Abstract
Background
Age-adjusted mortality rates for prostate cancer are higher for African American men compared with those of European ancestry. Recent data suggest that West African men also have elevated risk for prostate cancer relative to European men. Genetic susceptibility to prostate cancer could account for part of this difference.
Methods
We conducted a genome-wide association study (GWAS) of prostate cancer in West African men in the Ghana Prostate Study. Association testing was performed using multivariable logistic regression adjusted for age and genetic ancestry for 474 prostate cancer cases and 458 population-based controls on the Illumina HumanOmni-5 Quad BeadChip.
Results
The most promising association was at 10p14 within an intron of a long non-coding RNA (lncRNA RP11-543F8.2) 360 kb centromeric of GATA3 (p=1.29E−7). In sub-analyses, SNPs at 5q31.3 were associated with high Gleason score (≥7) cancers, the strongest of which was a missense SNP in PCDHA1 (rs34575154, p=3.66E−8), and SNPs at Xq28 (rs985081, p=8.66E−9) and 6q21 (rs2185710, p=5.95E−8) were associated with low Gleason score (<7) cancers. We sought to validate our findings in silico in the African Ancestry Prostate Cancer GWAS Consortium, but only one SNP, at 10p14, replicated at p<0.05. Of the 90 prostate cancer loci reported from studies of men of European, Asian or African American ancestry, we were able to test 81 in the Ghana Prostate Study, and 10 of these replicated at p<0.05.
Conclusion
Further genetic studies of prostate cancer in West African men are needed to confirm our promising susceptibility loci.
Keywords: prostate cancer, Africa, GWAS, case-control
Introduction
In the United States, age-adjusted incidence rates of prostate cancer in African-Americans are two-fold higher than those observed in men of European ancestry (Li et al. 2012). Although age-adjusted estimates of prostate cancer from African cancer registries suggest lower rates for African men (Center et al. 2012; Chu et al. 2011), international comparisons are complicated by population screening, overdiagnosis (Welch and Black 2010), and incomplete cancer registration (Parkin et al. 2010). Despite their challenges, mortality statistics (Ferlay et al. 2010) can provide a stable and robust cancer statistic (Welch and Black 2010), particularly for international comparisons. Age-adjusted mortality rates of prostate cancer show a pattern distinct from that of incidence (Center et al. 2012; Rebbeck et al. 2013), with equal or higher rates in Africa—and specifically, the West African region—compared with North America or Western Europe. Since West Africa is the principal ancestral origin of a substantial proportion of African-American men (Bryc et al. 2010; Torres et al. 2012), one may hypothesize that genetic susceptibility loci that vary in allele frequency by ancestral population could account for part of the observed differences in prostate cancer rates within the United States (US).
Recent genome-wide association studies (GWAS) have shown that genetic variants associated with prostate cancer—such as those at 8q24 (Amundadottir et al. 2006; Freedman et al. 2006; Haiman et al. 2007) and 17q21 (Haiman et al. 2011b)—are more common among African Americans compared with men of European ancestry. Whether the differences in susceptibility allele frequencies can partially explain differences in incidence across regions can best be answered by discovery of a larger fraction of the set of common variants associated with risk. To date, approximately 90 independent prostate cancer loci have been identified through GWAS, primarily in men of European background. Based on empiric analyses of existing data sets, it is estimated that these loci represent perhaps less than the total number of common variants, which could be even higher in men of African ancestry (Park et al. 2010). To investigate genetic susceptibility to prostate cancer in West Africa, we conducted a GWAS in the Ghana Prostate Study, a collaboration involving the US National Cancer Institute (NCI) and the University of Ghana.
Methods
Study Participants
Participants for analysis were recruited through the Ghana Prostate Study—a population-based component, and a clinical component. The population-based component was a probability sample designed using the 2000 Ghana Population and Housing Census data in an attempt to recruit approximately 1,000 men aged 50–74 years in the Greater Accra region (~3 million people) (Chokkalingam et al. 2012), which successfully recruited 1,037 healthy men between 2004 and 2006 with a response percentage of 98.8% (Chokkalingam et al. 2012). Consented individuals underwent an in-person interview, and within seven days had a digital rectal examination (DRE) and provided an over-night fasting blood sample for prostate specific antigen (PSA) testing, biomarker assays, and genetic analysis. Subjects who had a positive screen by PSA (>2.5 ng/ml) or DRE underwent a transrectal ultrasound-guided biopsy. A total of 73 histologically-confirmed prostate cancer cases were identified through the population-based screening component of the Ghana Prostate Study and are included in the case population analyzed herein. From the remaining 964 screen-negative individuals, 836 had at least 20 ug DNA extracted and available for analysis, and 500 of these were matched to cases for analysis by age (in 5 year categories).
In the Ghana Prostate Study, we recruited 676 prostate cancer cases at Korle Bu Teaching Hospital in Accra, Ghana between 2008 and 2012. All consented cases were interviewed and provided an overnight fasting blood sample. At the time of selection for this analysis we had recruited 582 prostate cancer cases, from which we selected 427 for analysis. Combined with the 73 cases diagnosed through the population-based component of the study, this yielded 500 available prostate cancer cases for analysis. Five technical replicates for each of three individuals served as quality control (QC) samples.
This Ghana Prostate Study was approved by institutional review boards in Ghana and at the National Cancer Institute.
Genotyping and Quality Control
DNA samples for this project were extracted from buffy coat samples using the Qiagen method according to the manufacturer’s instructions. Pre-genotyping quality control metrics excluded six case samples and two controls. 494 prostate cancer cases, 498 screen-negative controls, plus 15 distinct quality control samples were genotyped on the Illumina HumanOmni5-Quad BeadChip (Clarke et al. 2012).
The initial overall completion rate of genotype calls was 97.38%. After excluding 57,489 loci with no genotype call, the overall completion rate increased to 98.70%. A total of 70,872 loci were excluded due to low completion rates (<90%) and 1,393,418 single nucleotide polymorphisms (SNP) monomorphic in men from Ghana were further excluded. We advanced 2,837,019 SNPs for association analysis. For the quality control samples, which included three distinct samples each with five technical replicates, the concordance rate exceeded 99.99%. Twenty-seven (7 cases, 20 controls) samples were excluded due to low completion rate (lower than 94%) or extreme mean heterozygosity (lower than 13.5% or greater than 16.5%). A further five cases were excluded for having less than 80% African ancestry using the HapMap build 26 data (CEU, JPT+CHB, YRI) as the continental reference populations in a STRUCTURE analysis (Engelhardt and Stephens 2010). Principal components analysis (PCA) identified 16 individuals (five cases, 11 controls) with significant deviation of eigenvectors and thus, they were excluded (Reich et al. 2008). Finally, unexpected relatedness (1st–2nd degree), assessed using the GLU qc.ibds module (http://code.google.com/p/glu-genetics/) with an IBD0 threshold of 0.70, was detected for 11 pairs of full-sibling and one monozygotic twin; one individual randomly chosen from each related group was retained while two cases and nine controls were excluded. Note that one nuclear family involved three individuals and accounted for three related pairs, so, a total of 11 individuals were removed. In addition, one case sample was excluded due to incomplete phenotype with missing age. The final analytic data set included 474 prostate cancer cases and 458 screen-negative controls.
Statistical Analysis
Association tests for prostate cancer susceptibility were performed using multivariable unconditional logistic regression analyses (1 degree-of-freedom) adjusted for age and two eigenvectors (p<0.01 based on the null model) identified in PCA analysis. In addition, we stratified the analysis by Gleason score (≤6, ≥7). Follow-up analyses were conducted in the African Ancestry Prostate Cancer GWAS Consortium using a total 5,096 cases and 4,972 controls (Haiman et al. 2011a; Haiman et al. 2011b). Additional resources used for the analyses were dbSNP (Sherry et al. 2001), 1000 Genomes Project (Clarke et al. 2012), GENCODE (Harrow et al. 2012), HaploReg (Ward and Kellis 2012), RegulomeDB (Boyle et al. 2012), and Haploview 4.2 (Barrett et al. 2005).
