Summary
Multilocus sequence analysis of 417 strains of Yersinia pseudotuberculosis revealed that it is a complex of four populations, three of which have been previously assigned species status [Y. pseudotuberculosis sensu stricto (s.s.), Yersinia pestis and Yersinia similis] and a fourth population, which we refer to as the Korean group, which may be in the process of speciation. We detected clear signs of recombination within Y. pseudotuberculosis s.s. as well as imports from Y. similis and the Korean group. The sources of genetic diversification within Y. pseudotuberculosis s.s. were approximately equally divided between recombination and mutation, whereas recombination has not yet been demonstrated in Y. pestis, which is also much more genetically monomorphic than is Y. pseudotuberculosis s.s. Most Y. pseudotuberculosis s.s. belong to a diffuse group of sequence types lacking clear population structure, although this species contains a melibiose-negative clade that is present globally in domesticated animals. Yersinia similis corresponds to the previously identified Y. pseudotuberculosis genetic type G4, which is probably not pathogenic because it lacks the virulence factors that are typical for Y. pseudotuberculosis s.s. In contrast, Y. pseudotuberculosis s.s., the Korean group and Y. pestis can all cause disease in humans.
Introduction
Yersinia pseudotuberculosis has a broad host range and is capable of infecting a wide variety of animals, including humans. The disease caused by this bacterium may vary from a mild enteritis to extra-intestinal symptoms and septicemia (de Barcellos and de Castro, 1981; Sato et al., 1983; Riet-Correa et al., 1990; Welsh and Stair, 1993; Czernomysy-Furowicz, 1997; Hannu et al., 2003; Seimiya et al., 2005; Jalava et al., 2006; Shwimmer et al., 2007; Vincent et al., 2007; Iwata et al., 2008; Wessels et al., 2009). The severity of symptoms differs among strains, and Y. pseudotuberculosis has been subdivided into genetic groups designated G1-G6, dependent on the presence of the virulence plasmid (pYV), the high pathogenicity island (HPI), and the subtype of the Y. pseudotuberculosis-derived mitogen (YPM) (Fukushima et al., 2001). G4 isolates cannot ferment melibiose (melibiose-negative) and are not pathogenic: they express YPMb and lack the pYV and HPI. G5 isolates are also melibiose-negative and of low pathogenicity: they express YPMc, and contain the pYV as well as a truncated HPI (R-HPI). G1-G3 and G6 isolates are melibiose-positive and fully pathogenic: they contain the pYV, and variably harbour the HPI and YPMa (Fukushima et al., 2001). Although human cases of Y. pseudotuberculosis infections are usually sporadic (Sunahara et al., 2000; Vincent et al., 2008), several outbreaks have recently been reported from Finland and Russia (Nuorti et al., 2004; Anonymous, 2005; 2007; 2008; Jalava et al., 2006; Rimhanen-Finne et al., 2009). Humans are not the primary host for Y. pseudotuberculosis. These bacteria can be isolated from a wide variety of domestic and wild animals (Fukushima et al., 2001), as well as from the environment in Japan and occasionally in Europe (Fukushima et al., 1995). Yersinia pseudotuberculosis has also been subdivided on the basis of variable lipopolysaccharide O-side chain into 15 O-serotypes (O:1-O:15) and ten subtypes (O:1a-c, O:2a-c, O:4a-b and O:5a-b) (Bogdanovich et al., 2003). Most European isolates are of serotypes O:1-O:3 whereas serotypes O:4-O:15 are primarily found in Asia (Fukushima et al., 2001).
The description provided above is for the species Y. pseudotuberculosis defined on the basis of classical taxonomic criteria, including pairwise DNA-DNA re-association and biochemical tests. Isolates are routinely assigned to a species on the basis of biochemical tests, and little information is available on the genetic diversity within the different species of the genus Yersinia. Preliminary data indicates that the classical Yersinia species are not necessarily genetically homogenous, and isolates assigned to Yersinia enterocolitica, Yersinia frederiksenii, Yersinia kristensenii and Yersinia mollaretii have been found to belong to additional, previously unrecognized groups on the basis of multilocus sequence analysis (Kotetishvili et al., 2005). Recent updates on the taxonomy of Yersinia have recognized a total of 17 Yersinia species, three of which were first described in 2010 (Murros-Kontiainen et al., 2010a,b; Hurst et al., 2011).
It might be anticipated that population genetic analyses of Y. pseudotuberculosis would reveal greater details about its relationships to the closely related species, Yersinia pestis and Yersinia similis, or even define novel species. Yersinia pestis, the cause of plague, is a genetically monomorphic clade that evolved from Y. pseudotuberculosis 15 000 to 20 000 years ago (Achtman et al., 1999; 2004; Morelli et al., 2010). Yersinia similis is a recently described species (Sprague et al., 2008), which is biochemically similar to Y. pseudotuberculosis, similar enough that they cannot be distinguished by commercial kits which are widely used to identify Yersinia species, such as API 20E. Here we refer to these three species and their close relatives as the Y. pseudotuberculosis complex. A second recently described Yersinia species, Y. pekkanenii, also has similar biochemical reactions to Y. pseudotuberculosis (Niskanen et al., 2009). However, Y. pekkanenii does not cluster together with Y. pseudotuberculosis and Y. similis in 16S rDNA or MLSA analyses (Murros-Kontiainen et al., 2010b), and is therefore not a member of the Y. pseudotuberculosis complex.
Soon after multilocus sequence typing (MLST) was described as a method to study the population genetic structure of bacteria (Maiden et al., 1998; Maiden, 2006), we applied an MLST scheme based on five gene fragments to 36 isolates of Y. pestis and 12 isolates of Y. pseudotuberculosis (Achtman et al., 1999). Yersinia pestis was genetically monomorphic, and understanding its detailed evolution has required the use of diversity at the genomic level (Achtman et al., 2004; Morelli et al., 2010). However, our results at the time also showed that the 12 Y. pseudotuberculosis isolates fell into 11 unique combinations of alleles, so-called sequence types (STs), which indicates that MLST is suitable for investigating the population genetic structure of Y. pseudotuberculosis. We therefore reasoned that an extensive analysis of the population genetic structure of Y. pseudotuberculosis was warranted because prior subdivisions had been based predominantly on microbiological rather than genetic criteria. Here we report the results of such an analysis based on an extended MLST scheme and a large globally representative sample of the Y. pseudotuberculosis complex. An independent MLST scheme has also recently been published (Ch’ng et al., 2011), and we compare the results from both schemes.
Results
We developed a new MLST scheme for the Y. pseudotuberculosis complex that is based on fragments of seven housekeeping genes. These include four gene fragments (glnA, thrA, tmk, trpE) that were used in our previous analysis (Achtman et al., 1999) plus three others (adk, argA, aroA) (Table S1). The gene fragment sequences range in length from 338-465 bp, for a total of 2627 bp for concatenated sequences. We applied this MLST scheme to a diverse collection of 417 isolates from 29 countries and all continents (Table 1 and Table S2) in order to characterize the molecular epidemiology, population structure and relative importance of mutation versus recombination for the diversification of the Y. pseudotuberculosis complex. At the time this project began, Y. similis had not yet been defined, but two of the isolates we tested have since been assigned to Y. similis by 16S rRNA sequencing.
Table 1.
