Summary
Biofilm formation in Bacillus subtilis requires the differentiation of a subpopulation of cells responsible for the production of the extracellular matrix that structures the biofilm. Differentiation of matrix-producing cells depends, among other factors, on the FloT and YqfA proteins. These proteins are present exclusively in functional membrane microdomains of B. subtilis and are homologous to the eukaryotic lipid raft-specific flotillin proteins. In the absence of FloT and YqfA, diverse proteins normally localized to the membrane microdomains of B. subtilis are not functional. Here we show that the absence of FloT and YqfA reduces the level of the septal-localized protease FtsH. The flotillin homologues FloT and YqfA are occasionally present at the midcell in exponentially growing cells and the absence of FloT and YqfA negatively affects FtsH concentration. Biochemical experiments indicate a direct interaction between FloT/YqfA and FtsH. Moreover, FtsH is essential for the differentiation of matrix producers and hence, biofilm formation. This molecular trigger of biofilm formation may therefore be used as a target for the design of new biofilm inhibitors. Accordingly, we show that the small protein SpoVM, known to bind to and inhibit FtsH activity, inhibits biofilm formation in B. subtilis and other distantly related bacteria.
Introduction
A widely conserved feature of bacteria is their ability to grow attached to almost any given surface, developing multicellular aggregates commonly referred to as biofilms (Davey and O’Toole, 2000; Stewart and Franklin, 2008; Lopez et al., 2010). Although the strategies that bacteria use to form biofilms vary among species, production of an extracellular matrix is generally necessary to encase the microbial community (Costerton et al., 1999; Donlan, 2002; Fux et al., 2005; Karatan and Watnick, 2009). Matrix-encased microbial communities are often composed of heterogeneous populations of physiologically distinct yet genetically identical cell types that contribute to biofilm formation (Stewart and Franklin, 2008; Lopez et al., 2009a). For instance, biofilm formation in the model organism Bacillus subtilis requires the differentiation of numerous cell types. Among these cell types, the matrix-producing cells that are responsible for the production and secretion of the extracellular matrix are absolutely necessary for proper biofilm formation (Branda et al., 2004; Chai et al., 2008; Vlamakis et al., 2008).
In order to monitor the differentiation of distinct cell types, transcriptional reporters have been used in conjunction with fluorescence microscopy (Veening et al., 2008; Lopez et al., 2010). We have previously used this technique to observe that matrix-producing cells differentiate in response to the secretion of a self-produced signalling molecule called surfactin. Surfactin activates the membrane histidine kinase KinC (Lopez et al., 2009b,c). KinC phosphorylates and activates the Spo0A master regulator (LeDeaux et al., 1995; Jiang et al., 2000), which, in turn, triggers the genetic cascade responsible for the differentiation of matrix producers.
Importantly, KinC localizes to membrane lipid microdomains that are functionally similar to the lipid rafts of eukaryotic cells (Lopez and Kolter, 2010a). The membrane microdomains of B. subtilis harbour two flotillin-like proteins: YqfA and FloT (formerly YuaG). In eukaryotes, flotillin proteins localize to lipid rafts and orchestrate diverse signal transduction processes that are harboured within lipid rafts (Morrow and Parton, 2005; Brown, 2006; Browman et al., 2007). In bacteria, these membrane proteins can organize diverse proteins related to signal transduction and protein secretion (Lopez and Kolter, 2010a). yqfA was initially identified as a gene of unknown function when studying genes of B. subtilis whose expression is controlled by the extracytoplasmic function sigma factor SigW (Huang et al., 1999; Turner and Helmann, 2000; Wiegert et al., 2001; Cao et al., 2002; Butcher and Helmann, 2006). The second flotillin-like protein FloT was discovered in 1999 (Tavernarakis et al., 1999) and it has been referred to in subsequent studies as archetypical flotillin-like protein in bacteria (Huang et al., 1999; Tavernarakis et al., 1999; Cao et al., 2002; Malaga-Trillo et al., 2002; Moszer et al., 2002; Ding et al., 2005; Walker et al., 2008; Donovan and Bramkamp, 2009; Lopez and Kolter, 2010a; Lee et al., 2012). The absence of FloT and YqfA alters the localization of proteins that are normally found within the detergent-resistant membrane (DRM) microdomains of B. subtilis (Lopez and Kolter, 2010a). As a result, cells defective in FloT and YqfA do not produce extracellular matrix in response to surfactin, in part, due to mislocalization and misfunction of KinC (Lopez and Kolter, 2010a), among other physiological defects (Dempwolff et al., 2012; Lee et al., 2012).
Nevertheless, in the absence of a functional KinC, Spo0A can be phosphorylated by the action of four other histidine kinases (KinA, B, D and E) (Jiang et al., 2000). Different levels of phosphorylated Spo0A (Spo0A~P) can be achieved depending on the phosphorylation efficiency of these kinases. Lower levels of Spo0A~P induce matrix gene expression, whereas higher levels are necessary for sporulation gene expression (Fujita et al., 2005). The absence of KinC hinders the activation of matrix gene expression in response to the presence of surfactin (Lopez et al., 2009b), yet it does not affect the efficiency of sporulation (LeDeaux et al., 1995; Jiang et al., 2000).
The physiological defects associated with the absence of FloT and YqfA seem broader than simply a loss of KinC activity. A previous report showed that deletion of floT in B. subtilis reduced sporulation efficiency (Donovan and Bramkamp, 2009), suggesting that FloT and YqfA could influence other proteins involved in the activation of Spo0A. In this report we show that the deletion of yqfA and floT results in reduced levels of the FtsH protease, which has been shown to indirectly regulate the phosphorylation of Spo0A via phosphatase degradation (Lysenko et al., 1997; Zellmeier et al., 2003; Le and Schumann, 2009). We also show that FtsH is present at the midcell of exponentially growing cells (Wehrl et al., 2000) and that it occasionally coincides with FloT and YqfA. We found that FloT and YqfA directly interact with FtsH and their expression is important for FtsH functionality. Furthermore, we present evidence that the activity of FtsH is required for the differentiation of the subpopulation of matrix producers, and by extension, for biofilm formation. Indeed, inhibition of FtsH activity by exogenously added SpoVM peptide, a known in vitro inhibitor of FtsH, prevents biofilm formation (Cutting et al., 1997; Prajapati et al., 2000). We propose that inhibition of FtsH may represent a new strategy for the development of novel antimicrobials with additional activity against biofilm formation.
Results and discussion
The ΔyqfA ΔfloT mutant exhibits diminished biofilm formation and sporulation
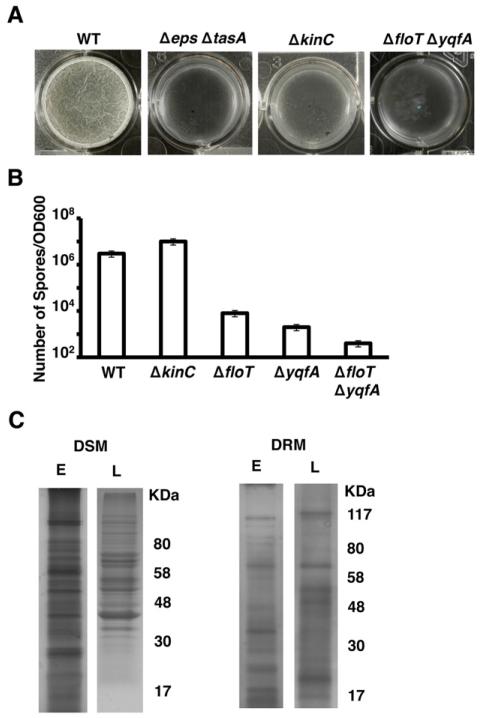
KinC localization to the functional membrane microdomains of B. subtilis requires the two flotillin homologue proteins FloT and YqfA (Lopez and Kolter, 2010a). Deletion of floT and yqfA not only results in mislocalization of KinC but also abolishes KinC activity, which prevents this strain from expressing matrix genes and forming a biofilm in response to the signal surfactin, similar to a ΔkinC mutant (Lopez et al., 2009b,c). However, the ΔfloT ΔyqfA mutant displayed a more severe defect in biofilm formation as well as an additional defect in sporulation as compared to the ΔkinC mutant. When both strains were grown in the biofilm-inducing medium (MSgg) for 24 h with no agitation, the ΔfloT ΔyqfA mutant was not able to form a biofilm in the form of a floating pellicle on the surface of the liquid. Figure 1A shows the top view of a wild-type pellicle of B. subtilis, which appears white, thick and wrinkled, whereas the extracellular matrix mutant fails to form surface pellicles (Δeps ΔtasA). The ΔfloT ΔyqfA flotillin-deficient mutant grew dispersed in the MSgg cultures, similar to the cultures of the matrix-deficient mutant (Fig. 1A). In contrast, the ΔkinC mutant was still able to form a thin, weak pellicle on the surface of the culture, leading us to conclude that the ΔfloT ΔyqfA mutant has a more severe defect in matrix production than the ΔkinC mutant.