Results
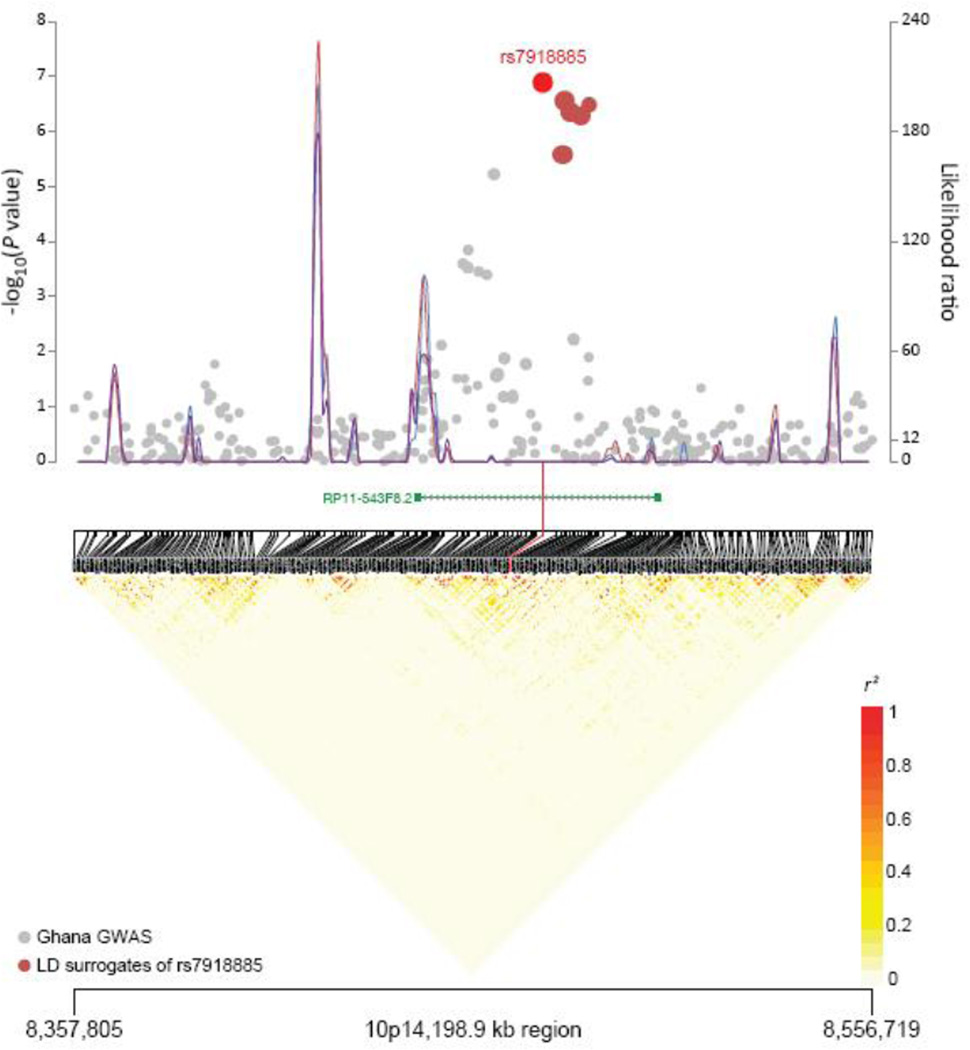
The results of the scan are shown in Table 1, which displays the 30 most promising SNPs associated with prostate cancer in the Ghana Prostate Study. Notable is a new locus marked by rs7918885 at 10p14 for prostate cancer risk (p value of 1.29E−7). Figure 1 depicts the eight correlated markers typed as part of our study, which are approximately 360 kb 5’ of GATA3; notably they localize to an intronic region of lncRNA gene RP11-543F8.2. Assessment of the correlated variants in 1000 Genomes Project data, restricted to the West African populations, did not reveal notable variants in the coding or splicing regions of the lncRNA RP11-543F8.2 (Supplementary Figure 1). Bioinformatic analyses of the eight correlated SNPs using the ENCODE resources point towards motif changes, DNase I hypersensitivity sites, and NHEK enhancer histone marks, but there was no overall significant enrichment for such elements (2012). Visual inspection of a quantile-quantile plot of the observed versus expected p values (Supplementary Figure 2), as well as the inflation factor of 1.01, indicated an absence of any systemic bias (e.g., residual population stratification). The results of the initial scan also suggested promising signals at 5q31.3, 3p26.1, and 8p23.2. Twenty-nine of our top 30 SNPs were analyzed in the African Ancestry Prostate Cancer GWAS Consortium; 12 SNPs were directly typed and 17 SNPs imputed using 1000 Genomes Project data (Clarke et al. 2012) (Supplementary Table 1). Only one of the 29 tested SNPs were statistically significant (p < 0.05) in the African American dataset, and that was rs2993385 located at 10p14.
Table 1.
The 30 most significant SNP associations with prostate cancer in the Ghana Prostate Study GWAS
| rs Number | Cytoband | Location1 | Alleles (referent | effect) |
Effect Allele Frequency (control | case) |
Per Effect Allele OR (95%CI) |
P value | In gene(s) or nearest gene(s) if distance specified2 |
|---|---|---|---|---|---|---|---|
| rs7918885 | 10p14 | 8474595 | T | G | 0.145 | 0.076 | 0.40 (0.28, 0.57) | 1.29E-07 | 358kb 5' of GATA3, RP11-543F8.2 (intron) |
| rs10905371 | 10p14 | 8480044 | A | G | 0.142 | 0.074 | 0.41 (0.29, 0.58) | 2.77E-07 | 363kb 5' of GATA3, RP11-543F8.2 (intron) |
| rs7896254 | 10p14 | 8486161 | G | A | 0.100 | 0.044 | 0.33 (0.21, 0.51) | 3.27E-07 | 369kb 5' of GATA3, RP11-543F8.2 (intron) |
| rs10905374 | 10p14 | 8481486 | G | A | 0.145 | 0.077 | 0.42 (0.29, 0.59) | 4.43E-07 | 364kb 5' of GATA3, RP11-543F8.2 (intron) |
| rs7096374 | 10p14 | 8484113 | C | T | 0.143 | 0.076 | 0.42 (0.30, 0.59) | 5.16E-07 | 368kb 5' of GATA3, RP11-543F8.2 (intron) |
| rs61749035 | 5q31.3 | 140718750 | G | T | 0.037 | 0.077 | 3.22 (2.00, 5.19) | 7.50E-07 | PCDHGA1 (intron), PCDHGA2 (missense) |
| rs114246623 | 3p26.1 | 6935066 | G | A | 0.015 | 0.078 | 4.93 (2.53, 9.59) | 7.70E-07 | GRM7-AS2 (AC066606.1) (intron) |
| rs147739031 | 8p23.2 | 3247408 | A | G | 0.005 | 0.039 | 11.48 (3.78, 34.90) | 9.96E-07 | CSMD1 (intron) |
| rs115850745 | 6p21.32 | 32655508 | A | G | 0.371 | 0.265 | 0.56 (0.44, 0.71) | 1.05E-06 | 19kb 3' of AL662789.1, 21kb 3' of HLA-DQB1 |
| rs10961884 | 9p22.3 | 15111573 | T | C | 0.023 | 0.086 | 3.96 (2.23, 7.02) | 1.11E-06 | 32kb 5' of U6.1033, 59kb 3' of TTC39B |
| rs115899206 | 6p21.2 | 40084864 | A | G | 0.015 | 0.052 | 4.91 (2.49, 9.68) | 1.59E-06 | 154kb 3' of RP11-552E20.1, 183kb 3' of MOCS1 |
| rs12477565 | 2q14.2 | 121081260 | G | T | 0.420 | 0.514 | 1.68 (1.35, 2.08) | 2.25E-06 | AC012363.13 (intron), 22kb 5' of INHBB |
| rs12251624 | 10p14 | 8479257 | T | C | 0.159 | 0.091 | 0.46 (0.33, 0.64) | 2.62E-06 | 362kb 5' of GATA3, RP11-543F8.2 (intron) |
| rs7090925 | 10p14 | 8479868 | A | G | 0.159 | 0.091 | 0.46 (0.33, 0.64) | 2.62E-06 | 363kb 5' of GATA3, RP11-543F8.2 (intron) |
| rs17097185 | 5q31.3 | 140711097 | C | G | 0.039 | 0.077 | 3.02 (1.88, 4.84) | 2.70E-06 | PCDHGA1 (missense) |
| rs114799364 | 1p36.13 | 18900554 | C | T | 0.073 | 0.038 | 0.33 (0.20, 0.53) | 2.72E-06 | 57kb 5' of PAX7 |
| rs4151685 | 5q31.3 | 140213805 | A | C | 0.044 | 0.089 | 2.80 (1.79, 4.38) | 3.52E-06 | PCDHA1, PCDHA2, PCDHA3, PCDHA4, PCDHA5, PCDHA6 (intron for all) |
| rs34575154 | 5q31.3 | 140166953 | A | G | 0.044 | 0.089 | 2.80 (1.79, 4.38) | 3.56E-06 | PCDHA1 (missense) |
| rs370971 | 20q13.12 | 42371095 | G | A | 0.428 | 0.518 | 1.65 (1.33, 2.04) | 4.92E-06 | 15kb 3' of GTSF1L |
| rs6878145 | 5q31.3 | 140718552 | A | G | 0.039 | 0.076 | 2.94 (1.83, 4.73) | 4.94E-06 | PCDHGA1 (intron), PCDHGA2 (missense) |
| rs2993385 | 10p14 | 8462439 | T | C | 0.178 | 0.110 | 0.50 (0.37, 0.68) | 5.94E-06 | 345kb 5' of GATA3, RP11-543F8.2 (intron) |
| rs1329536 | 6p12.3 | 47663326 | C | T | 0.051 | 0.096 | 2.54 (1.68, 3.84) | 6.20E-06 | GPR111 (UTR-3) |
| rs116776862 | 5q31.3 | 140177075 | G | A | 0.045 | 0.089 | 2.67 (1.72, 4.14) | 6.86E-06 | PCDHA1 (intron) |
| rs13432692 | 2p25.2 | 6085200 | C | T | 0.541 | 0.443 | 0.63 (0.51, 0.77) | 7.02E-06 | LOC150622 (AC073479.1) (intron) |
| rs75404762 | 13q12.3 | 30562130 | T | C | 0.015 | 0.043 | 4.49 (2.27, 8.90) | 7.04E-06 | 38kb 5' of LOC440131 (RP11-90M5.1) |
| rs73146440 | 7q11.22 | 67865802 | A | G | 0.038 | 0.012 | 0.19 (0.09, 0.41) | 7.53E-06 | 113kb 5' of RP5-945F2.3, 1.1Mb 5' of STAG3L4 |
| rs285198 | 20q13.12 | 42364734 | G | A | 0.431 | 0.519 | 1.64 (1.32, 2.03) | 7.87E-06 | 9.1kb 3' of GTSF1L, 19.4kb 3' of MYBL2 |
| rs12057381 | 1p32.2 | 57769516 | G | A | 0.132 | 0.068 | 0.45 (0.31, 0.64) | 8.44E-06 | DAB1 (intron) |
| rs116679801 | 5q31.3 | 140711954 | C | G | 0.037 | 0.071 | 3.00 (1.83, 4.91) | 8.49E-06 | PCDHGA1 (missense) |
| rs2056150 | 2p24.3 | 14603290 | G | A | 0.321 | 0.405 | 1.70 (1.34, 2.15) | 1.03E-05 | 62kb 5' of NCRNA00276, 170kb 5' of FAM84A |
Location is per GRCh37.p5.