Summary of 417 strains that were thought to be Y. pseudotuberculosis.
| Number |
||||||
|---|---|---|---|---|---|---|
| Continent | Strains | STs | Countries | Hosts | Host groupsa | O-serotypesb |
| Africa | 3 | 3 | 2 | 3 | 1 | 2 |
| Asia | 197 | 82 | 4 | 29 | 5c | 15c,d |
| Europe | 145 | 17 | 18 | 26 | 4c | 6c,d |
| North America | 6 | 3 | 2 | 4 | 2 | 2 |
| Oceania | 20 | 11 | 2 | 7 | 2 | 4 |
| South America | 41 | 2 | 2 | 4 | 1 | 2c |
| Unknown | 5 | 4 | 2c | 2c | 3 | |
| Total | 417 | 89 | 29 | 43c | 5c | 15c,d |
Host groups: human, mammal, environment, bird and fish.
Subtypes are not counted.
Includes strains with unknown information.
Also rough type strains.
Between six and sixteen alleles were found for each of the seven gene fragments, yielding a total of 89 STs among the 417 isolates. These data are freely available from a dedicated, curated database (http://mlst.ucc.ie/mlst/dbs/Ypseudotuberculosis), which also supports the entry of novel sequence information and/or alleles from additional isolates.
Clusters of genetic diversity
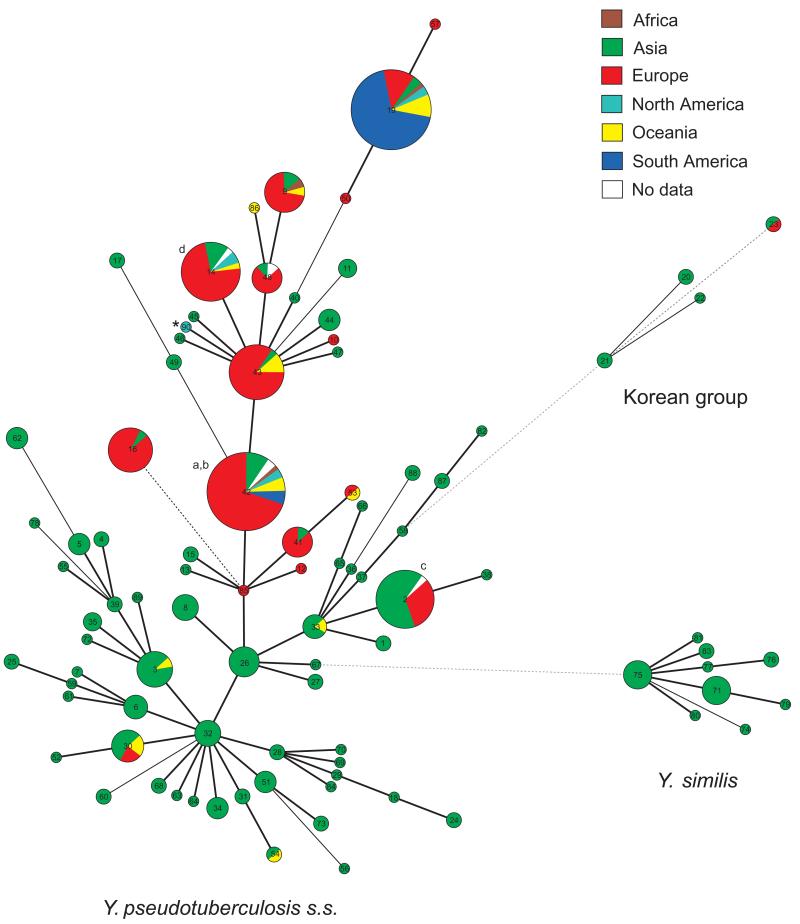
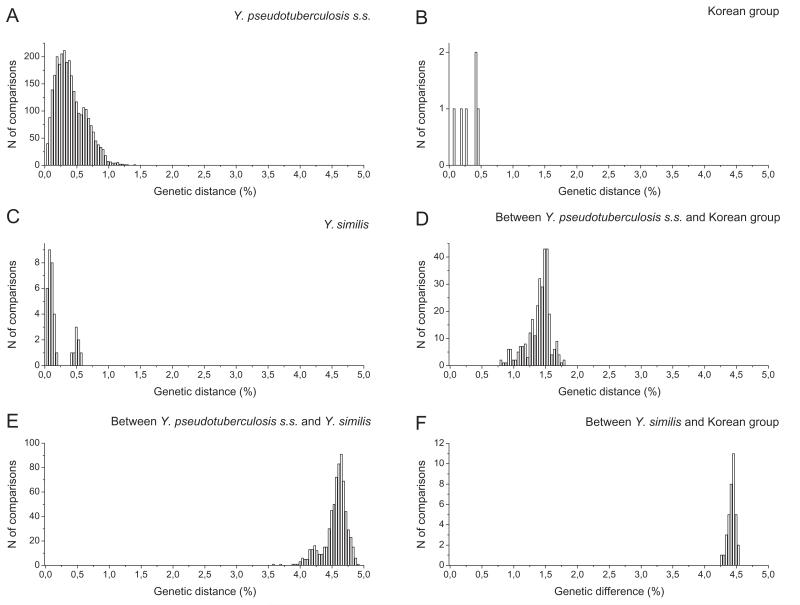
We investigated the genetic diversity of alleles in our sample of the Y. pseudotuberculosis complex by a minimal spanning tree and eBURST (Figs 1 and S1). These programs gave identical patterns of single locus links and delimited the same three major groups, which we refer to henceforth as Y. pseudotuberculosis s.s., Y. similis, and the Korean group (which has not been previously described). Yersinia pestis corresponds to one ST (ST90) within Y. pseudotuberculosis s.s. Within Y. pseudotuberculosis s.s., the pairwise distance between the concatenated sequences of individual STs ranged from 1-37 nucleotides (0.04-1.4%), with a similar range within the Korean group (2-12 nucleotides, 0.08-0.5%), and Y. similis (1-15 nucleotides, 0.04-0.6%) (Fig. 2A-C). In pairwise comparisons, individual Y. pseudotuberculosis s.s. isolates differed from those of the Korean group at 21-47 nucleotides (0.8-1.8%) and from Y. similis isolates at 94-129 nucleotides (3.6-4.9%) (Fig. 2D-E). The Korean group isolates differed from Y. similis isolates at 112-119 nucleotides (4.2-4.5%) (Fig. 2F). These nucleotide differences are accompanied by high population specific fixation index values (FST = 0.93, P < 0.01) and high pairwise genetic distances between populations (0.79-0.97) (Table 2). We conclude that the Y. pseudotuberculosis complex contains three species plus the previously unknown Korean group.
Fig. 1.
Minimal spanning tree of the Y. pseudotuberculosis complex coloured by continent. The asterisk marks ST90 which is specific for Y. pestis strain CO92 (Parkhill et al., 2001). Letters a-d indicate STs containing isolates whose genomes have been sequenced: a: IP32953 (Chain et al., 2004); b: PB1/+ (unpublished, NC_010634); c: IP31758 (Eppinger et al., 2007); and d: YPIII (unpublished, NC_010465).
Fig. 2.
Nucleotide mismatch distributions of the concatenated sequences for all STs within (A-C) and between (D-F) groups.
Table 2.
