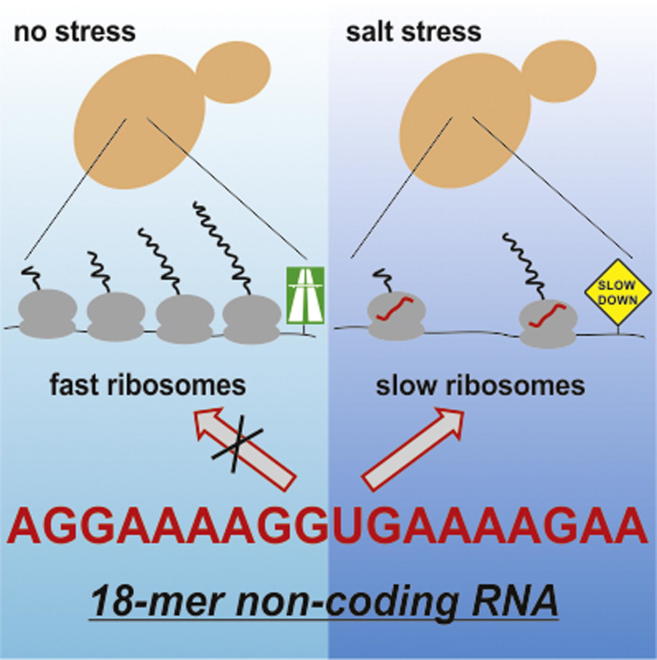
Summary
The structural and functional repertoire of small non-protein-coding RNAs (ncRNAs) is central for establishing gene regulation networks in cells and organisms. Here, we show that an mRNA-derived 18-nucleotide-long ncRNA is capable of downregulating translation in Saccharomyces cerevisiae by targeting the ribosome. This 18-mer ncRNA binds to polysomes upon salt stress and is crucial for efficient growth under hyperosmotic conditions. Although the 18-mer RNA originates from the TRM10 locus, which encodes a tRNA methyltransferase, genetic analyses revealed the 18-mer RNA nucleotide sequence, rather than the mRNA-encoded enzyme, as the translation regulator. Our data reveal the ribosome as a target for a small regulatory ncRNA and demonstrate the existence of a yet unkown mechanism of translation regulation. Ribosome-targeted small ncRNAs are found in all domains of life and represent a prevalent but so far largely unexplored class of regulatory molecules.
Graphical Abstract
Highlights
-
•
This study reveals the yeast ribosome as direct target for small regulatory ncRNAs
-
•
An 18-nt-long exon-derived RNA fragment from the TRM10 locus binds to ribosomes
-
•
This 18-mer ncRNA inhibits global protein biosynthesis in vivo and in vitro
-
•
This translation attenuation is crucial for adaption under hyperosmotic stress
Small noncoding RNAs involved in translation regulation typically target mRNAs. Pircher et al. reveal that the ribosome is a target for regulatory noncoding RNAs. An mRNA-derived 18-nucleotide-long RNA fragment directly interacts with ribosomes and inhibits global protein biosynthesis thus allowing specific stress adaption in yeast.
Introduction
Small non-protein-coding RNA (ncRNA) molecules are key players in controlling gene expression at multiple steps in all domains of life (Amaral et al., 2008; Hüttenhofer et al., 2005; Mattick, 2004; Tuck and Tollervey, 2011). In the past years, it became evident that ncRNAs represent a widespread class of regulatory molecules shaping cellular life (Aalto and Pasquinelli, 2012). The advantage of ncRNA regulators is their almost immediate availability because they act on the RNA level and thus do not need to be converted into a polypeptide in order to fulfill their cellular function. Translation represents the last step in gene expression, and its regulation allows a swift and reversible adaption to changing environmental conditions (Gebauer and Hentze, 2004). The list of validated ncRNAs regulating translation, such as micro RNAs (Huntzinger and Izaurralde, 2011; Krol et al., 2010) and small-interfering RNAs (Mello and Conte, 2004), is growing steadily; however, they almost exclusively target the mRNA rather than the ribosome, the key enzyme of protein biosynthesis. This is unexpected given the central position the ribosome plays in cell metabolism and the assumption that the protoribosome originated in the RNA world (Crick, 1968; Steitz and Moore, 2003) and thus likely depended on regulatory input from nonproteinous cofactors such as small metabolites or short ncRNAs. In contemporary biology, protein biosynthesis is a very energy-demanding process and therefore rigorously regulated in response to environmental changes. Controlled translation regulation enables a cell or an organism to fine-tune its proteome in time and space. Regulatory input is typically given by stress-induced modifications (e.g., phosphorylation) of essential initiation factors, by mRNA-binding proteins that can sense environmental changes (e.g., iron regulatory proteins), or by the action of mRNA-targeted microRNAs (Gebauer and Hentze, 2004). The yeast S. cerevisiae is one of the few characterized eukaryal organisms known to lack components of the RNA interference machinery and thus lives without microRNA and small interfering RNA (siRNA) translation regulation (Houseley and Tollervey, 2008).