Gene information curated from Haploreg, dbSNP, Ensembl, and GENCODE.
Figure 1. Association results, recombination plot and linkage disequilibrium structure for the 198.9kb region of 10p14.
Association results from a trend test in –log10Pvalues (y axis, left) of the SNPs are shown according to their chromosomal positions (x axis). The r2 values were computed from rs7918885 to all of the other plotted SNPs to differentially size data points according to their level of linkage disequilibrium. SNPs that are highly correlated with rs7918885 (n=6) were colored red. Linkage disequilibrium structure based on controls (n=458) was visualized by snp.plotter software. The line graph shows likelihood ratio statistics (y axis, right) for recombination hotspot by SequenceLDhot software and 3 different colors represent 3 tests of 100 controls without resampling. Physical locations are based on NCBI Build 37 of the human genome. Gene annotation was based on ENCODE/GENCODE version 12 from the UCSC Genome Browser.
Results from a sub-analysis, stratified by Gleason score (<7, 136 cases; ≥7, 317 cases), are shown in Table 2. In the Gleason score ≥7 stratum, the 5q31.3 locus reached genome-wide statistical significance (5E−8) with the lowest p value being 3.66E−8 for rs34575154, a missense SNP in PCDHA1. In addition, SNPs at 22q13.31, 7q31.31 and 2q14.2 had p values less than E−7. In the Gleason score <7 stratum, seven loci were observed to have p values less than E−7, and two SNPs were below genome-wide statistical significance: rs985081 at Xq28 (p=8.66E−9) and rs2185710 at 6q21 (p=5.95E−8). We analyzed these SNPs stratified by Gleason score in the African Ancestry Prostate Cancer GWAS Consortium yet none of them replicated at p<0.05, although there was evidence for between-study heterogeneity of effect for several of these SNPs within the African American data (Haiman et al. 2011a; Haiman et al. 2011b).
Table 2.
The most promising SNP associations with prostate cancer stratified by Gleason score in the Ghana Prostate Study GWAS, with estimates also from the African Ancestry Prostate Cancer GWAS Consortium
| Ghana Prostate Study | African Ancestry Prostate Cancer GWAS Consortium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gleason | rs Number | Cytoband | Location | Alleles (referent | effect) |
Effect Allele Frequency (control | case) |
Per Effect Allele OR (95%CI) |
P value | In gene(s) or nearest gene(s) if
distance specified |
Effect Allele Frequency (control | case) |
Per Effect Allele OR (95%CI) |
P value | P Het |
| Greater than or even to 7 | rs34575154 | 5q31.3 | 140166953 | A | G | 0.044 | 0.098 | 3.79 (2.31, 6.20) | 3.66E-08 | PCDHA1 (missense) | 0.048 | 0.050 | 1.05 (0.87, 1.27) | 0.582 | 0.025 |
| rs4151685 | 5q31.3 | 140213805 | A | C | 0.044 | 0.098 | 3.77 (2.30, 6.16) | 4.02E-08 | PCDHA1 (intron); PCDHA@ | 0.045 | 0.047 | 1.06 (0.87, 1.28) | 0.586 | 0.028 | |
| rs116776862 | 5q31.3 | 140177075 | G | A | 0.045 | 0.098 | 3.53 (2.18, 5.73) | 9.07E-08 | PCDHA1 (intron); PCDHA@ | 0.048 | 0.050 | 1.04 (0.86, 1.26)* | 0.675 | 0.028 | |
| rs7706544 | 5q31.3 | 140070148 | C | T | 0.051 | 0.104 | 3.34 (2.09, 5.34) | 1.94E-07 | HARS (intron) | 0.052 | 0.054 | 1.04 (0.87, 1.25) | 0.665 | 0.053 | |
| rs115338764 | 5q31.3 | 140041187 | G | A | 0.051 | 0.104 | 3.34 (2.09, 5.34) | 1.98E-07 | IK (intron) | 0.051 | 0.054 | 1.04 (0.87, 1.25)* | 0.660 | 0.053 | |
| rs61749035 | 5q31.3 | 140718750 | G | T | 0.037 | 0.084 | 3.82 (2.26, 6.47) | 2.07E-07 | PCDHGA2 (missense); PCDHA@ | 0.045 | 0.047 | 1.10 (0.91, 1.34)* | 0.339 | 0.022 | |
| rs6889768 | 5q31.3 | 139929877 | T | G | 0.039 | 0.085 | 3.70 (2.20, 6.24) | 3.81E-07 | SRA1 (3'UTR) | 0.035 | 0.041 | 1.18 (0.95, 1.46) | 0.134 | 0.196 | |
| rs6008813 | 22q13.31 | 46812585 | G | A | 0.389 | 0.490 | 1.90 (1.47, 2.44) | 4.34E-07 | CELSR1 (intron) | 0.371 | 0.387 | 1.06 (0.97, 1.16)* | 0.183 | 0.889 | |
| kgp22385671 | 5q31.3 | 140208753 | A | G | 0.044 | 0.092 | 3.59 (2.16, 5.97) | 4.47E-07 | PCDHA1 (intron); PCDHA@ | 0.048 | 0.049 | 1.04 (0.86, 1.26)* | 0.654 | 0.029 | |
| rs113425597 | 5q31.3 | 140187201 | G | C | 0.044 | 0.091 | 3.55 (2.13, 5.91) | 6.36E-07 | PCDHA1 (synonymous); PCDHA@ | 0.048 | 0.050 | 1.04 (0.86, 1.26)* | 0.675 | 0.028 | |
| rs12537079 | 7q31.31 | 117704706 | T | G | 0.146 | 0.071 | 0.35 (0.23, 0.53) | 6.95E-07 | 61kb 5' of AC003084.2; 119kb 5' of NAA38 | 0.204 | 0.195 | 0.97 (0.87, 1.07) | 0.540 | 0.818 | |
| rs6880234 | 5q31.3 | 140215956 | C | G | 0.044 | 0.092 | 3.53 (2.12, 5.88) | 7.49E-07 | PCDHA7 (missense); PCDHA@ | 0.046 | 0.047 | 1.05 (0.87, 1.28)* | 0.603 | 0.027 | |
| rs12477565 | 2q14.2 | 121081260 | G | T | 0.420 | 0.528 | 1.85 (1.44, 2.38) | 8.18E-07 | AC012363.13; 22kb 5' of INHBB | 0.514 | 0.526 | 1.06 (0.97, 1.16)* | 0.222 | 0.571 | |
| rs17097185 | 5q31.3 | 140711097 | C | G | 0.039 | 0.084 | 3.55 (2.10, 5.98) | 8.41E-07 | PCDHGA1 (missense) | 0.045 | 0.047 | 1.10 (0.90, 1.33)* | 0.342 | 0.022 | |
| rs17119623 | 5q31.3 | 139961837 | T | C | 0.050 | 0.101 | 3.17 (1.97, 5.10) | 1.19E-06 | APBB3; 13kb 5' of SLC35A4 | 0.047 | 0.049 | 1.04 (0.86, 1.26) | 0.661 | 0.292 | |
| rs6878145 | 5q31.3 | 140718552 | A | G | 0.039 | 0.082 | 3.43 (2.03, 5.80) | 1.93E-06 | PCDHGA2 (missense); PCDHA@ | 0.045 | 0.047 | 1.10 (0.91, 1.34)* | 0.339 | 0.022 | |
| rs7715021 | 5q31.3 | 139941943 | C | G | 0.051 | 0.098 | 3.10 (1.92, 5.01) | 2.09E-06 | APBB3 (missense) | 0.050 | 0.053 | 1.07 (0.89, 1.28)* | 0.487 | 0.313 | |
| Less than 7 | rs985081 | Xq28 | 148538099 | T | C | 0.025 | 0.141 | 3.08 (2.01, 4.73) | 8.66E-09 | 22kb 3' of IDS | 0.070 | 0.082 | 1.08 (0.98, 1.20) | 0.112 | 0.192 |
| rs2185710 | 6q21 | 113377048 | A | G | 0.166 | 0.295 | 2.77 (1.89, 4.05) | 5.95E-08 | 84kb 3' of U6 | 0.334 | 0.348 | 1.05 (0.97, 1.14)* | 0.229 | 0.573 | |
| rs66504230 | 6q14.3 | 86871444 | T | C | 0.090 | 0.186 | 3.15 (2.01, 4.95) | 1.77E-07 | 169kb 5' of U4 | 0.113 | 0.106 | 0.94 (0.84, 1.06)* | 0.344 | 0.030 | |
| rs114918764 | 4q26 | 117370706 | C | T | 0.002 | 0.045 | 34.07 (6.45, 179.92) | 3.65E-07 | 40kb 3' of RP11-55L3.1; 150kb 5' of MIR1973 | 0.002 | 0.002 | 0.93 (0.23, 3.70)* | 0.915 | 0.280 | |
| rs73043340 | 3p22.3 | 33126972 | C | T | 0.007 | 0.042 | 12.04 (3.98, 36.46) | 3.76E-07 | GLB1 (intron) | 0.085 | 0.085 | 0.98 (0.86, 1.13)* | 0.819 | 0.455 | |
| rs62477096 | 7q31.33 | 124936442 | A | G | 0.178 | 0.283 | 2.45 (1.70, 3.53) | 6.63E-07 | RP11-3B12.2; 366kb 3' of POT1 | 0.196 | 0.194 | 0.99 (0.90, 1.09)* | 0.814 | 0.619 | |
| rs6965492 | 7q31.33 | 124944982 | G | T | 0.179 | 0.286 | 2.49 (1.72, 3.61) | 6.92E-07 | RP11-3B12.2; 375kb 3' of POT1 | 0.191 | 0.190 | 1.00 (0.91, 1.09) | 0.947 | 0.484 | |
Imputed using 1000 Genomes Project data
Lastly, in our prostate cancer GWAS of African men we were able to assess 81 of the previously reported 90 prostate cancer susceptibility loci (Table 3), and observed that 10 of these SNPs were statistically significant (p<0.05), including SNPs within “Region 1” (rs7017300, rs10090154) and “Region 2” (rs13254738, rs16901979) of the 8q24 region (Haiman et al. 2007), as well as rs7210100 from the 17q21 region which was initially reported by the African Ancestry Prostate Cancer GWAS Consortium (Haiman et al. 2011b). Other SNPs previously associated with prostate cancer, mostly in populations of European ancestry, did not replicate in our African population.