Fixation index (FST) and pairwise genetic distances of individual and concatenated gene fragments.
| Population specific FST indices |
Population pairwise genetic distances |
|||||
|---|---|---|---|---|---|---|
| Gene | Y. pstb | Y. similis | Korean group | Y. pstb–Y. similis | Y. pstb–Korean | Y. similis–Korean |
| adk | 0.954 | 0.955 | 0.955 | 0.963 | 0.795 | 0.987 |
| argA | 0.931 | 0.934 | 0.934 | 0.941 | 0.825 | 0.992 |
| aroA | 0.983 | 0.982 | 0.980 | 0.986 | 0.973 | 0.955 |
| glnA | 0.846 | 0.843 | 0.847 | 0.848 | 0.840 | 0.800 |
| thrA | 0.867 | 0.871 | 0.867 | 0.890 | 0.595 | 0.952 |
| tmk | 0.867 | 0.872 | 0.872 | 0.888 | 0.574 | 1.000 |
| trpE | 0.975 | 0.976 | 0.975 | 0.981 | 0.611 | 0.991 |
| Concatenated sequences | 0.927 | 0.929 | 0.928 | 0.939 | 0.794 | 0.964 |
Y. pstb, Y. pseudotuberculosis s.s.
16S rRNA analysis
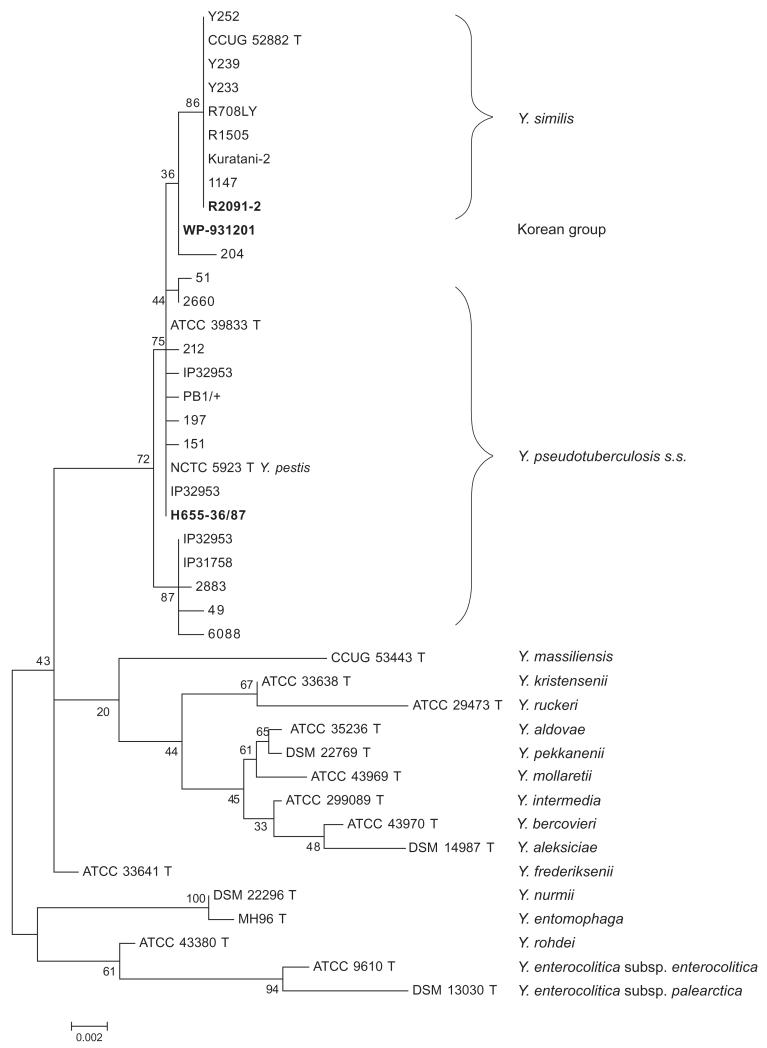
We equated one MLST cluster with Y. similis because it includes strains Kuratani-2 and R1505 isolates that had previously been assigned to Y. similis by Sprague and colleagues (2008), and seemed very coherent. To test this interpretation, we sequenced a 1440 bp fragment of the 16S rRNA gene from one representative each of Y. pseudotuberculosis s.s. (strain H655-36/87), the Y. similis group (strain R2091-2), and the Korean group (strain WP-931201) (Table S3). These sequences were compared with rRNA sequences of type strains of Yersinia spp. that were obtained from the NCBI Nucleotide database (Fig. 3). The 16S rRNA sequence of R2091-2 was identical to that of the Y. similis type strain (CCUG 52882T). It was also identical to those of seven other strains that had been assigned to Y. similis on the basis of their 16S sequences (Kim et al., 2003; Sprague et al., 2008). Strain WP-931201 from the Korean group clustered together with Y. pseudotuberculosis 204 (Kim et al., 2003) in a maximum likelihood tree, between Y. similis and Y. pseudotuberculosis s.s. (Fig. 3). Strain H655-36/87 from Y. pseudotuberculosis s.s. group clustered together with multiple other Y. pseudotuberculosis s.s. strains and the entire Y. pseudotuberculosis complex formed a clade with 72% bootstrap support. Similar results were obtained with a neighbour-joining tree (data not shown).
Fig. 3.
Phylogeny of 16S rRNA gene sequences (1436 bp) of isolates from diverse species of Yersinia. Phylogenies were calculated using the maximum likelihood method and the percentage of trees in which the associated taxa clustered together is shown next to the branches. Strains from Y. pseudotuberculosis, the Korean group, and Y. similis in bold text were sequenced in this study. The sources of other sequences are listed in Table S3.
MLST versus epidemiological associations
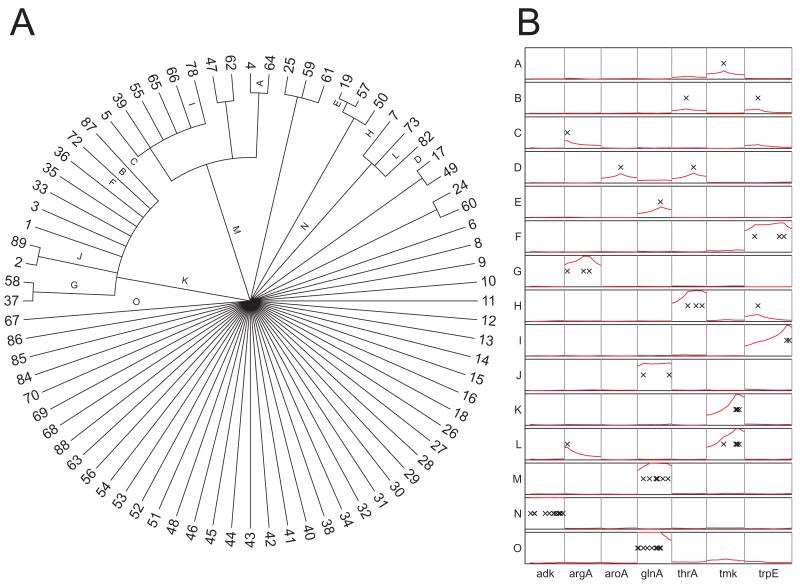
Two hundred and thirty-nine of the isolates subjected to MLST had previously been assigned to genetic groups G1-G6 (Fukushima et al., 2001) (Fig. S2). Yersinia pseudotuberculosis s.s. included isolates in G1-G3 and G5-G6, but not G4. All but one of the strains in the low-pathogenicity group G5 belonged to three clustered STs, ST50, ST19 and ST57, according to allelic analyses (Figs 1 and S1). The concatenated sequences of these three STs also clustered together according to ClonalFrame (Fig. 4), which constructs genealogies after stripping sequences of recombinational events (Didelot and Falush, 2007). These G5 strains were isolated from humans and domesticated animals (buffalo, cattle, deer, pigs) from all continents, belonged to serotype O:3, and did not ferment melibiose (Fig. 1 and Figs S3-S5).