Fig. 1. ΔfloT ΔyqfA mutant shows broader defect in biofilm formation and sporulation than the ΔkinC mutant.
A. Pellicle formation assay of different B. subtilis strains. Pictures show a top view of the pellicles formed on the surface of MSgg cultures incubated in 24-well plates at 30°C for 24 h. Positive and negative controls are represented by the wild-type strain (WT, DL1) and the matrix-deficient mutant (Δeps ΔtasA, DL7) respectively.
B. Viable spore counts comparing ΔfloT (DL1442) and ΔyqfA (DL1401) single mutants and ΔfloT ΔyqfA (AY93) double mutant in relation to ΔkinC (DL227) mutant and WT strain. Cultures were grown in shaking MSgg at 30°C for 48 h. The number of spores was correlated to the optical density of the MSgg cultures. Error bars indicate standard error of the means.
C. Membrane fractionation of WT cells according to differential sensitivity to detergent solubilization. SDS-PAGE analyses of the membrane fractions that are sensitive and resistant to detergent solubilization (DSM and DRM respectively). Samples were taken at early (E) and late (L) stages of growth in biofilm-inducing conditions. The protein pattern was analysed by Coomassie staining. The molecular weights are represented at the right of the gels.
We next asked if the ΔfloT ΔyqfA mutant also differed from the ΔkinC mutant in its sporulation efficiency. To test this, both strains were grown shaking in MSgg medium for 48 h and vegetative cells were subsequently heat-killed. Serial dilutions of the surviving spores were plated on fresh rich medium to induce their germination. The number of colonies resulting from the surviving spores relative to the optical density of the initial MSgg cultures was calculated (Fig. 1B). The ΔfloT ΔyqfA mutant was greatly defective in sporulation, while the ΔkinC mutant sporulated at a level similar to the wild-type strain. The sporulation defect observed in the ΔfloT ΔyqfA double mutant likely resulted from the combination of both gene deletions, because the ΔyqfA single mutant showed reduced sporulation efficiency similar to the effect previously described for the ΔfloT mutant (Fig. 1B) (Donovan and Bramkamp, 2009). Given that sporulation and matrix production are both processes regulated by Spo0A, we posited that the ΔfloT ΔyqfA mutant had other defects in addition to a non-functional KinC, which further inhibited the activation of the Spo0A genetic cascade.
To identify additional proteins that might influence the activation of Spo0A, we analysed the proteins that localize in the membrane microdomains along with FloT and YqfA. To this end, we purified the protein fraction associated with the DRM microdomains and analysed these samples as described previously (Zhang et al., 2005; Donovan and Bramkamp, 2009; Lopez and Kolter, 2010a). Briefly, cell extracts were treated with a mixture of non-ionic detergents and then separated by zonal centrifugation in sucrose gradients. This treatment resulted in two fractions: one that is sensitive to detergents (detergent-sensitive membrane fraction, DSM) and another fraction composed of larger membrane fragments that were more resistant to detergent disruption (detergent-resistant membrane fraction, DRM). Whereas it is important not to equate the pool of proteins present in the DRM fraction with raft-associated proteins, it is known that the DRM fraction is highly enriched in proteins associated with lipid rafts (Brown, 2002; 2006). Consequently, we analysed the DRM and DSM fractions from cultures in our biofilm-inducing medium at an early stage (2 h of incubation) or at a late stage of growth (24 h of incubation). Proteins associated with the DRM and the DSM fractions were analysed by SDS-PAGE. As was previously reported (Zhang et al., 2005; Donovan and Bramkamp, 2009; Lopez and Kolter, 2010a), there was a heterogeneous distribution of proteins in B. subtilis membranes (Fig. 1C). Moreover, substantial changes in the protein content of the DRM fraction were observed when we compared the cultures at early (E) or late (L) stage of growth in our experimental conditions (Fig. 1C). The DRM fraction at late stage of growth showed a similar banding pattern to what was previously reported (Lopez and Kolter, 2010a). To identify previously undetected proteins associated with the DRM, individual bands from the DRM fraction of 2 h cultures were excised from the gel and proteins were identified using mass spectrometry.
Mass spectrometry analysis of the prominent protein bands from the DRM fraction early stage of growth revealed a number of proteins involved in cell signalling and protein secretion, as previously described for more mature cultures (Lopez and Kolter, 2010a). For a complete list of proteins identified by mass spectrometry, see Table S1. Interestingly, the membrane-bound protease FtsH was found associated with the DRM fraction (Fig. S1). FtsH has been reported to indirectly affect the levels of phosphorylated Spo0A by degrading four regulatory phosphatase proteins, RapA, RapB, RapE and Spo0E, which feed into the Spo0A phosphorelay to ultimately decrease the levels of Spo0A~P (Le and Schumann, 2009). Accordingly, previous publications have shown that the ΔftsH mutant has a severe defect in sporulation, consistent with a decrease in the levels of Spo0A~P (Lysenko et al., 1997; Zellmeier et al., 2003). Because FtsH affects the activation of Spo0A, we hypothesized that the ΔfloT ΔyqfA mutant might have abrogated the functionality of FtsH and that this could be one of the reasons why the ΔfloT ΔyqfA mutant was defective in sporulation and matrix production. We therefore examined FtsH protein levels by Western blot analysis of cell extracts using polyclonal antibodies against the FtsH protease. In wild-type B. subtilis, FtsH was detectable in the DRM fraction and the signal was absent in the DRM fraction of the ΔfloT ΔyqfA mutant (Figs S1B and S2A). This result was important, but perhaps not surprising, as the protein profile from the DRM fraction of ΔfloT ΔyqfA mutant showed a general decrease in the overall protein content (Fig. S2B).
FloT and YqfA interact with FtsH in B. subtilis cells
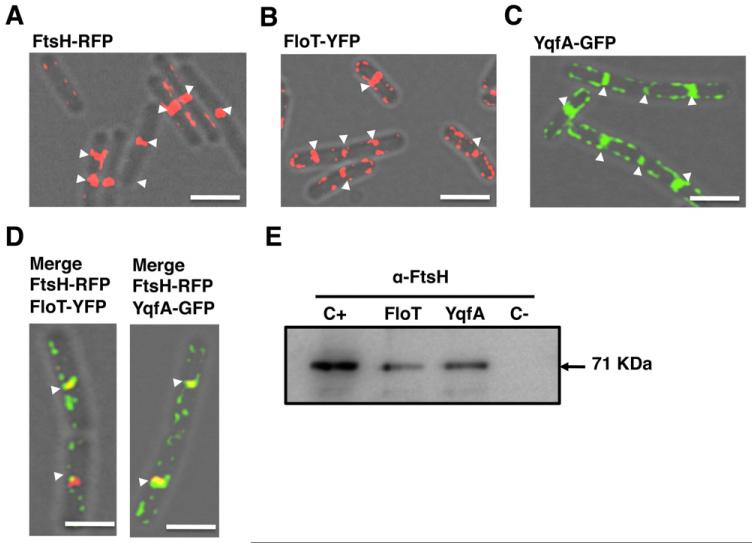
In order to determine if FtsH interacts with FloT and YqfA in the functional microdomains of B. subtilis, we first examined the subcellular distribution pattern of FtsH in relation to the flotillin homologue proteins FloT and YqfA using fluorescence microscopy. A previous report showed that FtsH preferentially localized to the septum of dividing cells (Wehrl et al., 2000), yet the distribution pattern of FloT and YqfA had not been investigated in exponentially growing cells. Accordingly, we constructed functional translational fusions of FloT or YqfA to fluorescent proteins under the control of their natural promoters (Lopez and Kolter, 2010a). In addition, we generated an FtsH–RFP translational fusion under the control of an IPTG inducible promoter. Using these strains, we analysed the distribution pattern of the three proteins in cells harvested at early stages of growth in MSgg medium. The translational fusion FtsH-RFP rendered a fully functional protein as shown in Figs S3, S4 and S11. As described previously, FtsH–RFP largely localized as a band at division septa (Fig. 2A). Fluorescence was also occasionally detected in discrete foci across the membrane (Fig. 2A; see also a larger field of view in Fig. S4). The fluorescence signal emitted by the FloT–YFP (Fig. 2B) and YqfA–GFP (Fig. 2C) was distributed as foci across the plasma membrane, and occasionally detected at the midcell with elevated fluorescence (white arrows in Fig. 2B and C; larger fields of view in Figs S5 and S6). Examination of cells harbouring both FtsH–RFP and FloT–YFP or FtsH–RFP and YqfA–GFP indicated that the proteins coincided at division septa in a significant number of cells (Fig. 2D; merge of the green and red signals is shown as yellow). Interference between green and red fluorescence signals was not detectable in our working conditions (Fig. S7).