Table 3.
The association of previously reported prostate cancer susceptibility SNPs from European and Asian populations in relation to prostate cancer in African men.
| SNP | Cytoband | Location1 | Nearest Gene2 | Study PMID(s) | Study Population Ancestry | Ghana SNP | Location | Reference | Effect Alleles |
Case | Control Effect Allele Frequencies |
P value | Per Allele OR (95%CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1218582 | 1q21.3 | 154834183 | KCNN3 (intron) | 23535732 | European | rs1218582 | 154834183 | G | A | 0.31 | 0.33 | 0.12 | 0.84 (0.67, 1.05) |
| rs4245739 | 1q32.1 | 204518842 | MDM4 (3'-URT) | 23535732 | European | rs4245739 | 204518842 | C | A | 0.78 | 0.77 | 0.82 | 0.97 (0.76, 1.25) |
| rs10187424 | 2p11.2 | 85794297 | VAMP8 (intronic); 5.6kb 3' of GGCX | 21743467 | European, Asian | rs10187424 | 85794297 | T | C | 0.66 | 0.72 | 0.17 | 0.86 (0.68, 1.07) |
| rs721048 | 2p15 | 63131731 | EHBP1 (intron) | 18264098 | European | rs7210483 | 63131731 | G | A | 0.00 | 0.00 | 0.90 | 0.79 (0.02, 38.16) |
| rs1465618 | 2p21 | 43553949 | THADA (intron) | 19767753 | European, Asian | rs1465618 | 43553949 | T | C | 0.95 | 0.94 | 0.13 | 1.43 (0.90, 2.28) |
| rs11902236 | 2p25.1 | 10117868 | GRHL1 (intron) | 23535732 | European | rs11902236 | 10117868 | C | T | 0.65 | 0.69 | 0.31 | 0.89 (0.72, 1.11) |
| rs12621278 | 2q31.1 | 173311553 | ITGA6 (intron) | 19767753 | European, African American, Asian | - | - | - | - | - | - |
| rs2292884 | 2q37.3 | 238443226 | MLPH (missense) | 21743057 | European | - | - | - | - | - | - |
| rs3771570 | 2q37.3 | 242382864 | FARP2 (intron) | 23535732 | European | rs3771570 | 242382864 | C | T | 0.01 | 0.00 | 0.030 | 5.39 (1.17, 24.77) |
| rs7629490 | 3p11.2 | 87241497 | 34kb 3' of MIR4795 | 21743057 | European | rs7629490 | 87241497 | C | T | 0.17 | 0.21 | 0.12 | 0.81 (0.62, 1.06) |
| rs2055109 | 3p11.2 | 87467332 | 142kb 3' of POU1F1 | 22366784 | Asian | - | - | - | - | - | - |
| rs2660753 | 3p12.1 | 87110674 | 70kb 3' of VGLL3 | 18264097 | European | - | - | - | - | - | - |
| rs9284813 | 3p12.1 | 87152169 | 112kb 5' of VGLL3 | 20676098 | Asian | rs9284813 | 87152169 | A | G | 0.46 | 0.43 | 0.39 | 1.09 (0.89, 1.35) |
| rs17181170 | 3p12.1 | 87173324 | 102kb 3' of MIR4795 | 19767753 | European, Asian | rs17181170 | 87173324 | G | A | 0.12 | 0.16 | 0.031 | 0.72 (0.54, 0.97) |
| rs7611694 | 3q13.2 | 113275624 | SIDT1 (intron) | 23535732 | European | rs7611694 | 113275624 | A | C | 0.34 | 0.37 | 0.29 | 0.89 (0.71, 1.11) |
| rs10934853 | 3q21.3 | 128038373 | EEFSEC (intron) | 19767754 | European | rs10934853 | 128038373 | C | A | 0.82 | 0.80 | 0.53 | 1.09 (0.83, 1.42) |
| rs6763931 | 3q23 | 141102833 | ZBTB38 (intron) | 21743467 | European | rs6763931 | 141102833 | G | A | 0.91 | 0.90 | 0.019 | 1.53 (1.07, 2.18) |
| rs10936632 | 3q26.2 | 170130102 | RP11-469J4.3; 6.5kb 5' of CLDN11 | 21743467 | European | rs109366323 | 170130102 | C | A | 0.15 | 0.18 | 0.051 | 0.75 (0.57, 1.00) |
| rs1894292 | 4q13.3 | 74349158 | AFM (intron) | 23535732 | European | rs1894292 | 74349158 | G | A | 0.26 | 0.27 | 0.76 | 0.96 (0.76, 1.22) |
| rs12500426 | 4q22.3 | 95514609 | PDLIM5 (intron) | 19767753 | European | rs12500426 | 95514609 | A | C | 0.61 | 0.62 | 0.90 | 0.99 (0.80, 1.22) |
| rs17021918 | 4q22.3 | 95562877 | PDLIM5 (intron) | 19767753 | European | rs17021918 | 95562877 | C | T | 0.21 | 0.21 | 0.71 | 1.05 (0.81, 1.36) |
| rs7679673 | 4q24 | 106061534 | AC004069.2; 5.5kb 5' of TET2 | 19767753 | European | rs7679673 | 106061534 | C | A | 0.60 | 0.63 | 0.010 | 0.75 (0.60, 0.93) |
| rs2121875 | 5p12 | 44365545 | FGF10 intronic | 21743467 | European | rs2121875 | 44365545 | C | A | 0.20 | 0.20 | 0.79 | 0.97 (0.74, 1.25) |
| rs2242652 | 5p15.33 | 1280028 | TERT (intron) | 21743467 | European | rs22426523 | 1280028 | G | A | 0.12 | 0.13 | 0.34 | 0.85 (0.61, 1.19) |
| rs12653946 | 5p15.33 | 1895829 | CTD-2194D22.4; 13kb 3' of IRX4 | 20676098 | Asian | rs12653946 | 1895829 | C | T | 0.40 | 0.42 | 0.25 | 0.88 (0.72, 1.09) |
| rs6869841 | 5q35.1 | 172939426 | 28kb 3' of CTB-164N12.1 | 23535732 | European | rs6869841 | 172939426 | C | T | 0.37 | 0.36 | 0.36 | 1.11 (0.89, 1.38) |
| rs3096702 | 6p21.32 | 32192331 | 486bp 5' of NOTCH4 | 23535732 | European | rs3096702 | 32192331 | A | G | 0.87 | 0.87 | 0.72 | 1.06 (0.77, 1.45) |
| rs130067 | 6p21.33 | 31118511 | CCHCR1 (missense) | 21743467 | European | - | - | - | - | - | - |
| rs2273669 | 6q21 | 109285189 | ARMC2 (intron) | 23535732 | European | rs2273669 | 109285189 | A | G | 0.37 | 0.35 | 0.22 | 1.15 (0.92, 1.42) |
| rs339331 | 6q22.1 | 117210052 | RFX6 (intron) | 20676098 | Asian | rs339331 | 117210052 | T | C | 0.17 | 0.23 | 0.002 | 0.67 (0.52, 0.87) |
| rs1933488 | 6q25.2 | 153441079 | RGS17 (intron) | 23535732 | European | rs1933488 | 153441079 | A | G | 0.43 | 0.44 | 0.78 | 0.97 (0.78, 1.20) |
| rs651164 | 6q25.3 | 160581374 | 1.6kb 3' of SLC22A1 | 19767753; 21743057 | European, Asian; European | rs651164 | 160581374 | A | G | 0.76 | 0.74 | 0.42 | 1.12 (0.85, 1.49) |
| rs9364554 | 6q25.3 | 160833664 | SLC22A3 (intron) | 18264097 | European | rs9364554 | 160833664 | C | T | 0.01 | 0.01 | 0.45 | 1.49 (0.53, 4.18) |
| rs10486567 | 7p15.2 | 27976563 | JAZF1 (intron) | 18264096 | European | rs10486567 | 27976563 | G | A | 0.25 | 0.26 | 0.27 | 0.88 (0.69, 1.11) |
| rs12155172 | 7p15.3 | 20994491 | AC006481.1 | 23535732 | European | rs12155172 | 20994491 | A | G | 0.88 | 0.88 | 0.85 | 0.97 (0.70, 1.34) |
| rs6465657 | 7q21.3 | 97816327 | LMTK2 (intron) | 18264097 | European | rs6465657 | 97816327 | C | T | 0.01 | 0.02 | 0.18 | 0.54 (0.22, 1.33) |
| rs1512268 | 8p21.2 | 23526463 | 9.7kb 3' of NKX3-1 | 19767753 | European, African American, Asian | - | - | - | - | - | - |
| rs11135910 | 8p21.2 | 25892142 | EBF2 (intron) | 23535732 | European | rs11135910 | 25892142 | C | T | 0.09 | 0.09 | 0.91 | 1.02 (0.70, 1.49) |
| rs13252298 | 8q24.21 | 128095156 | 689bp 5' of RP11-255B23.2 | 19767752 | European | rs13252298 | 128095156 | A | G | 0.01 | 0.01 | 0.70 | 1.27 (0.39, 4.14) |
| rs7841060 | 8q24.21 | 128096477 | 2kb 5' of RP11-255B23.2 | 19767755 | European | rs7841060 | 128096477 | T | G | 0.34 | 0.37 | 0.85 | 0.98 (0.79, 1.21) |