Fig. 4.
Clonal genealogy (A) and evolutionary events (B) reconstructed by ClonalFrame from concatenated sequences. The designations for branches in part A (A-O) are congruent with the designations of rows in part B. Each column in part B delineates the extent of one of seven MLST gene fragments. The height of the red line indicates the probability of recombination on a scale from 0 (row bottom) to 1 (row top). Each nucleotide substitution is represented by a black cross.
We did not detect any other strong patterns of specificity for geographical origin, host type, or O-serotypes within Y. pseudotuberculosis s.s. (Fig. 1 and Figs S3-S4). However, the diversity found in Asia differed from that in other continents. Of the European strains, 78% (113/145) were assigned to the six most frequent STs, whereas Asian strains were dispersed among a larger number of rarer STs (Fig. 1). 78% (131/168) of the Asian isolates were in STs with only 1-6 strains. Furthermore, 97% (139/144) of the European, 95% (19/20) of the Oceanian, and all African and American isolates were assigned to STs that are also prevalent in other continents but only 33% (56/168) of the isolates from Asia were found elsewhere (Fig. 1) The difference between Asia and other continents was significant (X2, P < 0.001). Altogether 31 STs containing more than one isolate were unique to Asia versus none for Europe or other continents (X2, P < 0.001). Other STs from Asia (25), Europe (5) and Oceania (1) were not informative because they contained only one isolate. Similar to the case with Europe, the diversity among the isolates from Brazil and other South American countries was also quite limited. Most South American isolates (93%; 38/41) were in ST19 and the remaining isolates were in ST42. However, the isolates from South America were primarily from cattle and buffalo in Brazil. Both ST42 and ST19 are globally distributed and 12 other STs were also isolated in two or more continents (Fig. 1).
The Korean group consisted of five isolates in ST20, ST21 and ST22, all of which were isolated in Korea (water: 4; human: 1). The Korean group also contains two isolates from Sweden (otter: 1) and Japan (human: 1) within a fourth ST, ST23 (Table S2, Fig. S3). The seven Korean group isolates belonged to genetic groups G3 and G6 and contained nine unique alleles in adk, glnA, trpE, thrA and aroA.
Yersinia similis were isolated from small mammals (mice, moles, voles and a marten) as well as the environment (water) (Fig. S3). All typed strains were melibiose negative and all except one of the 23 isolates belonged to the non-pathogenic group G4 (Figs S2 and S5). Thirteen alleles spread over all seven gene fragments were unique to Y. similis.
A wide range of O-serotypes were present among Y. similis strains and most of them were also present among Y. pseudotuberculosis s.s. strains including O:1c, O:5a, O:5b, O:6, O:7, O:10 and O:12 (Table S2, Fig. S4). However, serotype O:9 was unique to Y. similis and of the five serotype O:11 isolates, three were Y. similis and two were from the Korean group. The remaining five Korean group strains belonged to serotypes O:15, O:4 and O:4a. These data indicate a frequent exchange of LPS O-polysaccharide biosynthetic genes between Y. pseudotuberculosis s.s., Y. similis and the Korean group.
Recombination in Y. pseudotuberculosis s.s
The levels of nucleotide and allelic diversity within Y. pseudotuberculosis s.s. were low. Among the 387 Y. pseudotuberculosis s.s. strains, we only identified 4-11 alleles for each of the seven gene fragments, and the level of pairwise nucleotide diversity (π) for individual gene fragments ranged from 0.0008 to 0.0068 (Table 3). It is difficult to resolve true genealogies with such low levels of diversity. Indeed, the clonal genealogy of over half of the STs remained unresolved by ClonalFrame (Fig. 4A).
Table 3.
Nucleotide diversity of individual genes and concatenated sequences of Yersinia pseudotuberculosis s.s. strains (n = 387).
| Gene (size) | No. of alleles |
π s | π a | πa/πs | θa/θs | π | S |
|---|---|---|---|---|---|---|---|
| adk (389 bp) | 4 | 0.0037 | 0.0005 | 0.1386 | 0.0622 | 0.0012 | 12 |
| argA (361 bp) | 4 | 0.0037 | 0.0009 | 0.2493 | 0.1049 | 0.0016 | 4 |
| aroA (357 bp) | 7 | 0.0028 | 0.0001 | 0.2837 | 0.1648 | 0.0008 | 6 |
| glnA (338 bp) | 11 | 0.0164 | 0.0008 | 0.0513 | 0.0735 | 0.0044 | 21 |
| thrA (342 bp) | 11 | 0.0211 | 0.0001 | 0.0047 | 0.1838 | 0.0049 | 8 |
| tmk (375 bp) | 9 | 0.0270 | 0.0000 | 0.0015 | n.a. | 0.0068 | 10 |
| trpE (465 bp) | 7 | 0.0064 | 0.0000 | 0.0016 | 0.0450 | 0.0015 | 8 |
| Concatenated sequences (2627 bp) | 76 | 0.0112 | 0.0003 | 0.0304 | n.a. | 0.0029 | 69 |
n.a., could not be calculated due to a complex codon.
The ClonalFrame analyses estimated that mutation and recombination were approximately equally frequent within this dataset (ρ/θ 0.95; CI95 [95% credibility interval]: 0.38-2.06) and approximately equally important for genetic diversification (r/m = 1.34 CI95: 0.72-2.30). Examples of the evolutionary events reconstructed by ClonalFrame are shown in Fig. 4B for 15 selected branches of the clonal genealogy. Sequence changes along branches A to E seem to result only from mutations because the probability for recombination is very low for all substitutions as indicated by the red line. Branches F to J demonstrate examples where recombination is more likely (elevated red line), introducing two to three substitutions per recombination event. This number of changes corresponds roughly to the average genetic divergence (0.5%) between pairs of strains of Y. pseudotuberculosis s.s. (Fig. 2), which suggests that these fragments were imported from Y. pseudotuberculosis s.s. donors.
In contrast to branches A to J, 5-12 nucleotides were introduced by each event along branches K to O. These five atypical recombination events probably represent imports from the Korean group or from Y. similis because we identified identical sequences within those groups, or sequences differing by only one nucleotide. For instance, recombination events in tmk on branches K and L introduced five and six substitutions respectively, resulting in sequences that match sequences from the Korean group. Similarly, the import in glnA on branch M (nine substitutions) also came from the Korean group. On the other hand, the recombination events in adk on branch N (12 substitutions) and in glnA on branch O (11 substitutions) matched sequences within Y. similis. These examples provide evidence for interspecies recombination within the Y. pseudotuberculosis complex.
Discussion
Comparison of MLST schemes
Since completion of this work, an independent MLST scheme has been described (Ch’ng et al., 2011) in which 83 isolates were tested for seven housekeeping gene fragments. The two schemes are incompatible because different gene fragments were used. For example, of the 44 isolates that were tested with both schemes, 12 were assigned to six STs according to Ch’ng et al. and nine STs by our scheme. Conversely, 16 isolates were assigned to six STs by our scheme but belong to 14 STs according to Ch’ng et al. Only eight isolates are assigned to three pair-specific STs in both schemes (Table S6). Ch’ng and colleagues (2011) described a discrete clade within Y. pseudotuberculosis containing six isolates, designated clade B. Based on five isolates which were tested in both studies, this clade is Y. similis. The sixth isolate (OK5608) assigned to this clade by Ch’ng et al. belongs to genetic group G3, whereas we found that Y. similis is exclusively G4. In our hands, OK5608 is in ST49, within Y. pseudotuberculosis s.s. Contrary to Ch’ng et al., Y. similis (clade B) is not geographically restricted to Japan. Ch’ng et al. also showed that recombination and mutation are equally frequent within Y. pseudotuberculosis and that LPS O gene clusters recombine frequently between Y. pseudotuberculosis clades.