Fig. 2. FloT and YqfA interact with FtsH.
Overlays of fluorescence micrographs and transmitted light images of cells grown in liquid shaking MSgg at 30°C harvested in mid-exponential phase (approximately 8 h of incubation). Midcell fluorescence is labelled with white arrows. Scale bars are 2 μm.
A. FtsH–RFP translational fusion (AY224, false coloured in red). IPTG concentration required for protein induction was 1 mM.
B. FloT–YFP translational fusion (DL1295, false coloured in red).
C. YqfA–GFP translational fusion (DL1367, false coloured in green).
D. Colocalization of both signals in the double-labelled strains FloT–YFP FtsH–RFP and YqfA–GFP FtsH–RFP (AY240 and AY238) appears as yellow in the merge panels (YFP signal is false coloured in green and RFP signal false coloured in red).
E. Immunoblot assay using polyclonal antibodies against FtsH to detect FtsH in the protein samples pulled down with YqfA-His6 and FloT-His6 proteins. The arrow indicates the presence of a band with the size predicted for FtsH. Positive control (C+) is the wild-type membrane fraction. Negative control (C−) is what eluted from the nickel-charged columns when loaded with a sample of wild-type membrane fraction. Protein samples that were pulled down with FloT-His6 (JS202) and YqfA-His6 (JS201) are presented in lanes FloT and YqfA respectively.
To investigate if FloT and YqfA interact with FtsH at division septa, we attempted to co-purify FloT and YqfA with FtsH from cell extracts. We therefore constructed two B. subtilis strains producing C-terminal His6-tagged variants of FloT and YqfA flotillin proteins (FloT-His6 and YqfA-His6 respectively). These strains were grown to exponential phase (OD600 = 0.8) in MSgg medium. Cells were harvested and the membrane fraction purified and solubilized with 0.2% of DDM. Samples were loaded onto a column of nickel-charged resin (Qiagen) that selectivity binds His6-tagged proteins and the proteins that are directly or indirectly bound to them. The pool of proteins bound to the resin was eluted from the column using an imidazole-containing buffer and resolved by SDS-PAGE. Western blot analysis was carried out using polyclonal antibodies against FtsH. A protein band corresponding to FtsH was detected in the protein sample that co-eluted with FloT-His6 (Fig. 2E, lane ‘FloT’). FtsH was also detected in the protein sample that co-eluted with YqfA-His6 protein (Fig. 2E, lane named YqfA). As a positive control, we detected FtsH in the total membrane fraction purified from wild-type cells (Fig. 2E, lane C+). In contrast, as a negative control, FtsH was not detected in the elution fraction of purified membranes from otherwise wild-type cells that did not harbour a His6-tagged protein, suggesting that retention of FtsH on the column was dependent on FloT-His6 or YqfA-His6 (Fig. 2E, lane C−). Altogether, these results are consistent with the idea that a direct interaction occurs between FloT and YqfA with FtsH. Studies in Escherichia coli showed that FtsH must oligomerize to properly function (Bieniossek et al., 2006; 2009) and that its oligomerization requires the chaperone activity of two proteins, HflC and HflK (Schumann, 1999; Ito and Akiyama, 2005; Hinderhofer et al., 2009). Interestingly, these two proteins are structurally similar to FloT and YqfA as well as other flotillin-like proteins (Winter et al., 2007; Hinderhofer et al., 2009). As B. subtilis lacks HflC and HflK proteins in their genome, it is tempting to speculate that FloT and YqfA might be the functional replacement of HflC and HflK.
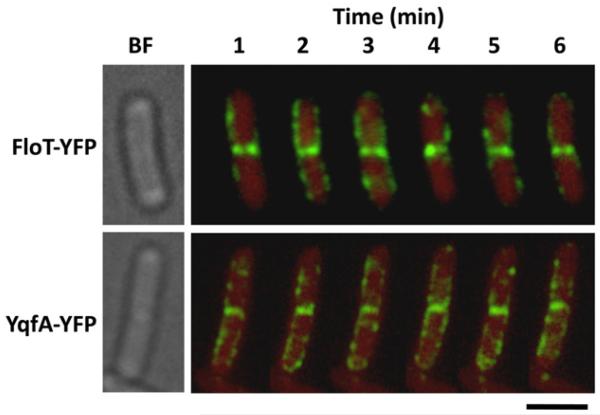
Because FtsH principally localizes to the septum of dividing cells (Wehrl et al., 2000), we reasoned that the interactions between FloT and YqfA with FtsH should be mainly localized at midcell. In this regard, we observed that, in the cases that flotillin-like proteins were detected in the septum, they behaved differently from the flotillins distributed throughout the membrane. Figure 3 shows time-lapse fluorescence microscopy of strains labelled with FloT–YFP or YqfA–GFP, to visualize FloT and YqfA foci every 60 s over a period of 6 min. The fluorescence signal that was distributed in foci across the membrane displayed a highly dynamic reorganization process, as the distribution pattern along the cell periphery continuously changed as previously reported for FloT (Donovan and Bramkamp, 2009). However, the fluorescence signal present at the septum for both FloT and YqfA flotillin proteins remained largely constant (Fig. 3). Quantitative measurements of the fluorescence signal detected in the time-lapse fluorescence microscopy experiments are shown in Figs S8-S10. As fluorescence signal was constantly detected at the septum of dividing cells during the time-lapse experiment, we reasoned that the continued presence of FloT and YqfA at the septum might be due to interactions between FloT and YqfA with FtsH (and probably with other proteins). It is currently unclear why FloT and YqfA behaved differently when located at the septum, but it is worth noting that the septal membrane has different lipid composition, and thus different physicochemical properties, to the cellular membrane (Kawai et al., 2004; Matsumoto et al., 2006; Donovan and Bramkamp, 2009) that might affect the behaviour of FloT and YqfA.
Fig. 3. Flotillins appear static at the midcell.
Time-lapse fluorescence analysis of the distribution pattern of FloT–YFP and YqfA–GFP foci. Cells were grown in liquid shaking MSgg at 30°C for 8 h. Exponentially growing cells were mounted on agarose-coated slides. The upper row shows the distribution of the FloT–YFP foci (DL1295, false-coloured green) within the same cell for 6 min. The bottom row shows the distribution of the YqfA–GFP foci (DL1367, false-coloured green) within the same cell for 6 min. Background is represented in red for better contrast of the fluorescent signal. Scale bar is 2 μm.
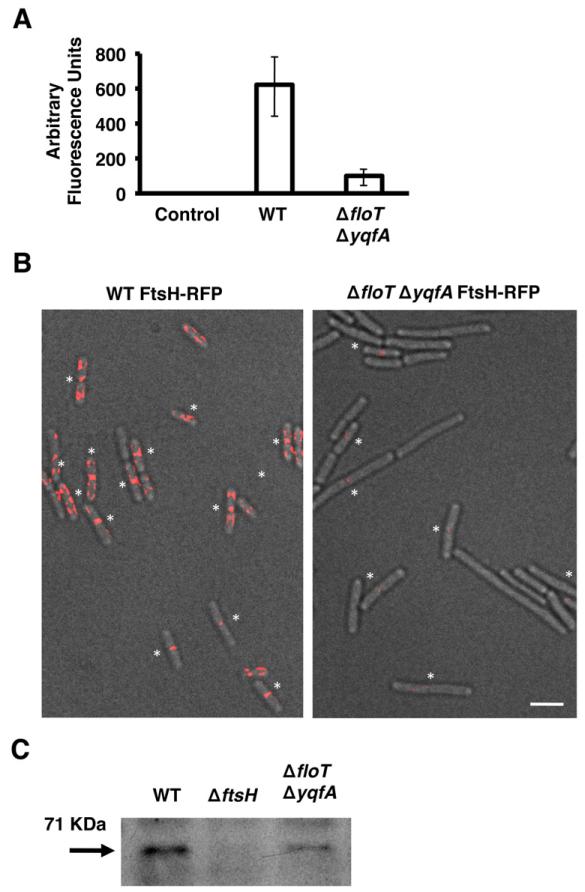
Thus far, our data indicated that FtsH and the flotillin homologues directly interact and that FtsH does not localize to the DRM microdomains in a strain lacking floT and yqfA (Fig. S2). In order to determine the dependence of FtsH localization on FloT and YqfA, we examined the localization of FtsH–RFP in a ΔfloT ΔyqfA double mutant. The absence of FloT and YqfA resulted in a decrease in the intensity of FtsH–RFP fluorescence signal. What faint signal we were able to detect appeared largely as foci near division septa (Fig. 4A and B). Immunoblot analysis using antibodies against FtsH confirmed that there were indeed lower levels of FtsH protein in the whole cell extracts of the ΔfloT ΔyqfA double mutant as compared to wild-type cells (Fig. 4C), confirming a functional link between FloT and YqfA and FtsH and possibly other septal-localized proteins (Dempwolff et al., 2012; Lee et al., 2012).