| rs16901979 | 8q24.21 | 128124916 | 30kb 5' of RP11-255B23.2; 303kb 5' of POU5F1B | 17401366 | European | rs169019793 | 128124916 | C | A | 0.56 | 0.50 | 0.022 | 1.27 (1.04, 1.57) |
| rs188140481 | 8q24.21 | 128191672 | 6.2kb 3' of RP11-255B23.4 | 23104005 | European | - | - | - | - | - | - |
| rs16902094 | 8q24.21 | 128320346 | RP11-382A18.1 | 19767754 | European | rs16902094 | 128320346 | A | G | 0.10 | 0.11 | 0.65 | 0.92 (0.66, 1.30) |
| rs620861 | 8q24.21 | 128335673 | RP11-382A18.1; 92kb 5' of POU5F1B | 19767752; 19767755 | European; European | rs620861 | 128335673 | G | A | 0.33 | 0.33 | 0.91 | 0.99 (0.79, 1.23) |
| rs6983267 | 8q24.21 | 128413305 | RP11-382A18.1; 15kb 5' of POU5F1B | 17401363; 18264097; 18264096; 19767752 | European and African American; European; European; European | rs6983267 | 128413305 | G | T | 0.02 | 0.02 | 0.51 | 1.28 (0.62, 2.63) |
| rs6999921 | 8q24.21 | 128440928 | RP11-382A18.1 | 21743057 | European | rs6999921 | 128440928 | A | G | 0.13 | 0.09 | 0.060 | 1.38 (0.99, 1.93) |
| rs1447293 | 8q24.21 | 128472320 | RP11-382A18.1 (intron) | 21743057 | European | rs1447293 | 128472320 | C | T | 0.08 | 0.06 | 0.053 | 1.51 (1.00, 2.29) |
| rs10090154 | 8q24.21 | 128532137 | 38kb 5' of RP11-382A18.1; 38kb 3' of LOC727677 | 19767752 | European | rs100901543 | 128532137 | T | C | 0.80 | 0.85 | 0.005 | 0.67 (0.51, 0.89) |
| rs7837688 | 8q24.21 | 128539360 | 45kb 5' of RP11-382A18.1 | 20676098 | Asian | rs7837688 | 128539360 | T | G | 0.94 | 0.96 | 0.063 | 0.64 (0.40, 1.02) |
| rs817826 | 9q31.2 | 110156300 | 26kb 3' of RP11-363D24.1; 62kb 5' of RAD23B | 23023329 | Asian | rs817826 | 110156300 | C | T | 0.68 | 0.65 | 0.47 | 1.09 (0.87, 1.36) |
| rs10993994 | 10q11.23 | 51549496 | TIMM23B; 55bp 5' of MSMB | 18264097; 18264096 | European; European | rs10993994 | 51549496 | T | C | 0.33 | 0.34 | 0.94 | 1.01 (0.81, 1.26) |
| rs3850699 | 10q24.32 | 104414221 | TRIM8 (intron) | 23535732 | European | rs3850699 | 104414221 | A | G | 0.41 | 0.44 | 0.45 | 0.92 (0.74, 1.14) |
| rs2252004 | 10q26.12 | 122844709 | 94kb 5' of RP11-159H3.2; 176kb 5' of WDR11 | 22366784 | Asian | rs2252004 | 122844709 | C | A | 0.65 | 0.63 | 0.52 | 1.07 (0.87, 1.33) |
| rs10749408 | 10q26.12 | 122967526 | 22kb 3' of RP11-159H3.2; 270kb 3' of FGFR2 | 22130093 | European | rs107494083 | 122967526 | C | T | 0.81 | 0.83 | 0.39 | 0.89 (0.68, 1.16) |
| rs11199874 | 10q26.12 | 123032519 | 87kb 3' of RP11-159H3.2; 205kb 3' of FGFR2 | 22130093 | European | rs111998743 | 123032519 | G | A | 0.11 | 0.12 | 0.30 | 0.84 (0.61, 1.17) |
| rs4962416 | 10q26.13 | 126696872 | CTBP2 (intron) | 18264096 | European | rs4962416 | 126696872 | T | C | 0.16 | 0.13 | 0.055 | 1.33 (0.99, 1.79) |
| rs7127900 | 11p15.5 | 2233574 | 32kb 3' of AC132217.2; 39kb 5' of MIR4686 | 19767753 | European | rs7127900 | 2233574 | A | G | 0.55 | 0.57 | 0.50 | 0.93 (0.75, 1.15) |
| rs1938781 | 11q12.1 | 58915110 | FAM111A (intron) | 22366784 | Asian | rs1938781 | 58915110 | A | G | 0.35 | 0.32 | 0.086 | 1.22 (0.97, 1.53) |
| rs10896438 | 11q13.3 | 68906570 | 8.1kb 5' of RP11-554A11.3; 48kb 5' of TPCN2 | 21531787 | European | rs10896438 | 68906570 | T | G | 0.20 | 0.23 | 0.96 | 0.99 (0.77, 1.29) |
| rs12793759 | 11q13.3 | 68974555 | 87kb 5' of MYEOV | 21531787 | European | rs127937593 | 68974555 | G | A | 0.09 | 0.08 | 0.21 | 1.26 (0.87, 1.82) |
| rs10896449 | 11q13.3 | 68994667 | 67kb 5' of MYEOV | 18264096; 21531787 | European; European | rs10896449 | 68994667 | A | G | 0.79 | 0.74 | 0.007 | 1.40 (1.10, 1.79) |
| rs7940107 | 11q13.3 | 69027770 | 34kb 5' of MYEOV | 21743057 | European | rs7940107 | 69027770 | G | A | 0.34 | 0.36 | 0.69 | 0.96 (0.77, 1.19) |
| rs11568818 | 11q22.2 | 102401661 | 176bp 5' of MMP7 | 23535732 | European | rs11568818 | 102401661 | T | C | 0.45 | 0.44 | 0.89 | 1.02 (0.83, 1.25) |
| rs10875943 | 12q13.12 | 49676010 | 8.9kb 5' of TUBA1C | 21743467 | European | rs108759433 | 49676010 | T | C | 0.75 | 0.71 | 0.11 | 1.21 (0.96, 1.53) |
| rs902774 | 12q13.13 | 53273904 | 17kb 3' of KRT8 | 21743057 | European | rs902774 | 53273904 | G | A | 0.09 | 0.08 | 0.087 | 1.39 (0.95, 2.02) |
| rs1270884 | 12q24.21 | 114685571 | 6kb 5' of RP11-139B1.1 | 23535732 | European | rs1270884 | 114685571 | A | G | 0.87 | 0.87 | 0.80 | 0.96 (0.71, 1.30) |
| rs9600079 | 13q22.1 | 73728139 | 14kb 3' of U7.97; 76kb 5' of KLF5 | 20676098 | Asian | rs9600079 | 73728139 | G | T | 0.54 | 0.62 | 0.002 | 0.71 (0.58, 0.88) |
| rs8008270 | 14q22.1 | 53372330 | FERMT2 (intron) | 23535732 | European | rs8008270 | 53372330 | T | C | 0.70 | 0.67 | 0.093 | 1.21 (0.97, 1.51) |
| rs7141529 | 14q24.1 | 69126744 | RAD51B | 23535732 | European | rs7141529 | 69126744 | T | C | 0.58 | 0.55 | 0.59 | 1.06 (0.85, 1.32) |
| rs1994198 | 15q21.1 | 46653167 | 9.2kb 5' of SNORD11.1; 670kb 5' of SQRDL | 22130093 | European | rs1994198 | 46653167 | C | T | 0.31 | 0.31 | 0.88 | 1.02 (0.81, 1.27) |
| rs684232 | 17p13.3 | 618965 | VPS53 | 23535732 | European | rs684232 | 618965 | T | C | 0.74 | 0.76 | 0.76 | 0.96 (0.76, 1.22) |
| rs11649743 | 17q12 | 36074979 | HNF1B (intron) | 18758462 | European | rs11649743 | 36074979 | G | A | 0.04 | 0.04 | 0.71 | 1.12 (0.63, 1.98) |
| rs4430796 | 17q12 | 36098040 | HNF1B (intron) | 17603485 | European | rs4430796 | 36098040 | G | A | 0.30 | 0.28 | 0.15 | 1.18 (0.94, 1.48) |
| rs7501939 | 17q12 | 36101156 | HNF1B (intron) | 19767753; 18264097; 17603485; 19318570 | European, Asian; European; European; European | rs7501939 | 36101156 | T | C | 0.40 | 0.38 | 0.64 | 1.05 (0.85, 1.29) |
| rs138213197 | 17q21.3 | 46805705 | HOXB13 (missense) | 23104005 | European | - | - | - | - | - | - |
| rs11650494 | 17q21.32 | 47345186 | 9.2kb 3' of RP1-62O9.3 | 23535732 | European | rs11650494 | 47345186 | G | A | 0.29 | 0.27 | 0.37 | 1.11 (0.88, 1.41) |
| rs7210100 | 17q21.33 | 47436749 | ZNF652 (intron) | 21602798 | African American | rs7210100 | 47436749 | G | A | 0.08 | 0.05 | 0.008 | 1.78 (1.16, 2.72) |
| rs1859962 | 17q24.3 | 69108753 | 186kb 3' of U7.34; 933kb 5' of KCNJ2 | 17603485 | European | rs1859962 | 69108753 | G | T | 0.76 | 0.78 | 0.40 | 0.90 (0.71, 1.15) |