Delineation of the Y. pseudotuberculosis complex
All isolates in this study had originally been assigned to Y. pseudotuberculosis, but MLST revealed that they represent three distinct groups. Most of the strains are Y. pseudotuberculosis s.s. and are closely related, but a few strains of group G4 are Y. similis, confirming prior conclusions based on three isolates (Sprague et al., 2008) and another few isolates belong to the Korean group, which has not been previously described (Fig. 1). Yersinia pestis is one ST within Y. pseudotuberculosis s.s as shown before (Achtman et al., 1999). Strains from Y. pseudotuberculosis s.s., Y. pestis, Y. similis and the Korean group, which we designate as the Y. pseudotuberculosis complex, form a clear clade on the basis of rRNA sequences (Fig. 3).
Although all our Y. similis strains were isolated in Japan, the Y. similis type strain was isolated in Germany (Sprague et al., 2008), suggesting that this species has a worldwide distribution. Yersinia similis strains were isolated from small mammals and the environment, but not from humans. Yersinia similis is thought not to be pathogenic for humans: it lacks the pYV (Fukushima et al., 2001) which is essential for the pathogenicity of Yersinia (Cornelis et al., 1998), and carries YPMb, whereas Y. pseudotuberculosis s.s. isolates belong to genetic types other than G4, are pYV-positive, possess YPMa or YPMc, and are potentially pathogenic.
The Korean group currently contains seven isolates in four STs. Its diversity will probably expand as additional members are identified. It is currently unclear whether the Korean group is a distinct species or a population in the process of speciation. The seven Korean group isolates belonged to genetic types G3 and G6, which are considered pathogenic (Fukushima et al., 2001). Two of the seven isolates were isolated from humans, suggesting a clinical relevance of these strains.
A wide range of O-serotypes were shared among Y. similis, Y. pseudotuberculosis s.s., and Korean group strains, probably reflecting transfer of LPS O-polysaccharide biosynthetic genes between species within the Y. pseudotuberculosis complex.
Y. pestis versus Y. pseudotuberculosis s.s
According to the minimal spanning tree, the closest relative to Y. pestis (ST 90) is ST43, which differs from ST90 only at trpE. Most ST43 isolates were from Europe but four were from Australia, New Zealand or China. ST43 was isolated from birds, mammals and humans. Twenty-five of the isolates from ST43 belong to serotype O:1, and the 19 which have been subtyped were all O:1b, supporting the hypothesis that Y. pestis descended from an O:1b Y. pseudotuberculosis (Skurnik et al., 2000). One exceptional ST43 strain is O:3. These data suggest that Y. pestis evolved from ST43. However, the minimal spanning tree identifies founder STs with multiple isolates as the evolutionary source of rarer STs and the straggly nature of Y. pseudotuberculosis s.s. suggests that many of the links might be spurious and reflect recombination between unrelated lineages (Turner et al., 2007). Among the STs of the currently published genomes of Y. pseudotuberculosis (ST2, ST14 and ST42; Fig. 1), the ST that is closest to ST90 is ST42, which differs from ST90 by two alleles and includes strain IP32953 (Chain et al., 2004). Like ST43, ST42 is global in distribution. It predominantly contains O:1 isolates, but also includes two O:13 and two O:2 strains. All of the 30 subtyped O:1 isolates in ST42 are O:1a, except for IP32953, which is O:1b. ST42 differs from ST43 at only one nucleotide in thrA.
Recombination and population structure in the Y. pseudotuberculosis complex
The evolution of Y. pseudotuberculosis was equally affected by recombination and mutation. Most recombination events within Y. pseudotuberculosis s.s. were intraspecific, but three imports were from the Korean group and two from Y. similis (Fig. 4). Interspecies recombination has also been previously observed in other species (Miller et al., 2006; Wilson et al., 2009; de Haan et al., 2010), but is thought to be less frequent than intraspecific recombination (Didelot and Maiden, 2010).
Recombination rates can vary substantially, even among closely related bacteria (Didelot and Maiden, 2010). In general, highly specialized pathogens show fewer signs of recombination than their non-specialized relatives (Achtman, 2008). For example, Y. pestis does not recombine (Achtman et al., 1999; Morelli et al., 2010) even though it is part of Y. pseudotuberculosis s.s., which does recombine extensively. It is possible that the lack of recombination in Y. pestis can account for the radical difference in population structures of the two species. Yersinia pestis shows strong geographical structuring of populations (Morelli et al., 2010), whereas only weak associations were observed within Y. pseudotuberculosis s.s. between geographical origin, host type, and/or O-serotype.
Weak geographical structuring in Y. pseudotuberculosis s.s
Yersinia pseudotuberculosis s.s. contains a cluster of STs consisting of ST19 plus its single locus variants ST50 and ST57 (Fig. 1, near one o’clock; Fig. 4, branch E). These isolates are O:3, melibiose-negative, and belong to genetic group G5. They have been isolated globally from buffalo, cattle, deer and pigs, and may correspond to the melibiosenegative Y. pseudotuberculosis O:3 that are common among asymptomatic domestic pigs in many European countries (Weber and Knapp, 1981; Niskanen et al., 2002; Ortiz Martínez et al., 2009; 2011) as well as in Japan (Tsubokura et al., 1984). It has been claimed that melibiose-negative Y. pseudotuberculosis O:3 strains of genetic group G5 are associated with lowered pathogenicity (Mair et al., 1979; Tsubokura et al., 1984; Nagano et al., 1997; Fukushima et al., 2001). However, these isolates harbour the pYV and the chromosomal inv gene (Nagano et al., 1997; Fukushima et al., 2001). They cause severe, sometimes fatal diarrhoea in cattle (Martins et al., 1998; Warth, 2010), and have also caused abortions in cattle and sheep and fatal enteric disease in squirrel monkeys (Mair et al., 1979; Buhles et al., 1981). They have also been isolated from humans with enteric symptoms (Tsubokura et al., 1984; Aleksić et al., 1995). It has been suggested that melibiose-negative Y. pseudotuberculosis O:3 evolved in Europe and reached other regions by co-transmission with livestock (Fukushima et al., 2001) because they do not seem to exist in wild animals.
A second example of weak geographic structure is that Y. pseudotuberculosis s.s. isolates from Asia belonged to STs throughout the entire minimal spanning tree whereas 97% of European isolates belonged to a few, global STs. This geographic correlation suggests that Y. pseudotuberculosis might have evolved in Asia, as did Y. pestis (Morelli et al., 2010).
Use of melibiose to differentiate Yersinia species
The ability to ferment melibiose has been used as a phenotypic marker of certain Yersinia species. Yersinia entomophaga, Y. intermedia, Y. pestis, Y. pseudotuberculosis s.s and Y. rohdei include strains that can ferment melibiose whereas other species do not (Anisimov et al., 2004; Bottone et al., 2005; Hurst et al., 2011). Fermentation of melibiose is a key test for differentiating Y. intermedia from Y. frederiksenii (Brenner et al., 1980; Bottone et al., 2005). However, Y. intermedia biotype 8 is incapable of fermenting melibiose, as are two other melibiose-negative variants (Martin et al., 2009). Within the Y. pseudotuberculosis complex, Sprague and colleagues (2008) suggested that melibiose fermentation could be used to differentiate Y. pseudotuberculosis from Y. similis. We confirm that the inability to ferment melibiose is uniform throughout Y. similis and that most Y. pseudotuberculosis s.s. isolates ferment melibiose. However, Y. pseudotuberculosis s.s. also included the melibiosenegative Y. pseudotuberculosis serotype O:3 strains in ST19, ST50 and ST57, rendering this phenotypic differentiation unreliable.