Fig. 4. The ΔfloT ΔyqfA mutant displays lower levels of FtsH.
A. Relative fluorescence of wild-type (WT) strain and ΔfloT ΔyqfA mutant labelled with the translational fusion FtsH–RFP (AY224 and DL1565 respectively). Cells were grown in liquid shaking MSgg at 30°C and harvested at mid-exponential phase (approximately 8 h of incubation; OD600 = 0.8). Quantification of relative fluorescence signal was assigned as fluorescence arbitrary units and presented in a graph. Error bars indicate standard error of the means.
B. Fluorescence micrographs overlayed on transmitted light images of WT and ΔfloT ΔyqfA mutant, both harbouring the translational fusion FtsH–RFP (false coloured in red). Asterisks indicate the fluorescence signal positioned in the septum of dividing cells for better visualization. Scale bar is 2 μm.
C. Immunoblot of FtsH protein in the indicated B. subtilis strains using polyclonal antibodies against FtsH. The arrow indicates the presence of a band with the size predicted for FtsH. Each lane contained 25 μg of total protein.
FtsH is required for the differentiation of the subpopulation of matrix producers and biofilm development
We wondered if the above results, suggesting that FloT and YqfA influence FtsH activity, could explain the defect in biofilm formation observed the ΔfloT ΔyqfA mutant. Previous research suggested that FtsH exerts a positive effect on the activation of Spo0A (Le and Schumann, 2009), yet its effect on the differentiation of matrix producers and thus, biofilm formation was not examined because the B. subtilis strains used in these studies were laboratory strains unable to produce the extracellular matrix necessary to form biofilms (Branda et al., 2001; McLoon et al., 2011).
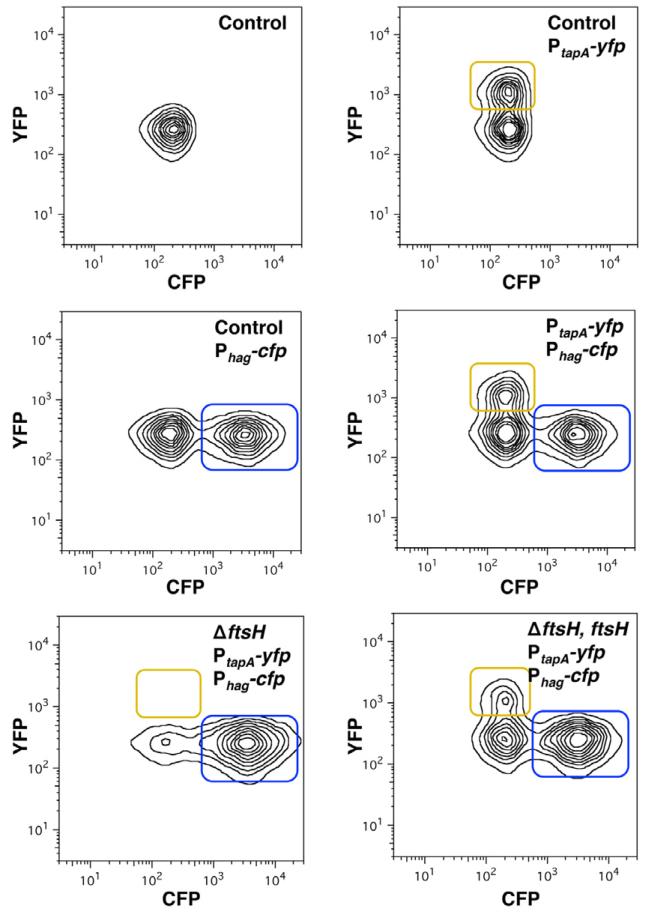
To test whether FtsH is required for the differentiation of the matrix producers, we monitored expression of matrix genes in the presence and absence of FtsH. Upon activation of Spo0A~P, B. subtilis cells transition from expressing motility genes to expressing genes involved in matrix production (Lopez and Kolter, 2010b). Thus, we measured the relative proportion of the subpopulations of motile cells and matrix producers using cultures of a double-labelled strain harbouring transcriptional fusions for structural components of the flagellum and matrix proteins. The Phag-cfp reporter is only expressed in the subpopulation of motile cells, whereas the PtapA-yfp fusion (formerly PyqxM-yfp) is exclusively expressed in matrix-producing cells (Chai et al., 2008; Vlamakis et al., 2008) A double-labelled strain harbouring the Phag-cfp and PtapA-yfp reporters was grown in standing biofilm-inducing medium MSgg cultures for 24 h. The subpopulations of motile cells and matrix producers were monitored by flow cytometry analysis of 50 000 cells (Fig. 5). The number of cells expressing each reporter was represented as contour isolines. The top left panel shows the background fluorescence in a strain harbouring no fluorescent reporters. Two additional controls were performed to distinguish the individual subpopulations in each channel using single-labelled strains and we observed about 38% of cells expressed the matrix reporter and 52% expressed motility reporter. When the double-labelled strain was monitored, both subpopulations appeared in each channel with approximately 30% and 50% of the population expressing either matrix or motility genes respectively (Fig. 5, right panel, middle row). In the absence of ftsH, about 67% of cells expressed the motility reporter, but there was no detectable expression of the PtapA-yfp matrix reporter (Fig. 5, bottom left panel), indicating that matrix producers did not differentiate in the absence of FtsH. The ΔftsH mutant complemented with an IPTG-inducible copy of the ftsH gene recovered the matrix fluorescence signal with about 21% of cells expressing matrix and 54% of cells expressing the motility reporter (Fig. 5, bottom right panel). The results suggested that FtsH has a role in assuring the fate of matrix-producing cells, perhaps by stabilizing the levels of active Spo0A (Spo0A~P).
Fig. 5. FtsH is necessary for the differentiation of matrix producers.
Flow cytometry monitoring the expression of the reporter PtapA-yfp (YFP fluorescence on the y-axis) and Phag-cfp (CFP fluorescence on the x-axis) from cells grown on MSgg medium. The number of cells is represented by isolines. The top left panel shows the control of background fluorescence for both CFP and YFP in a strain harbouring no fluorescent protein genes (DL1). Top right panel: the strain harbouring PtapA-yfp (DL382). The subpopulation expressing fluorescence above background is framed in yellow. Centre left panel: the strain harbouring Phag-cfp (DL1056) showed a subpopulation of cells highly expressing the reporter framed in blue. Centre right panel: the strain harbouring both reporters, PtapA-yfp and Phag-cfp (DL1079). Bottom left panel: the ΔftsH mutant strain harbouring both reporters, PtapA-yfp and Phag-cfp (DL1521). Bottom right panel: the ΔftsH mutant strain harbouring both reporters, PtapA-yfp and Phag-cfp, complemented with the gene ftsH controlled by an IPTG-inducible promoter (DL1523, induction with 1 mM IPTG).
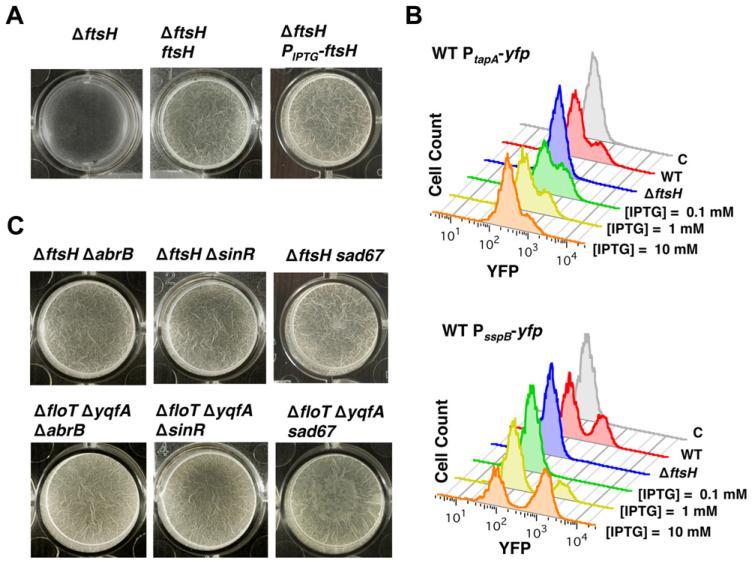
We predicted that the inability of the ΔftsH mutant to express matrix genes should abrogate biofilm formation. We tested this by allowing cultures to form floating pellicles on the surface of liquid MSgg (Figs 6A and S11). After 24 h of incubation, the ΔftsH mutant grew dispersed in the cultures and no pellicle formed. The ability to form pellicles could be restored in the ΔftsH mutant when complemented in trans with ftsH under the control of an IPTG-inducible promoter and partially restored when ftsH expression was under the control of its natural promoter (Figs 6A and S11). Flow cytometry was used to monitor expression of the PtapA-yfp reporter in the ΔftsH, PIPTG-ftsH strain. The ability of this strain to form pellicles was associated with the differentiation of the subpopulation of matrix producers (Fig. 6B). In addition, consistent with the sporulation defect that was previously described (Lysenko et al., 1997; Zellmeier et al., 2003), the ΔftsH mutant was unable to express the sporulation specific promoter PsspB-yfp unless the ΔftsH, PIPTG-ftsH strain was grown in the presence of IPTG (Fig. 6B).