Here, we set out to functionally characterize an mRNA exon-derived 18-residue-long ncRNA candidate that was picked up in our recent genomic screen for ribosome-bound small RNAs in S. cerevisiae (Zywicki et al., 2012). We show that this 18-mer RNA fragment is a functional ncRNA capable of adjusting translation rates by interacting with polysomes under hyperosmotic growth conditions.
Results
An mRNA-Derived 18-mer RNA Associates with Ribosomes In Vivo
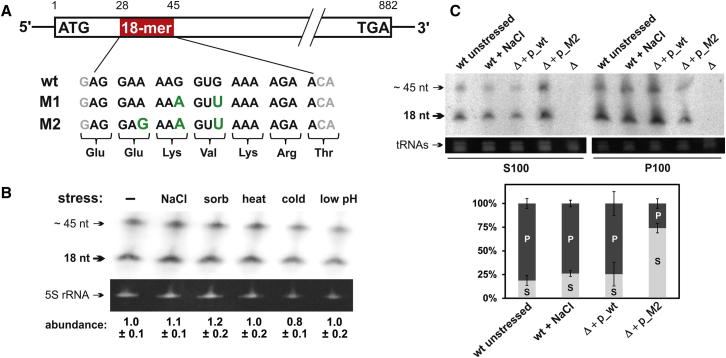
To identify potential alternate mechanisms of translation regulation in S. cerevisiae and to address the question whether small ncRNAs exist that directly target the ribosomes under specific growth conditions, we investigated the ribosome-associated RNome (size range ∼15–500 nucleotides) (Zywicki et al., 2012). In addition to known ribosome-bound ncRNAs (tRNAs, 7SL RNA), 20 mRNA exon-derived fragments, sized between 18 and 70 nucleotides, copurified with ribosomes. The most abundant mRNA fragment originates from the TRM10 locus, which encodes a tRNA methyltransferase (Jackman et al., 2003). The resulting TRM10 mRNA piece is 18 nucleotides long and is located 28 residues downstream of the translation start site (Figure 1A). The 18-mer RNA, and a putative ∼45-residue-long processing intermediate, was expressed in a stress-independent manner (Figure 1B). By comparing the northern blot signals for the 18-mer RNA in the pellet fraction of a 100,000 × g centrifugation (P100) of cell lysates, which contains the ribosomes, with the corresponding supernatant (S100) (see Supplemental Information available online for details), about 80% of the signal was detected in the P100 fraction. This demonstrates that the vast majority of cellular 18-mer RNA is associated with ribosomes in vivo (Figure 1C).
Figure 1.
The mRNA-Derived 18-mer RNA Associates with Ribosomes In Vivo
(A) Schematic representation of the S. cerevisiae TRM10 locus with its embedded 18-mer ncRNA candidate (shown in red). The wild-type (wt) sequence (bold; black) as well as two variants (M1, M2) of the 18-mer used in this study are shown below. The point mutations (green) were designed such that they generate synonymous codons in the context of the TRM10 open reading frame.
(B) Northern blot analyses on total RNA isolated from unstressed (−) or stressed cells (hyperosmotic stress: elevated NaCl or sorbitol [sorb]; heat, cold, or low pH stress). The presence of the 18-mer RNA and of a putative ∼45-nucleotide-long precursor is indicated by arrows. The ethidium-bromide-stained 5S rRNA serves as loading control. The relative abundance of the 18-mer RNA under different conditions was quantified relative to the 5S rRNA signal.
(C) The cellular distribution of the 18-mer RNA between the postribosomal supernatant (S100) and the ribosome-containing pellet (P100) fractions was assessed by northern blot analyses. Distribution in the unstressed wt strain was compared to the wt strain at high NaCl concentration, to the trm10Δ strain (Δ) or to the rm10Δ strain expressing the wt or the M2 mutant variant of the TRM10 locus from a plasmid (Δ + p_wt, Δ + p_M2). Ethidium-bromide-stained tRNAs serve as loading controls. The quantifications of three independent experiments is shown below the blots (P: P100 fraction; S: S100 fraction).