Conclusions
We define the Y. pseudotuberculosis complex as a superset of Y. pseudotuberculosis s.s., Y. pestis, the Korean group, and Y. similis. Except for Y. pestis, genetic diversity within each reflects an equal mixture of recombination and mutation, resulting in diffuse population structure within Y. pseudotuberculosis s.s. Yersinia pestis belongs to ST90 within Y. pseudotuberculosis s.s. Yersinia similis corresponds to a distinct clade of STs with genetic type G4 and may not be pathogenic for larger mammals. In contrast, Y. pseudotuberculosis s.s. includes multiple other genetic groups that are considered pathogenic for humans when they harbour a virulence plasmid. The Korean group resembles Y. pseudotuberculosis but is genetically somewhat distinct and may be in the process of becoming a distinct species.
Experimental procedures
Yersinia isolates
We studied a diverse collection of 417 strains that were initially thought to be Y. pseudotuberculosis, which were isolated from humans, mammals, birds, fish or the environment in 29 countries from all continents (Tables 1 and S2), plus one strain of Y. pestis. This collection includes all 15 O-serotypes. Data on virulence characteristics for 239 strains were from a previous study (Fukushima et al., 2001).
Multilocus sequence typing
Oligonucleotide primers for PCR amplification (Table S1) and sequencing (Table S4) were designed based on sequences of adk (adenylate kinase), argA (N-acetylglutamate synthase), aroA (3-phosphoshikimate-1-carboxylvinyltransferase), glnA (glutamine synthase), thrA (aspartokinase I/homoserine dehydrogenase I), tmk (thymidylate kinase) and trpE (anthranilate synthase component I) from Y. pseudotuberculosis IP32953 [Accession number NC006155 (Chain et al., 2004)]. Sequences were trimmed to a uniform length for each gene, which was determined from multiple alignment of all sequences. Sequences were trimmed using Bionumerics 5.10 (Applied Maths, Sint-Martens-Latem, Belgium) and each unique sequence was assigned a different allele number using the MLST database at University College Cork, which is accessible at http://mlst.ucc.ie./mlst/dbs/Ypseudotuberculosis. That MLST database provides publicly available downloads of all currently known sequences, allele designations and ST types as well as strain information.
Genetic analysis
We used DnaSP 5.10 (Librado and Rozas, 2009) with Jukes-Cantor correction to calculate the average numbers of nucleotide substitutions per site between paired sequences for synonymous (πs) and non-synonymous (πa) sites as well as total nucleotide diversity (π). The number of polymorphic sites and mismatch distributions were also calculated using DnaSP 5.10. A graph of mismatch distribution was drawn using Origin 7.5 (OriginLab Corporation, Northampton, MA, USA). Fixation indices (FST) were calculated using Arlequin 3.1 (Excoffier et al., 2005). Minimal spanning trees were constructed with Bionumerics 5.10 (Applied Maths) for the allelic characters by using a maximal crosslink distance of zero. An eBURST population snapshot was drawn by setting the group definition to zero of seven shared alleles (Feil et al., 2004).
16S rRNA analysis
16S rRNA genes were amplified by PCR and sequenced as described (Koort et al., 2005) using universal amplification primers F19-38 and R1541-1522 with additional sequencing primers F926 and R519 (Table S5). 16S rRNA was sequenced from one strain each of Y. pseudotuberculosis (strain H655-36/87, Accession No. JF826870), Y. similis (R2091-2, Accession No. JF826869) and the Korean group (WP-931201, Accession No. JF826868). Additional sequences of Yersinia spp. 16S rRNA genes were retrieved from the NCBI Nucleotide database at http://www.ncbi.nlm.nih.gov/nuccore (Table S3). Phylogenetic analyses were performed using the maximum likelihood method with MEGA5 (Tamura et al., 2011) and the Tamura-Nei model to estimate evolutionary distances (Tamura and Nei, 1993). Bootstraps were calculated on the basis of 1000 iterations. The tree with the highest log likelihood (−3125.1894) was chosen.
ClonalFrame analysis
We applied ClonalFrame (Didelot and Falush, 2007) to all STs from Y. pseudotuberculosis s.s. A total of 100 000 MCMC iterations were performed, of which the first half was discarded as burn-in. Mixing and convergence properties were found to be satisfactory based on comparisons of independent runs.
Supplementary Material
eBURST population snapshot of Y. pseudotuberculosis s.s., Y. similis and the Korean group. The blue dots represent predicted primary founders and yellow dots predicted subgroup founder with at least two descendent singlelocus variants.
Minimal spanning tree (maximum crosslink distance of one) of studied strains showing the genetic groups according to Fukushima and colleagues (2001).Other details are as in Fig. 1. In Y. similis ST75, strain Kuratani-2 is of genetic group G3 but contains YPMa although it does not harbour pYV or ferment melibiose.
Minimal spanning tree (maximum crosslink distance of one) of strains coloured by host group. Other details are as in Fig. 1.
Minimal spanning tree (maximum crosslink distance of one) of strains coloured by O-serotype. Other details are as in Fig. 1.
Minimal spanning tree (maximum crosslink distance of one) of strains coloured according to melibiose fermentation. Other details are as in Fig. 1.
Characteristics of the strains used in the analyses.
Acknowledgements
This work was funded by the Academy of Finland (grants 1104361 and 1114075 to M.S.), the Scientific Foundation of Ireland (05/FE1/B882 to M.A.), the Institut National de Veille Sanitaire (InVS, France to E.C.), and FAPESP (São Paulo Research Foundation) Proc. 2006/51434-7 and 2007/07784-6.
References
- Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci USA. 2004;101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksić S, Bockemühl J, Wuthe HH. Epidemiology of Y. pseudotuberculosis in Germany, 1983-1993. Contrib Microbiol Immunol. 1995;13:55–58. [PubMed] [Google Scholar]
- Anisimov AP, Lindler LE, Pier GB. Intraspecific diversity of Yersinia pestis. Clin Microbiol Rev. 2004;17:434–464. doi: 10.1128/CMR.17.2.434-464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous Yersiniosis – Russia (Siberia) 2005 Promed-mail 27 April 2005. Accession No. 20050427.1169.
- Anonymous Yersiniosis – Russia (Yamalo-Nenetsky) 2007 Promed-mail 1 October 2007. Accession No. 20071001.3240.
- Anonymous Yersiniosis – Russia (Krasnoyarsk) 2008 Promed-mail 18 July 2008. Accession No. 20080718.2184.