Fig. 6. Influence of FtsH on biofilm formation.
A. Pellicle formation of the ΔftsH mutant (DL1308, left panel), the ΔftsH mutant complemented with ftsH controlled by its native promoter (DL1433, middle panel) and ΔftsH mutant complemented with an IPTG-inducible promoter (DL1361, right panel). IPTG was added to a final concentration of 1 mM. Pictures show a top view of the pellicles formed on the surface of MSgg cultures incubated in 24-well plates at 30°C for 24 h.
B. Flow cytometry analysis of the same strains as in A harbouring PtapA-yfp reporter to monitor the differentiation of the subpopulation of matrix producers. Control strain harbouring no reporter fusion (grey profile). Wild-type (WT) profile shows a subpopulation of cells with high relative fluorescence, seen as the shoulder to the right of the main peaks (red profile) (DL382). The FtsH-defective mutant does not show the differentiation of this subpopulation (blue profile) (DL1404). Expression of ftsH under the control of an IPTG-inducible promoter led to a gradual expression of FtsH in the FtsH-defective mutant, which in turn caused a differentiation of matrix producers (different concentrations of IPTG are shown) (DL1461). Flow cytometry profiles of the reporter PsspB-yfp to detect sporulating cells. WT profile shows a subpopulation of cells with high relative fluorescence (red profile) (DL1089). The FtsH-defective mutant does not show the differentiation of this subpopulation (blue profile) (1349). Expression of ftsH under the control of an IPTG-inducible promoter led to a gradual expression of FtsH in the FtsH-defective mutant, which in turn caused a differentiation of sporulating cells (different concentrations of IPTG are shown) (DL1364).
C. Pellicle formation assay of the indicated strains of B. subtilis when incubated in MSgg at 30°C for 24 h. The sad67 variant was expressed under the control of an IPTG-inducible promoter with 1 μM IPTG.
The biofilm defect observed in the ΔftsH mutant appeared to be due to its inability to express matrix genes (see Figs 5 and 6B). If this was due to a signalling defect, we reasoned that we should be able to restore biofilm formation by uncoupling signalling and matrix gene expression. There are two repressor proteins, SinR and AbrB, which negatively regulate expression of the genes responsible for matrix production (Hamon et al., 2004; Chu et al., 2006; Chai et al., 2008; Murray et al., 2009) (Fig. S12). Thus, the absence of either SinR or AbrB repressors leads to constitutive expression of extracellular matrix. Deletion of either sinR or abrB in the ΔftsH mutant strain restored pellicle formation. Similarly, deletion of sinR or abrB suppressed the biofilm formation defect of strains that did not produce the flotillin proteins YqfA and FloT (Fig. 6C). These results indicated that both the ΔftsH and ΔfloT ΔyqfA mutants were physiologically capable of forming biofilms and their impairment in biofilm formation is likely due to an inhibition of the genetic cascade to matrix production.
The expression of the SinR and AbrB repressors is controlled by Spo0A~P and levels of Spo0A~P are indirectly regulated by the protease FtsH (Le and Schumann, 2009). Epistasis analyses were carried out in ΔftsH and ΔfloT ΔyqfA mutants by expressing a constitutively active variant of spo0A (sad67) produced under the control of an IPTG-inducible promoter. Sad67 does not require any upstream regulation to maintain high active levels of the Spo0A protein (Ireton et al., 1993). Pellicle formation was assayed in cells expressing sad67 and harbouring a deletion in either ftsH or floT and yqfA. Standing MSgg cultures incubated for 24 h showed pellicle formation in both strains, indicating that the artificial activation of Spo0A~P restored pellicle formation in the absence of FtsH or the flotillin homologue proteins (Fig. 6C).
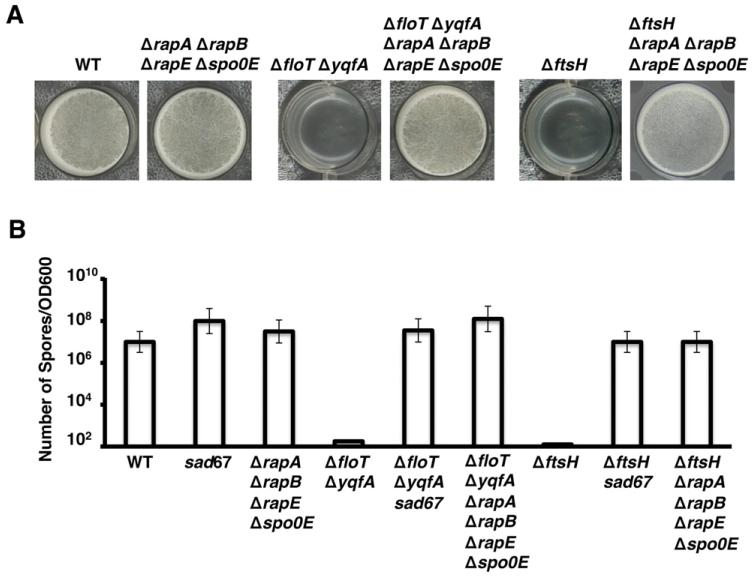
The absence of the FtsH protease increases the level of the phosphatase proteins, RapA, RapB, RapE and Spo0E, which ultimately decrease the levels of Spo0A~P (Le and Schumann, 2009). To bypass the reduction in the functionality of the FtsH in the ΔfloT ΔyqfA double mutant, we generated a strain lacking the pool of phosphatases degraded by FtsH using the ΔfloT ΔyqfA strain as genetic background. The resultant strain ΔfloT ΔyqfA ΔrapA ΔrapB ΔrapE Δspo0E was tested for its ability to form biofilm in biofilm-inducing conditions. This strain recovered the ability to form pellicles in standing MSgg cultures (Fig. 7A). Thus, the biofilm formation defect of the ΔfloT ΔyqfA double mutant can be bypassed by deleting the FtsH-regulated phosphatases that ultimately dephosphorylate Spo0A~P. Indeed, the ability to form pellicles in standing MSgg cultures was also recovered in the strain ΔftsH ΔrapA ΔrapB ΔrapE Δspo0E (Fig. 7A).
Fig. 7. Epistasis analysis of the ΔftsH mutant and ΔfloT ΔyqfA double mutant to restore biofilm formation.
A. Pellicle formation of the indicated strains of B. subtilis. Pictures show a top view of the pellicles formed on the surface of MSgg incubated in 24-well plates at 30°C for 24 h. Positive control is represented by the wild-type strain (WT) and negative controls are represented by the ΔfloT ΔyqfA double mutant (strain JS163) and the ΔftsH mutant (DL1308).
B. Viable spore counts comparing WT, ΔfloT ΔyqfA and ΔftsH strains when complemented with the sad67 variant (strains DL1148, DL1375 and DL1363 respectively) or with a deletion of the FtsH-regulated phosphatases (rapA, rapB, rapE and spo0E) (strains DL1430, DL1554 and DL1375 respectively). sad67 was expressed under the control of an IPTG-inducible promoter with 1 μM IPTG. Cultures were grown in shaking MSgg at 30°C for 48 h. Number of spores was correlated to the optical density of the cultures. Error bars indicate standard error of the means.
Several studies have described that the ΔftsH mutant is unable to sporulate (Lysenko et al., 1997; Zellmeier et al., 2003; Le and Schumann, 2009), and similar results are presented in this work for the ΔfloT ΔyqfA double mutant (see Fig. 1). As wild-type levels of sporulation could be restored in the ΔftsH mutant when complemented with sad67 (Le and Schumann, 2009), we wondered if expression of sad67 could restore sporulation to the ΔfloT ΔyqfA double mutant. Indeed, we found that expression of sad67 in ΔfloT ΔyqfA mutant cells restored sporulation efficiency to near wild-type levels (Fig. 7B). Importantly, wild-type levels of sporulation were also observed in the ΔfloT ΔyqfA ΔrapA ΔrapB ΔrapE Δspo0E and ΔftsH ΔrapA ΔrapB ΔrapE Δspo0E strains. Wild-type levels of sporulation were also observed in the ΔftsH sad67 strain and ΔftsH ΔrapA ΔrapB ΔrapE Δspo0E strains (Fig. 7B). Altogether, the various phenotypes arising from the absence of the FtsH protease, including biofilm formation and sporulation, were restored in the ΔftsH or ΔfloT ΔyqfA double mutants either by inhibiting the pool of FtsH-regulated phosphatases that decrease the levels of Spo0A~P or alternatively, by expressing a spo0A variant (sad67), whose activation is not influenced by the phosphorelay in which the FtsH-regulated phosphatases are involved.