| rs7241993 | 18q23 | 76773973 | 11kb 3' of SALL3 | 23535732 | European | rs7241993 | 76773973 | C | T | 0.58 | 0.58 | 0.85 | 1.02 (0.83, 1.26) |
| rs8102476 | 19q13.2 | 38735613 | 6.3kb 3' of PPP1R14A | 19767754 | European | rs8102476 | 38735613 | C | T | 0.16 | 0.16 | 0.51 | 0.91 (0.69, 1.20) |
| rs11672691 | 19q13.2 | 41985587 | 35kb 3' of C19orf69 | 23065704 | European | rs11672691 | 41985587 | A | G | 0.06 | 0.06 | 0.92 | 0.98 (0.64, 1.49) |
| rs2735839 | 19q13.33 | 51364623 | 602bp 5' of KLK3 | 18264097 | European | rs2735839 | 51364623 | A | G | 0.67 | 0.67 | 0.64 | 1.05 (0.85, 1.30) |
| rs103294 | 19q13.42 | 54797848 | 2kb 3' of LILRA3 | 23023329 | Asian | rs103294 | 54797848 | C | T | 0.08 | 0.06 | 0.20 | 1.31 (0.87, 1.96) |
| rs2427345 | 20q13.33 | 61015611 | 9kb 3' of RP5-908M14.5 | 23535732 | European | rs2427345 | 61015611 | C | T | 0.56 | 0.55 | 0.81 | 1.03 (0.83, 1.27) |
| rs6062509 | 20q13.33 | 62362563 | ZGPAT (intron) | 23535732 | European | rs6062509 | 62362563 | G | T | 0.94 | 0.95 | 0.79 | 0.94 (0.60, 1.48) |
| rs5759167 | 22q13.2 | 43500212 | 6.5kb 5' of BIK | 21390317 | European | rs5759167 | 43500212 | G | T | 0.17 | 0.19 | 0.53 | 0.92 (0.70, 1.20) |
| rs742134 | 22q13.2 | 43518275 | BIK (intron) | 21743057 | European | rs742134 | 43518275 | A | G | 0.85 | 0.81 | 0.055 | 1.32 (0.99, 1.76) |
| rs5945572 | Xp11.22 | 51229683 | 3.2kb 3' of NUDT11 | 18264098 | European | rs59455723 | 51229683 | A | G | 0.88 | 0.90 | 0.31 | 0.88 (0.68, 1.13) |
| rs2405942 | Xp22.2 | 9814135 | SHROOM2 (intron) | 23535732 | European | - | - | - | - | - | - |
| rs5919432 | Xq12 | 67021550 | 77kb 5' of AR | 21743467 | European | rs59194323 | 67021550 | C | T | 0.14 | 0.14 | 0.56 | 1.08 (0.84, 1.39) |
Location is per GRCh37.p5.
Gene information curated from Haploreg, dbSNP, Ensembl, and GENCODE.
Imputed using 1000 Genomes Project data. Table includes SNPs with published p values <5x10-8 for overall, advanced or aggressive prostate cancer. Loci detected in European populations only were pruned using 1000 Genomes Project data with pairwise r2 > 0.3 and the SNP with lower p value preferably chosen to be listed in the table.
Discussion
In this GWAS of prostate cancer in West African men, there is evidence for an association between a possible new locus at 10p14 and prostate cancer. In an additional sub-analysis, we observed SNPs at 5q31.3 associated with high Gleason score (≥7) prostate cancers, and of SNPs at Xq28 and 6q21 in relation to low Gleason score (<7) prostate cancers; however, these observations require further confirmation because of the small sample size reported herein. Because of the noted racial disparities in prostate cancer incidence between men of European and African descent, this unique analysis may provide insight into prostate cancer pathogenesis, particularly for African populations. Lastly, of the 90 prostate cancer loci reported from studies of men of European, Asian or African American ancestry, we were able to test 81 in the Ghana Prostate Study, and 10 of these replicated at p<0.05, differences that may be partly ascribed to distinct genomic architecture (including different patterns of underlying linkage disequilibrium), heterogeneous prostate cancer populations, and uncharacterized gene-environment interactions.
The strongest associated SNP, rs7918885, localizes to 10p14 and is approximately 360 kb 5’ of GATA3 within an intron of the lncRNA gene RP11-543F8.2. An assessment of highly correlated SNPs using the 1000 Genomes Project data did not reveal splice or exonic variants of RP11-543F8.2. In the NHGRI catalog of GWAS SNPs, the closest SNP associated with any cancer resides 227kb away (Chung and Chanock 2011; Hindorff et al. 2009). The locus is marked by rs10795668 in a colorectal GWAS but its underlying biology is still unknown (Tomlinson et al. 2008). We also note that there is minimal linkage disequilibrium between that rs10795668 and rs7918885 (r2=0.001 in scan control set); moreover, there are several recombination-hot spots between the two markers.
Although we detected a promising association between the 10p14 SNPs and prostate cancer in this study of West African men, the highest p value did not quite reach the genome-wide threshold of 5×10−8. It is possible that the signal we have detected at 10p14 is due to chance, but more likely insufficient power to detect a low effect SNP (Donnelly 2008; Kraft and Hunter 2009). We had 64% power to detect rs7918885 under 5E-8 and based on the observed allele frequencies and odds ratio.
We did attempt to replicate the findings presented herein using a large meta-analytic dataset comprised of African Americans—the African Ancestry Prostate Cancer GWAS Consortium—, but only one SNP (rs2993385 at 10p14) replicated at p<0.05. There are three primary, non-mutually exclusive reasons why genetic signals may not be observed in the ancestrally-related, but distinct, African American population. The first reason is that the genomic architecture of African American men is distinct from that of African men. A recent analysis estimated that the African American population has continuously received gene flow from European populations over 14 generations (Jin et al. 2012) that has resulted in the African American population having mean ancestral percentages of 75% West African, 20% European, and 5% Native American (Bryc et al. 2010; Tishkoff et al. 2009; Zakharia et al. 2009). Individuals who self-identify as African American are highly variable in degree of West African admixture (Bryc et al. 2010; Henn et al. 2010; Jin et al. 2012; Tishkoff et al. 2009). There are also large differences in allele frequencies (Adeyemo and Rotimi 2010; Ntzani et al. 2012), private SNPs (Campbell and Tishkoff 2008; Conrad et al. 2006), patterns of homozygosity (Pemberton et al. 2012), and LD structure (Campbell and Tishkoff 2008; Conrad et al. 2006) between African and African American populations. In addition, African ancestry of African Americans is most similar to the non-Bantu Niger-Kordofanian population of West Africa (Bryc et al. 2010; Tishkoff et al. 2009), tribes of which overlap with the geographical location of the Ghana Prostate Study.