- de Barcellos DE, de Castro AF. Isolation of Yersinia pseudotuberculosis from diarrhoea in pigs. Br Vet J. 1981;137:95–96. doi: 10.1016/s0007-1935(17)31793-1. [DOI] [PubMed] [Google Scholar]
- Bogdanovich T, Carniel E, Fukushima H, Skurnik M. Use of O-antigen gene cluster-specific PCRs for the identification and O-genotyping of Yersinia pseudotuberculosis and Yersinia pestis. J Clin Microbiol. 2003;41:5103–5112. doi: 10.1128/JCM.41.11.5103-5112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottone EJ, Bercovier H, Mollaret HH. Genus XLI. Yersinia. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. 2nd edn. Springer; New York, USA: 2005. pp. 838–848. [Google Scholar]
- Brenner D, Bercovier H, Ursing J, Alonso J, Steigerwalt A, Fanning G, et al. Yersinia intermedia: a new species of Enterobacteriaceae composed of rhamnosepositive, melibiose-positive, raffinose-positive strains (formerly called Yersinia enterocolitica or Yersinia enterocolitica-like) Curr Microbiol. 1980;4:207–212. [Google Scholar]
- Buhles WC, Jr, Vanderlip JE, Russell SW, Alexander NL. Yersinia pseudotuberculosis infection: study of an epizootic in squirrel monkeys. J Clin Microbiol. 1981;13:519–525. doi: 10.1128/jcm.13.3.519-525.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng SL, Octavia S, Xia Q, Duong A, Tanaka MM, Fukushima H, Lan R. Population structure and evolution of pathogenicity of Yersinia pseudotuberculosis. Appl Environ Microbiol. 2011;77:768–775. doi: 10.1128/AEM.01993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, Neyt C, et al. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czernomysy-Furowicz D. An outbreak of foal yersiniosis in Poland: pathological and bacteriological examination. Zentralbl Bakteriol. 1997;286:542–546. doi: 10.1016/s0934-8840(97)80058-8. [DOI] [PubMed] [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Maiden MC. Impact of recombination on bacterial evolution. Trends Microbiol. 2010;18:315–322. doi: 10.1016/j.tim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppinger M, Rosovitz MJ, Fricke WF, Rasko DA, Kokorina G, Fayolle C, et al. The complete genome sequence of Yersinia pseudotuberculosis IP31758, the causative agent of Far East scarlet-like fever. PLoS Genet. 2007;3:e142. doi: 10.1371/journal.pgen.0030142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima H, Gomyoda M, Tsubokura M, Aleksić S. Isolation of Yersinia pseudotuberculosis from river waters in Japan and Germany using direct KOH and HeLa cell treatments. Zentralbl Bakteriol. 1995;282:40–49. doi: 10.1016/s0934-8840(11)80795-4. [DOI] [PubMed] [Google Scholar]
- Fukushima H, Matsuda Y, Seki R, Tsubokura M, Takeda N, Shubin FN, et al. Geographical heterogeneity between Far Eastern and Western countries in prevalence of the virulence plasmid, the superantigen Yersinia pseudotuberculosis-derived mitogen, and the high-pathogenicity island among Yersinia pseudotuberculosis strains. J Clin Microbiol. 2001;39:3541–3547. doi: 10.1128/JCM.39.10.3541-3547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan CP, Kivistö RI, Hakkinen M, Corander J, Hänninen ML. Multilocus sequence types of Finnish bovine Campylobacter jejuni isolates and their attribution to human infections. BMC Microbiol. 2010;10:200. doi: 10.1186/1471-2180-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannu T, Mattila L, Nuorti JP, Ruutu P, Mikkola J, Siitonen A, Leirisalo-Repo M. Reactive arthritis after an outbreak of Yersinia pseudotuberculosis serotype O:3 infection. Ann Rheum Dis. 2003;62:866–869. doi: 10.1136/ard.62.9.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst MR, Becher SA, Young SD, Nelson TL, Glare TR. Yersinia entomophaga sp. nov. isolated from the New Zealand grass grub Costelytra zealandica. Int J Syst Evol Microbiol. 2011;61:844–849. doi: 10.1099/ijs.0.024406-0. [DOI] [PubMed] [Google Scholar]
- Iwata T, Une Y, Okatani AT, Kato Y, Nakadai A, Lee K, et al. Virulence characteristics of Yersinia pseudotuberculosis isolated from breeding monkeys in Japan. Vet Microbiol. 2008;129:404–409. doi: 10.1016/j.vetmic.2007.11.029. [DOI] [PubMed] [Google Scholar]
- Jalava K, Hakkinen M, Valkonen M, Nakari UM, Palo T, Hallanvuo S, et al. An outbreak of gastrointestinal illness and erythema nodosum from grated carrots contaminated with Yersinia pseudotuberculosis. J Infect Dis. 2006;194:1209–1216. doi: 10.1086/508191. [DOI] [PubMed] [Google Scholar]
- Kim W, Song MO, Song W, Kim KJ, Chung SI, Choi CS, Park YH. Comparison of 16S rDNA analysis and rep-PCR genomic fingerprinting for molecular identification of Yersinia pseudotuberculosis. Antonie Van Leeuwenhoek. 2003;83:125–133. doi: 10.1023/a:1023301924932. [DOI] [PubMed] [Google Scholar]
- Koort J, Murros A, Coenye T, Eerola S, Vandamme P, Sukura A, Björkroth J. Lactobacillus oligofermentans sp. nov., associated with spoilage of modifiedatmosphere-packaged poultry products. Appl Environ Microbiol. 2005;71:4400–4406. doi: 10.1128/AEM.71.8.4400-4406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotetishvili M, Kreger A, Wauters G, Morris JG, Jr, Sulakvelidze A, Stine OC. Multilocus sequence typing for studying genetic relationships among Yersinia species. J Clin Microbiol. 2005;43:2674–2684. doi: 10.1128/JCM.43.6.2674-2684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Maiden MC. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair NS, Fox E, Thal E. Biochemical, pathogenicity and toxicity studies of type III strains of Yersinia pseudotuberculosis isolated from the cecal contents of pigs. Contrib Microbiol Immunol. 1979;5:359–365. [PubMed] [Google Scholar]
- Martin L, Leclercq A, Savin C, Carniel E. Characterization of atypical isolates of Yersinia intermedia and definition of two new biotypes. J Clin Microbiol. 2009;47:2377–2380. doi: 10.1128/JCM.02512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins CH, Bauab TM, Falcão DP. Characteristics of Yersinia pseudotuberculosis isolated from animals in Brazil. J Appl Microbiol. 1998;85:703–707. doi: 10.1111/j.1365-2672.1998.00579.x. [DOI] [PubMed] [Google Scholar]
- Miller WG, Englen MD, Kathariou S, Wesley IV, Wang G, Pittenger-Alley L, et al. Identification of hostassociated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology. 2006;152:245–255. doi: 10.1099/mic.0.28348-0. [DOI] [PubMed] [Google Scholar]
- Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 2010;42:1140–1143. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murros-Kontiainen AE, Fredriksson-Ahomaa M, Korkeala H, Johansson P, Rahkila R, Björkroth J. Yersinia nurmii sp. nov. Int J Syst Evol Microbiol. 2010a doi: 10.1099/ijs.0.024836-0. doi:10.1099/ijs.0.024836-0. [DOI] [PubMed] [Google Scholar]
- Murros-Kontiainen AE, Johansson P, Niskanen T, Fredriksson-Ahomaa M, Korkeala H, Björkroth J. Yersinia pekkanenii sp. nov. Int J Syst Evol Microbiol. 2010b doi: 10.1099/ijs.0.019984-0. doi:10.1099/ijs.0.019984-0. [DOI] [PubMed] [Google Scholar]