The TRM10 18-mer ncRNA Binds to Polysomes under Hyperosmotic Stress and Promotes Cell Growth
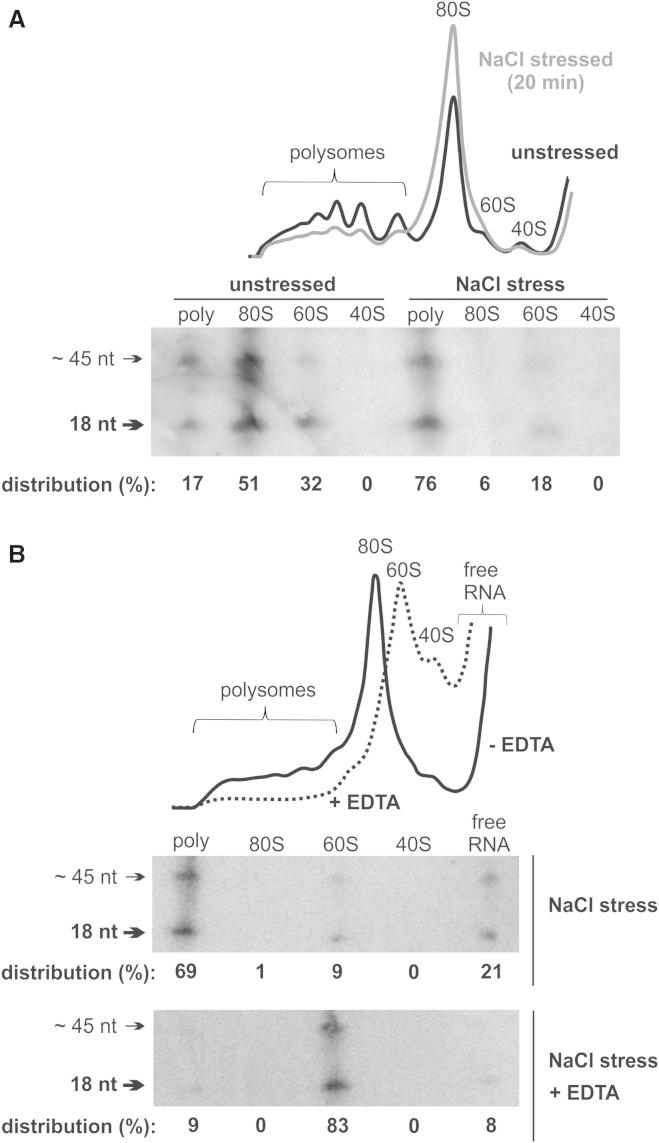
To gain insight into the in vivo function of this 18-mer ncRNA candidate, the growth characteristics of a TRM10 knockout strain (trm10Δ) were investigated under nine different growth conditions. In line with previous reports (Gustavsson and Ronne, 2008; Jackman et al., 2003), the lack of the TRM10-encoded methyltransferase had no growth phenotype under most conditions (Figures S1A–S1F). However, under hyperosmotic stress conditions in the presence of elevated concentrations of NaCl or sorbitol, the trm10Δ strain showed a slow growth phenotype (Figures S1G and S1H). To test if and how the TRM10-derived 18-mer fragment plays a role in this phenomenon, polysome profiling and genetic analyses were performed. Even though the total portion of ribosome-associated 18-mer RNA remains constant in unstressed and high-salt-stressed cells (Figure 1C), polysome profiling revealed significant differences. Polysome profiling identified nontranslating 80S ribosomes in unstressed and polysomes in salt-stressed cells as main targets (Figure 2A). More than 80% of 18-mer was associated with 80S ribosomes and 60S subunits in unstressed cells, and only a minor portion entered the actively translating polysome pool. However, the fractional distribution changed markedly upon salt addition. Under these hyperosmotic conditions, the 18-mer RNA relocates and almost 80% was present in the polysomes, whereas it was almost completely absent in the 80S ribosome fraction (Figure 2A). Although the data shown in Figures 1C and 2A indicate a direct interaction between ribosomes and the 18-mer RNA, they do not unequivocally exclude the possibility of an mRNA-association mechanism. To clarify this, polysome profiling was performed in the presence of EDTA, conditions known to remove and dissociate translating polysomes from mRNAs (del Prete et al., 2007). If the 18-mer was mRNA bound, it is expected to shift into the pool of free RNA on top of the gradient, whereas it should remain in heavier fractions, when the 18-mer RNA was ribosome associated. Northern blot analysis revealed that the 18-mer RNA sediments primarily in the 60S ribosomal subunit fraction and does not accumulate in the free RNA pool (Figure 2B). These data demonstrate that 18-mer RNA targets the 60S subunit in vivo.