Synthetic SpoVM protein inhibited FtsH protease and biofilm formation
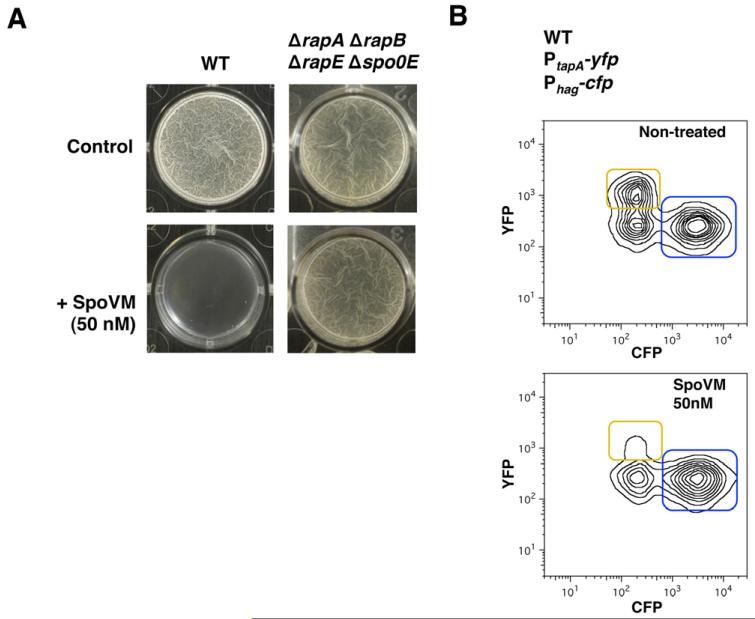
The interconnection between FtsH protease activity and matrix production presented in this study led us to explore new possibilities to develop anti-biofilm compounds by targeting FtsH protease activity. Previous studies have reported that the small protein SpoVM, a 26-amino-acid-long protein that is normally present in the forespore membrane of B. subtilis (Cutting et al., 1997; van Ooij and Losick, 2003; Ramamurthi et al., 2006; 2009; Wang et al., 2009), binds to and inhibits FtsH protease in vitro (Cutting et al., 1997; Prajapati et al., 2000). Thus, we tested the ability of SpoVM to inhibit biofilm formation in cultures of B. subtilis. To this end, we synthetically engineered SpoVM peptide, and sub-growth-inhibitory concentrations (50 nM) of a stock buffered solution of SpoVM were added to the pellicle formation assay in MSgg medium. We observed that nanomolar concentrations of the peptide were sufficient to inhibit pellicle formation after 24 h (Fig. 8A). Interestingly, the presence of nanomolar concentrations of SpoVM did not affect the ability of the ΔrapA ΔrapB ΔrapE Δspo0E mutant to form biofilm (Fig. 8A). Biofilm inhibition was also observed in solid MSgg when SpoVM was added to the MSgg agar as evidenced by flatter colony morphology (Fig. S13). Single-cell analysis for gene expression in the double-labelled strain Phag-cfp, PtapA-yfp treated with nanomolar concentrations of the SpoVM peptide showed that B. subtilis was unable to express matrix-specific genes in the presence of SpoVM. Unlike untreated samples, the majority of the SpoVM-treated cells remained as motile cells (Fig. 8B). Furthermore, sub-growth-inhibitory concentrations of SpoVM inhibited biofilm formation in other bacterial species such as Staphylococcus aureus and Pseudomonas aeruginosa (Fig. S14). Whether this inhibition occurs in an FtsH-dependent manner remains to be determined. Nevertheless, the small protein SpoVM, an amphipathic alpha helical FtsH inhibitor (Cutting et al., 1997), is an appealing molecule that shows great potency and versatility against biofilms of diverse organisms could be further exploited to develop novel anti-biofilm agents.
Fig. 8. SpoVM protein inhibits differentiation of matrix producers and biofilm formation.
A. Pellicle formation of B. subtilis wild-type (WT) or rapA, rapB, rapE and spo0E mutant in the presence or absence (control) of the protein SpoVM (50 nM).
B. Flow cytometry monitoring the expression of the reporter PtapA-yfp (YFP fluorescence on the y-axis) and Phag-cfp (CFP fluorescence on the x-axis) from B. subtilis cells grown on the pellicle formation assay. Fluorescence for both CFP and YFP in a strain harbouring both reporters, PtapA-yfp and Phag-cfp which correspond to the matrix producers and motile cells, framed in yellow and blue respectively. Non-treated (top panel) or treated with the SpoVM protein 50 nM (bottom panel).
Experimental procedures
Strains, media and culture conditions
Strains used in this study were B. subtilis strain NCIB3610 (Branda et al., 2001), S. aureus strains SC-01 (Beenken et al., 2003) and P. aeruginosa PA14 (O’Toole and Kolter, 1998a). Additional laboratory strains of E. coli DH5α and B. subtilis 168 were used for cloning purposes. A complete strain list is shown in Table S2.
To monitor cell differentiation during cell division, B. subtilis was incubated in liquid MSgg without shaking at 30°C. Incubation times varied depending on the requirements of the experiment. Specific conditions are presented in the figure legends. Generally, pellicle formation assays required incubation times of 24 h at 30°C. Cells harvested at early stages of growth in biofilm-inducing conditions required 2 h of incubation time at 30°C, while cells harvested at late stages of growth in biofilm-inducing conditions required 24 h of incubation at 30°C. To monitor cell division in shaking cultures, B. subtilis was incubated in liquid MSgg at 200 r.p.m. at 30°C. Incubation times are specified for each experiment in the figure legends. Biofilm formation assay for the strain B. subtilis 3610 was carried out as follows. Overnight cultures grown in LB were diluted 1:100 in biofilm-inducing medium MSgg (Branda et al., 2001). For pellicle formation assays, 1 ml of culture was dispensed in polystyrene well plates and incubated overnight at 30°C.
To grow biofilms of the strain S. aureus SC-01, a preculture grown overnight in TSB liquid medium was diluted 1:00 in TSB + glucose 0.5% + NaCl 3% (Beenken et al., 2003). One millilitre of the culture was dispensed in polystyrene well plates and incubated overnight at 37°C. To grow biofilms of P. aeruginosa, a preculture grown overnight in TB medium (O’Toole and Kolter, 1998b) was diluted 1:100. Two hundred microlitres of the culture was dispensed in polystyrene well plates and incubated overnight at 37°C. Biofilms formed by S. aureus and P. aeruginosa were stained with crystal violet for better visualization, according to the protocol described by O’Toole and Kolter (1998b).
Selective media were prepared in LB agar using antibiotics at the following final concentrations: ampicillin 100 μg ml−1, kanamycin 50 μg ml−1, chloramphenicol 5 μg ml−1, tetracycline 5 μg ml−1, spectinomycin 100 μg ml−1 and erythomycin 2 μg ml−1 + lincomycin 25 μg ml−1 for MLS. When required, MSgg culture medium was supplemented with threonine 1%. When needed, IPTG was added at concentrations 1 mM for the overexpression of ftsH, floT and yqfA and 1 μM for the overexpression of sad67. When required, SpoVM was added to the medium at a concentration of 50 nM.
Strain construction and reporters
Deletion mutants were generated using long flanking homology PCR (Wach, 1996) (using the primers listed in Table S3). Markerless gene deletions were used to generate the ΔfloT ΔyqfA double mutant. Upstream and downstream regions of the floT and yqfA genes were joined by long flanking homology PCR (Wach, 1996) and cloned into the temperature-sensitive vector pMAD. Gene deletion occurs by a sequential process of double recombination. Isolation of the mutants was achieved by counterselection, as described in Arnaud et al. (2004). Transcriptional reporters used in this study were previously constructed and published (Vlamakis et al., 2008; Lopez et al., 2009c,d; Aguilar et al., 2010; Lopez and Kolter, 2010a). Translational fusions used in this study were generated using long flanking homology PCR. Unless specified differently in the figure legends, transcriptional fusions are expressed under the control of their natural promoters. Translational fusions were cloned in pKM008 or pKM003 vectors, and integration into the bacterial genome occurs at the amyE locus. pKM008 or pKM003 vectors were kindly provided by Prof. Dr David Rudner (Harvard Medical School, Boston, MA, USA) as was the pDR183 vector which allows the integration of the translational fusions into the lacA locus. pDG1663 was used for integration into the thrC locus (Guerout-Fleury et al., 1996). In all cases, plasmids were linearized to favour a double recombination process and added to a culture of B. subtilis strain 168 grown in conditions that promotes the activation of natural competence (Hardwood and Cutting, 1990). After 2 h of incubation, cells were plated in the corresponding selection media.
