blood. 2003;102:4369–4376. doi: 10.1182/blood-2003-05-1762. [DOI] [PubMed] [Google Scholar]
- [12].Peterson LF, Boyapati A, Ahn EY, et al. Acute myeloid leukemia with the 8q22;21q22 translocation:secondary mutational events and alternative t(8;21) transcripts. Blood. 2007;110:799–805. doi: 10.1182/blood-2006-11-019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Muller AM, Duque J, Shizuru JA, et al. Complementing mutations in core binding factor leukemias:from mouse models to clinical applications. Oncogene. 2008;27:5759–5773. doi: 10.1038/onc.2008.196. [DOI] [PubMed] [Google Scholar]
- [14].Liu Y, Chen W, Gaudet J, et al. Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO’ s activity. Cancer Cell. 2007;11:483–497. doi: 10.1016/j.ccr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dekelver RC, Yan M, Ahn EY, et al. Attenuation of AML1-ETO cellular dysregulation correlates with increased leukemogenic potential. Blood. 2013;121:3714–3717. doi: 10.1182/blood-2012-11-465641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- [17].Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Muller U, Steinhoff U, Reis LF, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- [19].O’Connell RM, Saha SK, Vaidya SA, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kotenko SV, Gallagher G, Baurin VV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- [21].Sheppard P, Kindsvogel W, Xu W, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- [22].de Weerd NA, Nguyen T. The interferons and their receptors—distribution and regulation. Immunol Cell Biol. 2012;90:483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- [24].Ank N, Iversen MB, Bartholdy C, et al. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- [25].Kujawski LA, Talpaz M. The role of interferon-alpha in the treatment of chronic myeloid leukemia. Cytokine Growth Factor Rev. 2007;18:459–471. doi: 10.1016/j.cytogfr.2007.06.015. [DOI] [PubMed] [Google Scholar]
- [26].Damasio EE, Clavio M, Masoudi B, et al. Alpha-interferon as induction and maintenance therapy in hairy cell leukemia: a long-term follow-up analysis. Eur J Haematol. 2000;64:47–52. doi: 10.1034/j.1600-0609.2000.90014.x. [DOI] [PubMed] [Google Scholar]
- [27].Kiladjian JJ, Chomienne C, Fenaux P. Interferon-alpha therapy in bcr-abl-negative myeloproliferative neoplasms. Leukemia. 2008;22:1990–1998. doi: 10.1038/leu.2008.280. [DOI] [PubMed] [Google Scholar]
- [28].Anguille S, Lion E, Willemen Y, et al. Interferon-alpha in acute myeloid leukemia: an old drug revisited. Leukemia. 2011;25:739–748. doi: 10.1038/leu.2010.324. [DOI] [PubMed] [Google Scholar]
- [29].Burel SA, Harakawa N, Zhou L, et al. Dichotomy of AML1-ETO functions: growth arrest versus block of differentiation. Mol Cell Biol. 2001;21:5577–5590. doi: 10.1128/MCB.21.16.5577-5590.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shia WJ, Okumura AJ, Yan M, et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood. 2012;119:4953–4962. doi: 10.1182/blood-2011-04-347476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Peterson LF, Wang Y, Lo MC, et al. The multi-functional cellular adhesion molecule CD44 is regulated by the 8;21 chromosomal translocation. Leukemia. 2007;21:2010–2019. doi: 10.1038/sj.leu.2404849. [DOI] [PubMed] [Google Scholar]
- [32].Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- [33].Der SD, Zhou A, Williams BR, et al. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ahn EY, Yan M, Malakhova OA, et al. Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proc Natl Acad Sci USA. 2008;105:17103–17108. doi: 10.1073/pnas.0802696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kim T, Kim TY, Song YH, et al. Activation of interferon regulatory factor 3 in response to DNA-damaging agents. J Biol Chem. 1999;274:30686–30689. doi: 10.1074/jbc.274.43.30686. [DOI] [PubMed] [Google Scholar]
- [36].Tamura T, Ishihara M, Lamphier MS, et al. An IRF-1-dependent pathway of DNA damage-induced apoptosis in mitogen-activated T lymphocytes. Nature. 1995;376:596–599. doi: 10.1038/376596a0. [DOI] [PubMed] [Google Scholar]
- [37].Alcalay M, Meani N, Gelmetti V, et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest. 2003;112:1751–1761. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Honda K, Yanai H, Takaoka A, et al. Regulation of the type I IFN induction: a current view. Int Immunol. 2005;17:1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- [39].Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- [40].Sleijfer S, Bannink M, Van Gool AR, et al. Side effects of interferon-alpha therapy. Pharm World Sci. 2005;27:423–431. doi: 10.1007/s11096-005-1319-7. [DOI] [PubMed] [Google Scholar]
- [41].Arnaud P. The interferons: pharmacology, mechanism of action, tolerance and side effects. Rev Med Interne. 2002;23(Suppl. 4):449s–458s. doi: 10.1016/s0248-8663(02)00659-8. [DOI] [PubMed] [Google Scholar]
- [42].Benjamin R, Khwaja A, Singh N, et al. Continuous delivery of human type I interferons (alpha/beta) has significant activity against acute myeloid leukemia cells in vitro and in a xenograft model. Blood. 2007;109:1244–1247. doi: 10.1182/blood-2006-02-002915. [DOI] [PubMed] [Google Scholar]
- [43].Osborn BL, Olsen HS, Nardelli B, et al. Pharmacokinetic and pharmacodynamic studies of a human serum albumin-interferon-alpha fusion protein in cynomolgus monkeys. J Pharmacol Exp Ther. 2002;303:540–548. doi: 10.1124/jpet.102.037002. [DOI] [PubMed] [Google Scholar]
- [44].Dummer R, Mangana J. Long-term pegylated interferon-alpha and its potential in the treatment of melanoma. Biologics. 2009;3:169–182. doi: 10.2147/btt.2009.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.