The second reason is screening. In the United States the use of DRE and PSA screening has been widespread and has resulted in over-diagnosis (Welch and Black 2010) and a concurrent change in Gleason score distribution. SEER data indicates that, of the black men diagnosed with prostate cancer during 2003–2008, 1% were reported with Gleason 2-4, 48% with Gleason 5-6, and 51% with Gleason 7-10. Respective percentages for the 474 Ghanaian cases in the study were 1%, 29% and 70%. The greater proportion of advanced grades may be expected given that only 70 cases in the final analysis were detected through the population screening component of our study; a majority of the remaining 404 were symptomatic cases who presented at the clinic; the prevalence of PSA screening in Ghana, during the study period, was low (4% vs. >50% in US). Thus the majority of our case population is comprised of symptomatic disease rather than screen-detected cancers, which is especially important for this malignancy given it is expected to develop in 80% of men by age 80 years (Franks 1954; Haas et al. 2008; Sakr et al. 1993).
The third reason could be related to uncharacterized gene-environment interactions (Hemminki et al. 2006; Perez-Losada et al. 2011). In support of a strong environmental component is the fact prostate cancer mortality rates in the US are geographically variable both between and within racial groups 1, yet neither migration patterns (Tolnay 2003) nor genetic diversity (Zakharia et al. 2009) of the African American population can account for such patterns and trends.
Differences in population genetics, environments, and disease spectra can account for differences in observed associations with complex diseases across ancestrally diverse groups (Ioannidis 2007; Ntzani et al. 2012). For example, the African Ancestry Prostate Cancer GWAS Consortium replicated approximately half of the 49 SNPs previously associated and validated in populations of European ancestry (Haiman et al. 2011a). Other studies of European sub-populations (Gaj et al. 2012; Vijai et al. 2011), Asian populations (Liu et al. 2012; Takata et al. 2010) and populations of African descent (Chang et al. 2005; Xu et al. 2011) have reported similar results. Conversely, prostate cancer risk loci discrete to specific non-Caucasian populations have also been described (Akamatsu et al. 2012; Batra et al. 2011; Haiman et al. 2011b; Takata et al. 2010; Wang et al. 2012). In addition, functional studies support the idea that some of these differences by ancestry may have a biological grounding (Grisanzio et al. 2012). In summary, we present evidence for a promising novel prostate cancer locus at 10p14 in a West African population in our initial GWAS. Further studies of prostate cancer in West African men are required for validation of this locus as well as those associated with low or high Gleason score prostate cancers. Further efforts are required to recruit greater numbers of cases in high quality epidemiologic studies to investigate the underlying genetics—and in turn gene-environment interactions—of a complex disease such as prostate cancer.
Supplementary Material
Acknowledgements
The authors thank Ms. Vicky Okyne for her expert help in coordinating the study; consultants/resident urologists, pathologists, nurses, and interviewers of Korle-Bu Hospital and University of Ghana Medical School for their assistance with subject enrollment, screening, and clinical examination; the study participants for their contribution toward a better understanding of prostate disease; A. DeMarzo and G. Netto of Johns Hopkins University for pathology review; Ms. Violet Devairakkam, Ms. Norma Kim, and Mr. John Heinrich of Research Triangle Institute (RTI) for their expert study management; Prof. Rosalind A. Eeles and her team for cross-checking published prostate cancer loci specified in Table 3; and members of the African Ancestry Prostate Cancer GWAS Consortium for looking-up our most promising associations from our African GWAS. This research was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health.
FUNDING
Intramural Program of the National Cancer Institute, National Institutes of Health, Department of Health and Human Services including Contract No. HHSN261200800001E.
Footnotes
There are no financial disclosures from any of the authors.
References
- Database of Single Nucleotide Polymorphisms (dbSNP) Bethesda (MD): National Center for Biotechnology Information, National Library of Medicine. (dbSNP Build ID 137); Available from: http://www.ncbi.nlm.nih.gov/SNP/ [Google Scholar]
- An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. doi: http://www.nature.com/nature/journal/v489/n7414/abs/nature11247.html#supplementaryinformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemo A, Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics. 2010;13:72–79. doi: 10.1159/000218711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu S, Takata R, Haiman CA, Takahashi A, Inoue T, Kubo M, Furihata M, Kamatani N, Inazawa J, Chen GK, Le Marchand L, Kolonel LN, Katoh T, Yamano Y, Yamakado M, Takahashi H, Yamada H, Egawa S, Fujioka T, Henderson BE, Habuchi T, Ogawa O, Nakamura Y, Nakagawa H. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nature Genetics. 2012;44:426–429. doi: 10.1038/ng.1104. S1. [DOI] [PubMed] [Google Scholar]
- Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Balter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Batra J, Lose F, Chambers S, Gardiner RA, Aitken J, Yaxley J, Clements JA, Spurdle AB Australian Prostate Cancer B. A replication study examining novel common single nucleotide polymorphisms identified through a prostate cancer genome-wide association study in a Japanese population. American Journal of Epidemiology. 2011;174:1391–1395. doi: 10.1093/aje/kwr271. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Research. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo JM, Wambebe C, Tishkoff SA, Bustamante CD. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA. African genetic diversity: Implications for human demographic history, modern human origins, and complex disease mapping. Annual Review of Genomics and Human Genetics. 2008;vol 9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center MM, Jemal A, Lortet-Tieulent J, Ward E, Ferlay J, Brawley O, Bray F. International variation in prostate cancer incidence and mortality rates. European Urology. 2012;61:1079–1092. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- Chang BL, Isaacs SD, Wiley KE, Gillanders EM, Zheng SL, Meyers DA, Walsh PC, Trent JM, Xu J, Isaacs WB. Genome-wide screen for prostate cancer susceptibility genes in men with clinically significant disease. Prostate. 2005;64:356–361. doi: 10.1002/pros.20249. [DOI] [PubMed] [Google Scholar]
- Chokkalingam AP, Yeboah ED, Demarzo A, Netto G, Yu K, Biritwum RB, Tettey Y, Adjei A, Jadallah S, Li Y, Chu LW, Chia D, Niwa S, Partin A, Thompson IM, Roehrborn C, Hoover RN, Hsing AW. Prevalence of BPH and lower urinary tract symptoms in West Africans. Prostate Cancer Prostatic Dis. 2012;15:170–176. doi: 10.1038/pcan.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu LW, Ritchey J, Devesa SS, Quraishi SM, Zhang H, Hsing AW. Prostate cancer incidence rates in Africa. Prostate Cancer. 2011;2011:947870. doi: 10.1155/2011/947870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Human Genetics. 2011;130:59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- Clarke L, Zheng-Bradley X, Smith R, Kulesha E, Xiao C, Toneva I, Vaughan B, Preuss D, Leinonen R, Shumway M, Sherry S, Flicek P, Genomes Project C. The 1000 Genomes Project: data management and community access. Nat Methods. 2012;9:459–62. doi: 10.1038/nmeth.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nature Genetics. 2006;38:1251–60. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- Engelhardt BE, Stephens M. Analysis of population structure: a unifying framework and novel methods based on sparse factor analysis. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Franks LM. Latent carcinoma of the prostate. Journal of Pathology and Bacteriology. 1954;68:603–616. doi: 10.1002/path.1700680233. [DOI] [PubMed] [Google Scholar]
- Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, Oakley-Girvan I, Whittemore AS, Cooney KA, Ingles SA, Altshuler D, Henderson BE, Reich D. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj P, Maryan N, Hennig EE, Ledwon JK, Paziewska A, Majewska A, Karczmarski J, Nesteruk M, Wolski J, Antoniewicz AA, Przytulski K, Rutkowski A, Teumer A, Homuth G, Starzynska T, Regula J, Ostrowski J. Pooled sample-based GWAS: a cost-effective alternative for identifying colorectal and prostate cancer risk variants in the Polish population. PLoS One. 2012;7:e35307. doi: 10.1371/journal.pone.0035307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisanzio C, Werner L, Takeda D, Awoyemi BC, Pomerantz MM, Yamada H, Sooriakumaran P, Robinson BD, Leung R, Schinzel AC, Mills I, Ross-Adams H, Neal DE, Kido M, Yamamoto T, Petrozziello G, Stack EC, Lis R, Kantoff PW, Loda M, Sartor O, Egawa S, Tewari AK, Hahn WC, Freedman ML. Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11252–11257. doi: 10.1073/pnas.1200853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas GP, Delongchamps N, Brawley OW, Wang CY, de la Roza G. The worldwide epidemiology of prostate cancer: perspectives from autopsy studies. Can J Urol. 2008;15:3866–3871. [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, Rybicki BA, Isaacs WB, Ingles SA, Stanford JL, Diver WR, Witte JS, Chanock SJ, Kolb S, Signorello LB, Yamamura Y, Neslund-Dudas C, Thun MJ, Murphy A, Casey G, Sheng X, Wan P, Pooler LC, Monroe KR, Waters KM, Le Marchand L, Kolonel LN, Stram DO, Henderson BE. Characterizing genetic risk at known prostate cancer susceptibility loci in African Americans. PLoS Genet. 2011a;7:e1001387. doi: 10.1371/journal.pgen.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, Rybicki BA, Isaacs WB, Ingles SA, Stanford JL, Diver WR, Witte JS, Hsing AW, Nemesure B, Rebbeck TR, Cooney KA, Xu J, Kibel AS, Hu JJ, John EM, Gueye SM, Watya S, Signorello LB, Hayes RB, Wang Z, Yeboah E, Tettey Y, Cai Q, Kolb S, Ostrander EA, Zeigler-Johnson C, Yamamura Y, Neslund- Dudas C, Haslag-Minoff J, Wu W, Thomas V, Allen GO, Murphy A, Chang BL, Zheng SL, Leske MC, Wu SY, Ray AM, Hennis AJ, Thun MJ, Carpten J, Casey G, Carter EN, Duarte ER, Xia LY, Sheng X, Wan P, Pooler LC, Cheng I, Monroe KR, Schumacher F, Le Marchand L, Kolonel LN, Chanock SJ, Berg DV, Stram DO, Henderson BE. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nature Genetics. 2011b;43:570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, Greenway SC, Stram DO, Le Marchand L, Kolonel LN, Frasco M, Wong D, Pooler LC, Ardlie K, Oakley-Girvan I, Whittemore AS, Cooney KA, John EM, Ingles SA, Altshuler D, Henderson BE, Reich D. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki K, Lorenzo Bermejo J, Forsti A. The balance between heritable and environmental aetiology of human disease. Nat Rev Genet. 2006;7:958–965. doi: 10.1038/nrg2009. [DOI] [PubMed] [Google Scholar]
- Henn BM, Gravel S, Moreno-Estrada A, Acevedo-Acevedo S, Bustamante CD. Fine-scale population structure and the era of next-generation sequencing. Human Molecular Genetics. 2010;19:R221–R226. doi: 10.1093/hmg/ddq403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP. Non-replication and inconsistency in the genome-wide association setting. Human Heredity. 2007;64:203–213. doi: 10.1159/000103512. [DOI] [PubMed] [Google Scholar]
- Jin W, Wang S, Wang H, Jin L, Xu S. Exploring Population Admixture Dynamics via Empirical and Simulated Genome-wide Distribution of Ancestral Chromosomal Segments. American Journal of Human Genetics. 2012;91:849–862. doi: 10.1016/j.ajhg.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, Hunter DJ. Genetic risk prediction--are we there yet? New England Journal of Medicine. 2009;360:1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- Li J, German R, King J, Joseph D, Thompson T, Wu XC, Ajani U, Tai E. Recent trends in prostate cancer testing and incidence among men under age of 50. Cancer Epidemiol. 2012;36:122–127. doi: 10.1016/j.canep.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Liu M, Wang J, Xu Y, Wei D, Shi X, Yang Z. Risk loci on chromosome 8q24 are associated with prostate cancer in northern Chinese men. Journal of Urology. 2012;187:315–321. doi: 10.1016/j.juro.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Ntzani EE, Liberopoulos G, Manolio TA, Ioannidis JPA. Consistency of genome-wide associations across major ancestral groups. Human Genetics. 2012;131:1057–1071. doi: 10.1007/s00439-011-1124-4. [DOI] [PubMed] [Google Scholar]
- Park JH, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin DM, Ferlay J, Curado MP, Bray F, Edwards B, Shin HR, Forman D. Fifty years of cancer incidence: CI5 I-IX. International Journal of Cancer. 2010;127:2918–2927. doi: 10.1002/ijc.25517. [DOI] [PubMed] [Google Scholar]
- Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. Genomic Patterns of Homozygosity in Worldwide Human Populations. American Journal of Human Genetics. 2012;91:275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Losada J, Castellanos-Martin A, Mao JH. Cancer evolution and individual susceptibility. Integr Biol (Camb) 2011;3:316–328. doi: 10.1039/c0ib00094a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck TR, Devesa SS, Chang BL, Bunker CH, Cheng I, Cooney K, Eeles R, Fernandez P, Giri VN, Gueye SM, Haiman CA, Henderson BE, Heyns CF, Hu JJ, Ingles SA, Isaacs W, Jalloh M, John EM, Kibel AS, Kidd LR, Layne P, Leach RJ, Neslund-Dudas C, Okobia MN, Ostrander EA, Park JY, Patrick AL, Phelan CM, Ragin C, Roberts RA, Rybicki BA, Stanford JL, Strom S, Thompson IM, Witte J, Xu J, Yeboah E, Hsing AW, Zeigler-Johnson CM. Global patterns of prostate cancer incidence, aggressiveness, and mortality in men of african descent. Prostate Cancer. 2013;2013:560857. doi: 10.1155/2013/560857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Price AL, Patterson N. Principal component analysis of genetic data. Nat Genet. 2008;40:491–492. doi: 10.1038/ng0508-491. [DOI] [PubMed] [Google Scholar]
- Sakr WA, Haas GP, Cassin BF, Pontes JE, Crissman JD. The frequency of carcinoma and intraepithelial neoplasia of the prostate in young male patients. Journal of Urology. 1993;150:379–385. doi: 10.1016/s0022-5347(17)35487-3. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, Tsunoda T, Inazawa J, Kamatani N, Ogawa O, Fujioka T, Nakamura Y, Nakagawa H. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nature Genetics. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, Ibrahim M, Juma AT, Kotze MJ, Lema G, Moore JH, Mortensen H, Nyambo TB, Omar SA, Powell K, Pretorius GS, Smith MW, Thera MA, Wambebe C, Weber JL, Williams SM. The Genetic Structure and History of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolnay SE. The African American "Great Migration" and beyond. Annual Review of Sociology. 2003;29:209–232. [Google Scholar]
- Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K, Jaeger E, Fielding S, Rowan A, Vijayakrishnan J, Domingo E, Chandler I, Kemp Z, Qureshi M, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Penegar S, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop DT, Gray R, Maher ER, Lucassen A, Kerr D, Evans DG, Consortium C, Schafmayer C, Buch S, Volzke H, Hampe J, Schreiber S, John U, Koessler T, Pharoah P, van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellvi-Bel S, Ruiz-Ponte C, Carracedo A, Castells A, Consortium E, Forsti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JW, Cheng KK, Sham PC, Luk J, Agundez JA, Ladero JM, de la Hoya M, Caldes T, Niittymaki I, Tuupanen S, Karhu A, Aaltonen L, Cazier JB, Campbell H, Dunlop MG, Houlston RS. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nature Genetics. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- Torres JB, Doura MB, Keita SO, Kittles RA. Y chromosome lineages in men of west African descent. PLoS One. 2012;7:e29687. doi: 10.1371/journal.pone.0029687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijai J, Kirchhoff T, Gallagher D, Hamel N, Guha S, Darvasi A, Lencz T, Foulkes WD, Offit K, Klein RJ. Genetic architecture of prostate cancer in the Ashkenazi Jewish population. British Journal of Cancer. 2011;105:864–869. doi: 10.1038/bjc.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Liu F, Hsing AW, Wang X, Shao Q, Qi J, Ye Y, Wang Z, Chen H, Gao X, Wang G, Chu LW, Ding Q, OuYang J, Gao X, Huang Y, Chen Y, Gao YT, Zhang ZF, Rao J, Shi R, Wu Q, Zhang Y, Jiang H, Zheng J, Hu Y, Guo L, Lin X, Tao S, Jin G, Sun J, Lu D, Zheng SL, Sun Y, Mo Z, Yin C, Zhang Z, Xu J. Replication and cumulative effects of GWAS-identified genetic variations for prostate cancer in Asians: a case-control study in the ChinaPCa consortium. Carcinogenesis. 2012;33:356–360. doi: 10.1093/carcin/bgr279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch HG, Black WC. Overdiagnosis in Cancer. Journal of the National Cancer Institute. 2010;102:605–613. doi: 10.1093/jnci/djq099. [DOI] [PubMed] [Google Scholar]
- Xu Z, Bensen JT, Smith GJ, Mohler JL, Taylor JA. GWAS SNP Replication among African American and European American men in the North Carolina-Louisiana prostate cancer project (PCaP) Prostate. 2011;71:881–891. doi: 10.1002/pros.21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharia F, Basu A, Absher D, Assimes TL, Go AS, Hlatky MA, Iribarren C, Knowles JW, Li J, Narasimhan B, Sidney S, Southwick A, Myers RM, Quertermous T, Risch N, Tang H. Characterizing the admixed African ancestry of African Americans. Genome Biology. 2009;10 doi: 10.1186/gb-2009-10-12-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.