Overexpression of ftsH occurred under the control of the IPTG-inducible promoter Phyperspank using the plasmid pDR111 (Britton et al., 2002; Nakano et al., 2003; Erwin et al., 2005). Additional subcloning into pDR183 allowed the integration of the construct Phyperspank-ftsH into the lacA locus. The concentration of IPTG used to induce the expression of ftsH was 1 mM. Constructions were transferred to the strain NCIB3610 by SPP1 phage transduction (Yasbin and Young, 1974; Novick, 1991). Briefly, donor strain was grown in TY medium (LB + 10 mM MgSO4 + 10 μM MnSO4). A diluted sample of a SPP1 phage stock was added to the culture and, after 30 min of incubation, 3 ml of soft TY agar was added to the culture. Phage halos arised after incubation overnight at 37°C. Soft agar was resuspended in TY liquid medium and supernatant was passed through a 0.22 μm syringe filter. This supernatant was used to infect a culture of the NCIB3610 or mutant derivative recipient strain grown in TY medium. A full protocol of this process is available in the literature (Garcia-Betancur et al., 2012).
Image capture and analysis
Unless different conditions were specified in figure legends, samples were fixed with paraformaldehyde treatment before single-cell analysis. Cells were resuspended in 1 ml of 4% paraformaldehyde solution and incubated at room temperature for 7 min. After washing, samples were resuspended in PBS buffer. Samples were repeatedly washed prior to single-cell analysis. Images were processed using Leica Application Suite V3.7 software. Microscopy images were taken on a Leica DMI6000B microscope equipped with a Leica CRT6000 illumination system. The microscope was equipped with a HCX PL APO oil immersion objective with 100 × 1.47 magnification that was used in this study. The microscope was also equipped with a colour camera Leica DFC630FX and temperature-control system. Image processing was performed using Leica Application Suite Advance Fluorescence Software and Photoshop. Deconvolution of fluorescence signal was performed using AutoQuant™ Deconvolution algorithms software, from Media Cibernetics. Signals were detected using the following filters: GFP signal was detected using an excitation filter BP480/40 and an emission filter BP527/30; YFP signal was detected using an excitation filter BP500/20 and an emission filter BP535/30; RFP signal was detected using an excitation filter BP546/40 and a emission filter BP600/40. Excitation times for GFP, YFP and RFP signals were between 100 and 200 ms. Transmitted light images were taken with 36 ms of excitation time. Pellicles formed in microtitre plates were photographed using a Nikon D100 camera coupled to a Kaiser RB5000 illumination system. Pictures were processed using Photoshop software. Fluorescence intensity analyses were performed in cells growing in MSgg agar for 24 h at 30°C using a Luminiscent Image Analyzer ImageQuant® LAS4000 (General Electric) coupled with ImageQuant-TL software for quantification of fluorescence.
Flow cytometry
For flow cytometric analysis, cells were dispersed from biofilms with 12 sonication pulses (power output 0.7 and cycle 50%). After dispersion, cells were fixed with a treatment of 4% paraformaldehyde, washed and resuspended in PBS buffer. Dilution of samples 1:100 was necessary prior to flow cytometry analyses. Further sonication treatment was required to separate single cells in the sample. In this case, samples were subjected to three consecutive series of 12 pulses (power output 50% and cycle 0.7 s). Flow cytometry analysis was carried out in a BD Fortessa flow cytometer (BD Biosciences). For YFP fluorescence, we used a laser excitation of 488 nm coupled with 530/30 and 505LP sequential filters. For CFP fluorescence, we used laser excitation at 405 nm coupled with 408/40 and 460LP sequential filters. The photomultiplier voltage was set between 400 and 500 V. No gates were required during the analysis of the samples. Every sample was analysed measuring 50 000 events using FACS Diva (BD Biosciences) software to capture the data. Further data analysis was performed in FlowJo 9.2 (http://www.flowjo.com).
Spore counting
Strains were grown in shaking MSgg at 30°C for 48 h. The optical density of each culture was recorded to represent the total cell density in Figs 1B and 8B. For the quantification of spores in the cultures, a sample from each culture was normalized to 1 ml of a final optical density (OD600 nm) of 1. Vegetative cells were killed by incubating samples at 80°C for 30 min. Serial dilutions were plated from the normalized preparation and colony forming units were counted. Colony forming units grown from viable spores were represented in relation to the optical density of the culture, which the sample was extracted. This protocol was adapted from previous publications (Aguilar et al., 2010).
Cell membrane fractionation and Western blot analysis
To purify the membrane fraction at late time points during cell growth, 30 ml of MSgg was inoculated and cells incubated for 24 h at 30°C to a final OD600 of about 3.0. Cultures were centrifuged and cells resuspended in PBS buffer. To purify the membrane fraction at early time points during cell growth, 500 ml of MSgg culture was inoculated to an OD600 of 0.1. After 2 h of incubation time at 30°C and a final OD600 of about 0.2, cells were harvested and processed. Cell suspensions were treated with lysozyme 1 mg ml−1 at 37°C for 30 min. Cell debris was eliminated by normal centrifugation (13 000 r.p.m. for 2 min). Supernatant was subjected to ultra-centrifugation (75 000 r.p.m. for 40 min) to separate the membrane fraction. Next, the membrane fraction was resuspended in PBS buffer and treated with the CellLytic MEM protein extraction kit (Sigma-Aldrich) to purify proteins associated with detergent-resistant fractions. CellLytic MEM protein extraction kit separates two samples containing the DRM and the DSM fractions. Both samples were run on SDS-PAGE and proteins were detected by Coomassie blue staining. Coomassie-stained bands were excised from the gel and analysed by mass spectrometry (Thermo Scientific LTQ Orbitrap XL). Immunoblot was carried out as previously described (Lopez et al., 2009b) using polyclonal antibody against FtsH, kindly provided by Prof. Dr Thomas Wiegert (Institute of Genetics, University of Bayreuth, Germany). When specified, the protein content was adjusted to 25 μg of total protein per lane by using Nanodrop® Spectrophotometer ND-1000 to quantify the protein concentration of the samples.
Pull down assays
The two strains overexpressing the His-tagged variants of the flotillin FloT-His6 and YqfA-His6 proteins were grown in 100 ml of MSgg cultures to exponential phase (OD600 of 0.8). The cell pellet was harvested and the membrane fraction was purified according to the protocol described in the above section. The membrane fraction was solubilized in 5 ml of Buffer S (HEPES 20 mM, glycerol 20%, DTT 1 mM, DDM 0.02%) and loaded into a column of Ni-NTA superflow (Qiagen). The column was washed with 20 ml of PBS buffer DDM 0.02%. Proteins bound to the column were eluted using 2 ml of PBS buffer DDM 0.02% + imidazol 250 mM. Elution fraction was collected and the proteins precipitated by adding three volumes of acetone and further incubation overnight at −20°C. Samples were centrifuged and intensely washed with acetone. Protein samples were resuspended in 0.1 ml of PBS buffer and tested for the presence of FtsH by Western blot analysis, using polyclonal antibodies against FtsH. Immunoblot was carried out as previously described (Lopez et al., 2009b).
Supplementary Material
Acknowledgements
We thank all members of the Institute of Molecular Infection Biology (IMIB), especially Isa Westedt for technical assistance. We thank Prof. Dr Thomas Wiegert (University of Bayreuth, Germany) for kindly providing FtsH antibody and the strain harbouring the deletion ftsH∷tet. This work was funded by the Young Investigator Program of the Research Center of Infectious Diseases (ZINF) of the University of Würzburg, Germany and the grant LO 1804/2-1 from The German Research Foundation DFG. G.K. is recipient of Schrödinger fellowship (FWF). J.C.G.B. is recipient of PhD fellowship from the Graduate School of Life Sciences (GSLS) of the University of Wuerzburg. K.S.R. acknowledges funding from the Intramural Research Program of the NIH National Cancer Institute Center for Cancer Research.
Footnotes
Conflict of interest: The authors declared no conflict of interest.