Figure 2.
The 18-mer RNA Binds to 60S Ribosomal Subunits
(A) Northern blot analyses on ribosome-associated RNA obtained from density gradient (top) fractions of cells grown in rich medium (unstressed; black trace) or in high-salt medium (NaCl stress; gray trace). Equal portions of each gradient fraction were loaded for analyses. The fractional distribution of the 18-mer RNA in the different fractions is given at the bottom of the blot.
(B) Polysome profiling of ribosomal particles isolated from cells grown under high-salt conditions was performed in the absence (black trace) or in the presence of EDTA (dotted trace). The fractional distribution of the 18-mer RNA between polysomes, 80S, 60S, and 40S, and the pool of free RNAs was determined by northern blot analysis. The quantification is always given below the blot. The experiments shown in (A) and (B) were repeated at least three times, whereas the SDs were less than 14%.
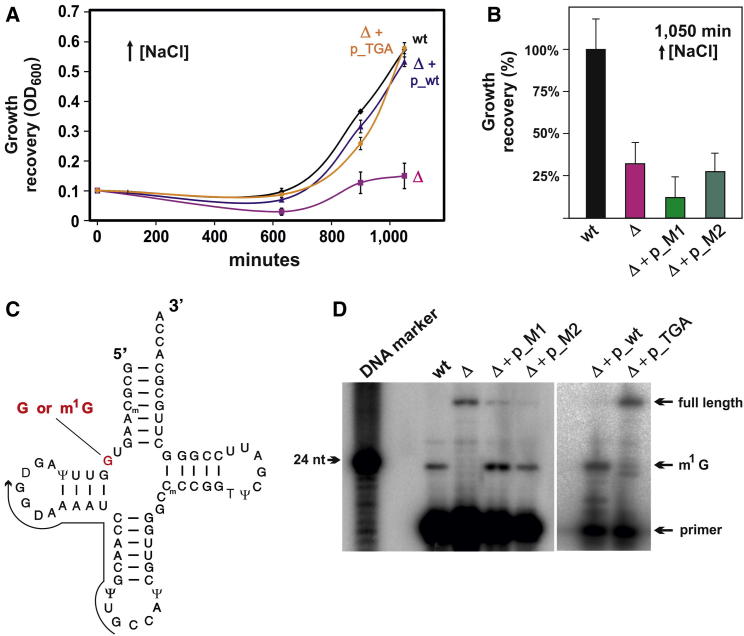
To explore the role of the ribosome-bound 18-mer RNA fragment during cell growth under stress conditions, genetic complementation experiments were performed. Northern blot analyses showed that the TRM10-derived 18-mer RNA was expressed and associated with ribosomes also when the gene was transcribed from the plasmid (Figure 1C). As expected, no northern blot signal for the 18-mer RNA was evident in the trm10Δ strain thus demonstrating that this RNA fragment derives from the TRM10 gene. To monitor growth under stringent high-salt conditions, the strains were first grown to stationary phase in stress medium, diluted with fresh stress medium, and subsequently allowed to resume growth. Complementation experiments under these harsh stress conditions demonstrated that the markedly reduced growth of the trm10Δ strain could be rescued by expressing the TRM10 locus from a plasmid (Figure 3A). To distinguish whether the absence of the TRM10-encoded tRNA methyltransferase or the mRNA-derived 18-mer RNA fragment is responsible for the growth defects, we either introduced a UGA stop codon or introduced synonymous mutations (M1, M2) within the mRNA 18-mer region (Figure 1A). These experiments showed that cells expressing a nontranslatable TRM10 mRNA had no growth defect at elevated salt concentrations (Figure 3A), even though no active tRNA methyltransferase was produced (Figures 3C and 3D). The reciprocal experiment, when two (M1) or three (M2) synonymous codons were introduced into the TRM10 gene, showed the opposite effect. Despite the fact that an active tRNA methyltransferase was expressed (Figure 3D), the cells failed to resume growth in high-salt medium (Figure 3B). These data reveal the lack of the mRNA-derived ncRNA candidate and not the mRNA-encoded tRNA methyltransferase as the cause for the observed growth defects at high-salt concentrations. The M1 and M2 mutants could not recover growth thus demonstrating the sequence specific mode of action of the TRM10 18-mer RNA. In support of this, the M2 mutant version of the 18-mer RNA was also largely absent from ribosomes (Figure 1C). Therefore, in order to promote growth under hyperosmotic conditions, the 18-mer RNA needs to physically interact with ribosomes in vivo. The 18-mer RNA acts specifically under hyperosmotic conditions, because its absence did not result in a growth phenotype during heat or cold shock (Figure S2).