- Nagano T, Ichimura K, Haji N, Nagao K, Someya K, Kiyohara T, et al. Characteristics and pathogenicity of non-melibiose-fermenting strains of Yersinia pseudotuberculosis O3. Microbiol Immunol. 1997;41:175–183. doi: 10.1111/j.1348-0421.1997.tb01188.x. [DOI] [PubMed] [Google Scholar]
- Niskanen T, Fredriksson-Ahomaa M, Korkeala H. Yersinia pseudotuberculosis with limited genetic diversity is a common finding in tonsils of fattening pigs. J Food Prot. 2002;65:540–545. doi: 10.4315/0362-028x-65.3.540. [DOI] [PubMed] [Google Scholar]
- Niskanen T, Laukkanen R, Murros A, Björkroth J, Skurnik M, Korkeala H, Fredriksson-Ahomaa M. Characterisation of non-pathogenic Yersinia pseudotuberculosis-like strains isolated from food and environmental samples. Int J Food Microbiol. 2009;129:150–156. doi: 10.1016/j.ijfoodmicro.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Nuorti JP, Niskanen T, Hallanvuo S, Mikkola J, Kela E, Hatakka M, et al. A widespread outbreak of Yersinia pseudotuberculosis O:3 infection from iceberg lettuce. J Infect Dis. 2004;189:766–774. doi: 10.1086/381766. [DOI] [PubMed] [Google Scholar]
- Ortiz Martínez P, Fredriksson-Ahomaa M, Sokolova Y, Roasto M, Berzins A, Korkeala H. Prevalence of enteropathogenic Yersinia in Estonian, Latvian, and Russian (Leningrad region) pigs. Foodborne Pathog Dis. 2009;6:719–724. doi: 10.1089/fpd.2008.0251. [DOI] [PubMed] [Google Scholar]
- Ortiz Martínez P, Fredriksson-Ahomaa M, Pallotti A, Rosmini R, Houf K, Korkeala H. Variation in the prevalence of enteropathogenic Yersinia in slaughter pigs from Belgium, Italy, and Spain. Foodborne Pathog Dis. 2011;8:445–450. doi: 10.1089/fpd.2009.0461. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- Riet-Correa F, Gil-Turnes C, Reyes JC, Schild AL, Mendez MC. Yersinia pseudotuberculosis infection of buffaloes (Bubalus bubalis) J Vet Diagn Invest. 1990;2:78–79. doi: 10.1177/104063879000200117. [DOI] [PubMed] [Google Scholar]
- Rimhanen-Finne R, Niskanen T, Hallanvuo S, Makary P, Haukka K, Pajunen S, et al. Yersinia pseudotuberculosis causing a large outbreak associated with carrots in Finland, 2006. Epidemiol Infect. 2009;137:342–347. doi: 10.1017/S0950268807000155. [DOI] [PubMed] [Google Scholar]
- Sato K, Ouchi K, Taki M. Yersinia pseudotuberculosis infection in children, resembling Izumi fever and Kawasaki syndrome. Pediatr Infect Dis. 1983;2:123–126. doi: 10.1097/00006454-198303000-00011. [DOI] [PubMed] [Google Scholar]
- Seimiya YM, Sasaki K, Satoh C, Takahashi M, Yaegashi G, Iwane H. Caprine enteritis associated with Yersinia pseudotuberculosis infection. J Vet Med Sci. 2005;67:887–890. doi: 10.1292/jvms.67.887. [DOI] [PubMed] [Google Scholar]
- Shwimmer A, Freed M, Blum S, Khatib N, Weissblit L, Friedman S, Elad D. Mastitis caused by Yersinia pseudotuberculosis in Israeli dairy cattle and public health implications. Zoonoses Public Health. 2007;54:353–357. doi: 10.1111/j.1863-2378.2007.01072.x. [DOI] [PubMed] [Google Scholar]
- Skurnik M, Peippo A, Ervela E. Characterization of the O-antigen gene clusters of Yersinia pseudotuberculosis and the cryptic O-antigen gene cluster of Yersinia pestis shows that the plague bacillus is most closely related to and has evolved from Y. pseudotuberculosis serotype O:1b. Mol Microbiol. 2000;37:316–330. doi: 10.1046/j.1365-2958.2000.01993.x. [DOI] [PubMed] [Google Scholar]
- Sprague LD, Scholz HC, Amann S, Busse HJ, Neubauer H. Yersinia similis sp. nov. Int J Syst Evol Microbiol. 2008;58:952–958. doi: 10.1099/ijs.0.65417-0. [DOI] [PubMed] [Google Scholar]
- Sunahara C, Yamanaka Y, Yamanishi S. Sporadic cases of Yersinia pseudotuberculosis serotype 5 infection in Shodo Island, Kagawa Prefecture. Jpn J Infect Dis. 2000;53:74–75. [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr121. doi:10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubokura M, Otsuki K, Kawaoka Y, Maruyama T. Characterization and pathogenicity of Yersinia pseudotuberculosis isolated from swine and other animals. J Clin Microbiol. 1984;19:754–756. doi: 10.1128/jcm.19.6.754-756.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KM, Hanage WP, Fraser C, Connor TR, Spratt BG. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 2007;7:30. doi: 10.1186/1471-2180-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent P, Salo E, Skurnik M, Fukushima H, Simonet M. Similarities of Kawasaki disease and Yersinia pseudotuberculosis infection epidemiology. Pediatr Infect Dis J. 2007;26:629–631. doi: 10.1097/INF.0b013e3180616d3c. [DOI] [PubMed] [Google Scholar]
- Vincent P, Leclercq A, Martin L, Yersinia Surveillance Network. Duez JM, Simonet M, Carniel E. Sudden onset of pseudotuberculosis in humans, France, 2004-05. Emerg Infect Dis. 2008;14:1119–1122. doi: 10.3201/eid1407.071339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth JF. Current panoramic view and aspects of bovine yersiniosis caused by Yersinia pseudotuberculosis in Parana State-Brazil; Yersinia 2010 - 10th International Symposium; 2010.p. 54. [Google Scholar]
- Weber A, Knapp W. Demonstration of Yersinia enterocolitica and Yersinia pseudotuberculosis in fecal samples of healthy slaughter swine depending on the season. Zentralbl Veterinarmed B. 1981;28:407–413. [PubMed] [Google Scholar]
- Welsh RD, Stair EL. Yersinia pseudotuberculosis bovine abortion. J Vet Diagn Invest. 1993;5:109–111. doi: 10.1177/104063879300500127. [DOI] [PubMed] [Google Scholar]
- Wessels ME, Payne JH, Bannerman RP. Oculoglandular syndrome caused by Yersinia pseudotuberculosis in a dairy goat. J Comp Pathol. 2009;141:190–194. doi: 10.1016/j.jcpa.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Wilson DJ, Gabriel E, Leatherbarrow AJ, Cheesbrough J, Gee S, Bolton E, et al. Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol Biol Evol. 2009;26:385–397. doi: 10.1093/molbev/msn264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eBURST population snapshot of Y. pseudotuberculosis s.s., Y. similis and the Korean group. The blue dots represent predicted primary founders and yellow dots predicted subgroup founder with at least two descendent singlelocus variants.
Minimal spanning tree (maximum crosslink distance of one) of studied strains showing the genetic groups according to Fukushima and colleagues (2001).Other details are as in Fig. 1. In Y. similis ST75, strain Kuratani-2 is of genetic group G3 but contains YPMa although it does not harbour pYV or ferment melibiose.
Minimal spanning tree (maximum crosslink distance of one) of strains coloured by host group. Other details are as in Fig. 1.
Minimal spanning tree (maximum crosslink distance of one) of strains coloured by O-serotype. Other details are as in Fig. 1.
Minimal spanning tree (maximum crosslink distance of one) of strains coloured according to melibiose fermentation. Other details are as in Fig. 1.
Characteristics of the strains used in the analyses.