References
- Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. MBio. 2010;1:00035-10. doi: 10.1128/mBio.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C, Schalch T, Bumann M, Meister M, Meier R, Baumann U. The molecular architecture of the metalloprotease FtsH. Proc Natl Acad Sci USA. 2006;103:3066–3071. doi: 10.1073/pnas.0600031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C, Niederhauser B, Baumann UM. The crystal structure of apo-FtsH reveals domain movements necessary for substrate unfolding and translocation. Proc Natl Acad Sci USA. 2009;106:21579–21584. doi: 10.1073/pnas.0910708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browman DT, Hoegg MB, Robbins SM. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 2007;17:394–402. doi: 10.1016/j.tcb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Brown DA. Isolation and use of rafts. Curr Protoc Immunol. 2002;Chapter 11 doi: 10.1002/0471142735.im1110s51. Unit 11.10. [DOI] [PubMed] [Google Scholar]
- Brown DA. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology. 2006;21:430–439. doi: 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- Butcher BG, Helmann JD. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol Microbiol. 2006;60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- Cao M, Kobel PA, Morshedi MM, Wu MF, Paddon C, Helmann JD. Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J Mol Biol. 2002;316:443–457. doi: 10.1006/jmbi.2001.5372. [DOI] [PubMed] [Google Scholar]
- Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg E. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Cutting S, Anderson M, Lysenko E, Page A, Tomoyasu T, Tatematsu K, et al. SpoVM, a small protein essential to development in Bacillus subtilis, interacts with the ATP-dependent protease FtsH. J Bacteriol. 1997;179:5534–5542. doi: 10.1128/jb.179.17.5534-5542.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey ME, O’Toole AG. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempwolff F, Möller MH, Graumann PL. Synthetic motility and cell shape defects for deletions of flotillin/reggie paralogs in Bacillus subtilis and interplay with NfeD proteins. J Bacteriol. 2012;194:4652–4661. doi: 10.1128/JB.00910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Jiang M, Jiang W, Su Y, Zhou H, Hu X, Zhang Z. Expression, purification, and characterization of recombinant human flotillin-1 in Escherichia coli. Protein Expr Purif. 2005;42:137–145. doi: 10.1016/j.pep.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan C, Bramkamp M. Characterizationand subcellular localization of a bacterial flotillin homologue. Microbiology. 2009;155:1786–1799. doi: 10.1099/mic.0.025312-0. [DOI] [PubMed] [Google Scholar]
- Erwin KN, Nakano S, Zuber P. Sulfate-dependent repression of genes that function in organosulfur metabolism in Bacillus subtilis requires Spx. J Bacteriol. 2005;187:4042–4049. doi: 10.1128/JB.187.12.4042-4049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Gonzalez-Pastor JE, Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Garcia-Betancur JC, Yepes A, Schneider J, Lopez D. Single-cell analysis of Bacillus subtilis biofilms using fluorescence microscopy and flow cytometry. J Vis Exp. 2012;(60):e3796. doi: 10.3791/3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier NP. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Hamon MA, Stanley NR, Britton RA, Grossman AD, Lazazzera BA. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol Microbiol. 2004;52:847–860. doi: 10.1111/j.1365-2958.2004.04023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwood CR, Cutting SM. Molecular Biological Methods for Bacillus. Wiley; New York: 1990. [Google Scholar]
- Hinderhofer M, Walker CA, Friemel A, Stuermer CA, Moller HM, Reuter A. Evolution of prokaryotic SPFH proteins. BMC Evol Biol. 2009;9:10. doi: 10.1186/1471-2148-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Gaballa A, Cao M, Helmann JD. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W. Mol Microbiol. 1999;31:361–371. doi: 10.1046/j.1365-2958.1999.01180.x. [DOI] [PubMed] [Google Scholar]
- Ireton K, Rudner DZ, Siranosian KJ, Grossman AD. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- Ito K, Akiyama Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K. Cardiolipin domains in Bacillus subtilis marburg membranes. J Bacteriol. 2004;186:1475–1483. doi: 10.1128/JB.186.5.1475-1483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le AT, Schumann W. The Spo0E phosphatase of Bacillus subtilis is a substrate of the FtsH metalloprotease. Microbiology. 2009;155:1122–1132. doi: 10.1099/mic.0.024182-0. [DOI] [PubMed] [Google Scholar]
- LeDeaux JR, Yu N, Grossman AD. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:861–863. doi: 10.1128/jb.177.3.861-863.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Kingston AW, Helmann JD. Glutamate dehydrogenase affects resistance to cell wall antibiotics in Bacillus subtilis. J Bacteriol. 2012;194:993–1001. doi: 10.1128/JB.06547-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Kolter R. Functional microdomains in bacterial membranes. Genes Dev. 2010a;24:1893–1902. doi: 10.1101/gad.1945010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Kolter R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev. 2010b;34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev. 2009a;33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci USA. 2009b;106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Losick R, Kolter R. Paracrine signaling in a bacterium. Genes Dev. 2009c;23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol. 2009d;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko E, Ogura T, Cutting SM. Characterization of the ftsH gene of Bacillus subtilis. Microbiology. 1997;143(Pt 3):971–978. doi: 10.1099/00221287-143-3-971. [DOI] [PubMed] [Google Scholar]
- McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. Tracing the domestication of a biofilmforming bacterium. J Bacteriol. 2011;193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaga-Trillo E, Laessing U, Lang DM, Meyer A, Stuermer CA. Evolution of duplicated reggie genes in zebrafish and goldfish. J Mol Evol. 2002;54:235–245. doi: 10.1007/s00239-001-0005-1. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Kusaka J, Nishibori A, Hara H. Lipid domains in bacterial membranes. Mol Microbiol. 2006;61:1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- Morrow IC, Parton RG. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic. 2005;6:725–740. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- Moszer I, Jones LM, Moreira S, Fabry C, Danchin A. SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res. 2002;30:62–65. doi: 10.1093/nar/30.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EJ, Strauch MA, Stanley-Wall NR. SigmaX is involved in controlling Bacillus subtilis biofilm architecture through the AbrB homologue Abh. J Bacteriol. 2009;191:6822–6832. doi: 10.1128/JB.00618-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Kuster-Schock E, Grossman AD, Zuber P. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci USA. 2003;100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- van Ooij C, Losick R. Subcellular localization of a small sporulation protein in Bacillus subtilis. J Bacteriol. 2003;185:1391–1398. doi: 10.1128/JB.185.4.1391-1398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998a;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998b;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Prajapati RS, Ogura T, Cutting SM. Structural and functional studies on an FtsH inhibitor from Bacillus subtilis. Biochim Biophys Acta. 2000;1475:353–359. doi: 10.1016/s0304-4165(00)00089-1. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Clapham KR, Losick R. Peptide anchoring spore coat assembly to the outer fore-spore membrane in Bacillus subtilis. Mol Microbiol. 2006;62:1547–1557. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann W. FtsH - a single-chain charonin? FEMS Microbiol Rev. 1999;23:1–11. doi: 10.1111/j.1574-6976.1999.tb00389.x. [DOI] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Driscoll M, Kyrpides NC. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem Sci. 1999;24:425–427. doi: 10.1016/s0968-0004(99)01467-x. [DOI] [PubMed] [Google Scholar]
- Turner MS, Helmann JD. Mutations in multidrug efflux homologs, sugar isomerases, and antimicrobial biosynthesis genes differentially elevate activity of the sigma(X) and sigma(W) factors in Bacillus subtilis. J Bacteriol. 2000;182:5202–5210. doi: 10.1128/jb.182.18.5202-5210.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Walker CA, Hinderhofer M, Witte DJ, Boos W, Moller HM. Solution structure of the soluble domain of the NfeD protein YuaF from Bacillus subtilis. J Biomol NMR. 2008;42:69–76. doi: 10.1007/s10858-008-9261-3. [DOI] [PubMed] [Google Scholar]
- Wang KH, Isidro AL, Domingues L, Eskandarian HA, McKenney PT, Drew K, et al. The coat morphogenetic protein SpoVID is necessary for spore encasement in Bacillus subtilis. Mol Microbiol. 2009;74:634–649. doi: 10.1111/j.1365-2958.2009.06886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrl W, Niederweis M, Schumann W. The FtsH protein accumulates at the septum of Bacillus subtilis during cell division and sporulation. J Bacteriol. 2000;182:3870–3873. doi: 10.1128/jb.182.13.3870-3873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegert T, Homuth G, Versteeg S, Schumann W. Alkaline shock induces the Bacillus subtilis sigma(W) regulon. Mol Microbiol. 2001;41:59–71. doi: 10.1046/j.1365-2958.2001.02489.x. [DOI] [PubMed] [Google Scholar]
- Winter A, Kamarainen O, Hofmann A. Molecular modeling of prohibitin domains. Proteins. 2007;68:353–362. doi: 10.1002/prot.21355. [DOI] [PubMed] [Google Scholar]
- Yasbin RE, Young FE. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellmeier S, Zuber U, Schumann W, Wiegert T. The absence of FtsH metalloprotease activity causes overexpression of the sigmaW-controlled pbpE gene, resulting in filamentous growth of Bacillus subtilis. J Bacteriol. 2003;185:973–982. doi: 10.1128/JB.185.3.973-982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Li Z, Tsudome M, Ito S, Takami H, Horikoshi K. An alkali-inducible flotillin-like protein from Bacillus halodurans C-125. Protein J. 2005;24:125–131. doi: 10.1007/s10930-004-1519-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.