Figure 3.
Growth Characteristics of the TRM10 Knockout Strain
(A and B) Growth of the wt strain was compared to the TRM10 knockout strain trm10Δ (Δ) in high NaCl medium in the stringent “redilution assay.” The growth characteristics of the trm10Δ strain carrying the TRM10 gene on a plasmid (Δ + p_wt) was furthermore compared to strains that were complemented with the start-codon mutant plasmid (Δ + p_TGA), or plasmids harboring synonymous codon mutations in the TRM10 open reading frame (Δ + p_M1; Δ + p_M2). In (B), the cell density of a wt culture that was rediluted to OD600 0.1 and subsequently incubated in high NaCl medium for 1,050 min (black bar) was set to 100%. Each experimental point was done in triplicates, and the growth curves were repeated three times. The mean and the SDs are shown. See also Figures S1 and S2.
(C) Schematic representation of S. cerevisiae tRNAGly secondary structure is shown, whereas the guanosine at position 9 is colored in red and is either unmethylated (G) or methylated at nucleobase position 1 (m1G). Activity of the TRM10-encoded tRNA methyltransferase was assessed by monitoring the methylation status at G9 of tRNAGly by primer extension analysis (arrow indicates primer binding site).
(D) In wt cells, G9 is fully methylated resulting in a reverse transcriptase stop one nucleotide before the methylation site, thus resulting in a 24-nucleotide-long product. In the trm10Δ strain (Δ) or in the trm10Δ strain expressing the untranslatable start-codon mutant of TRM10 from a plasmid (Δ + p_TGA), G9 is unmethylated, and reverse transcription proceeds until the tRNA 5′ end (full length). Expressing the TRM10-gene-containing synonymous mutations (Δ + p_M1 and Δ + p_M2) in the open reading frame within the 18-mer region from a plasmid, the tRNA methyltransferase was active, thus resulting in an almost quantitative m1G modification. A radiolabeled 24-mer DNA oligo served as length marker (left lane). Two separate representative polyacrylamide gels are depicted.
The 18-mer ncRNA Inhibits Protein Biosynthesis In Vivo and In Vitro
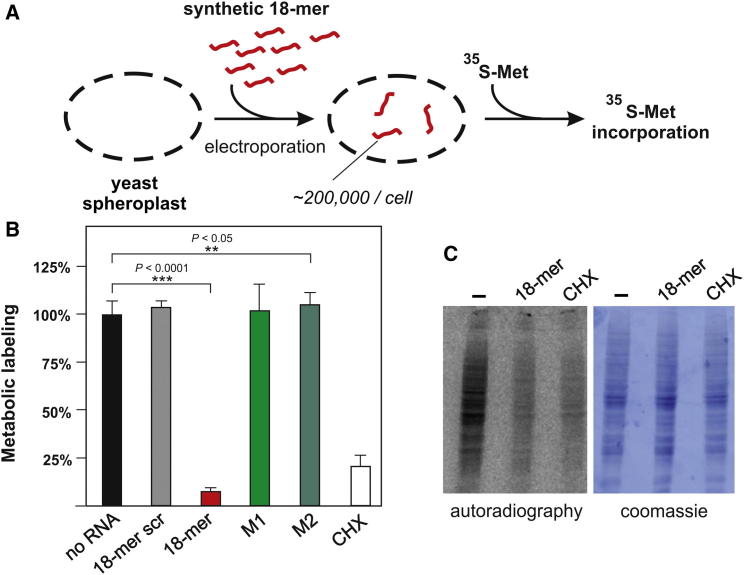
To study the role of the TRM10 18-mer fragment in vivo, we adapted a metabolic labeling approach using yeast spheroplasts (Russell et al., 1991) by measuring the 35S-Met incorporation into newly made proteins (Figure 4A). Introducing the 18-mer RNA into the spheroplasts by electroporation resulted in an almost complete inhibition of protein biosynthesis, whereas a scrambled 18-mer RNA control had no effect (Figure 4B). Gel electrophoresis and autoradiography indicated global translation inhibition, thus arguing for a general downregulation of protein synthesis in the presence of the 18-mer RNA (Figure 4C). Quantification of the uptake efficiency of the synthetic 18-mer into spheroplasts indicated the presence of about 200,000 molecules per cell, thus roughly equaling the ribosome concentration (Warner, 1999). Also, in this assay, the inhibitory function of the 18-mer RNA was sequence specific because RNA strands containing two (M1) or three (M2) mutations (Figure 1A) were unable to affect metabolic labeling of cellular proteins (Figure 4B). Additionally, also the secondary structure context of the 18-mer sequence within the introduced RNA strand influenced the inhibitory potential (Figure S3).
Figure 4.
The Effect of the TRM10-Derived 18-mer on In Vivo Protein Biosynthesis
(A) Protein synthesis in yeast spheroplasts was monitored by a metabolic labeling procedure in the presence or absence of synthetic 18-mer RNA (red). The synthetic RNA was introduced into the spheroplasts via electroporation, which results in ∼200,000 molecules/cell (see Supplemental Experimental Procedures). The amounts of newly synthesized proteins were assessed after addition of 35S-Met and TCA precipitation.
(B) The incorporation of 35S-methionine into the proteome of yeast spheroplasts in the absence of added RNA (no RNA) was taken as 100% and compared to spheroplasts harboring the synthetic 18-mer RNA, a scrambled 18-mer (18-mer scr), or 18-mer RNA variants carrying point mutations (M1, M2). Cycloheximide (CHX) served as a translation inhibition control. The mean and the SD of at least three metabolic labeling experiments are shown. p values were determined by a two-tailed unpaired Student’s t test. See also Figure S3.
(C) Newly synthesized proteins after the 35S-methionine spike in the absence of RNA (−) or in the presence of the 18-mer RNA or cycloheximide were visualized by SDS PAGE and subsequent autoradiography. The Coomassie-stained gel serves as a loading control.
These results described above indicate that the TRM10 18-mer RNA targets protein biosynthesis. To corroborate these findings, in vitro translation reactions were performed using S. cerevisiae cell extracts. Addition of the 18-mer RNA, but not the M2 mutant, clearly reduced in vitro protein synthesis to a similar extent as the known translation elongation inhibitor cycloheximide (Figure 5A). Translation inhibition by the 18-mer RNA was dose dependent with an apparent IC50 of 2.5 μM (Figures 5B and 5C). This value is in a physiologically reasonable range considering the in vivo concentrations of the 18-mer RNA (1.1 μM; Figure S4) and of yeast ribosomes (∼8 μM) (Petelenz-Kurdziel et al., 2011; Warner, 1999). Notably, the yeast 18-mer RNA was also able to inhibit in vitro translation in a wheat germ system with a slightly increased IC50 of 7 μM but affected mammalian and bacterial protein biosynthesis to a lesser extent (Figure S5). To gain insight whether the 18-mer interferes with translation initiation or elongation, the S. cerevisiae in vitro translation assay was slightly modified. Complete reactions were assembled in the absence of radiolabeled methionine and 18-mer RNA at the regular temperature of 23°C, thus allowing translation initiation and elongation. Subsequently, the reactions were placed on ice and 35S methionine and 13 μM synthetic 18-mer were added (Figure 5D; condition II). At low temperature, translation initiation is massively inhibited (Al-Fageeh and Smales, 2006; Hofmann et al., 2012), whereas already initiated ribosomes can continue protein synthesis. Under these conditions that prevent reinitiation, the 18-mer RNA had no inhibitory effect on in vitro translation activity (Figure 5D). On the other hand, when the 18-mer and 35S methionine were added in an experimental set-up that also monitors translation initiation (condition I, Figure 5D), significant translation inhibition was observed. These findings thereby suggest the 18-mer RNA to interfere with the initiation phase of protein biosynthesis.
Figure 5.
The 18-mer RNA Inhibits In Vitro Translation
(A) Addition of synthetic wt 18-mer RNA but not the M2 oligonucleotide (13 μM f.c.) results in in vitro translation inhibition in S. cerevisiae. The sample in the absence of any synthetic RNA (−) served as positive translation control and was set to 100%, whereas reactions containing CHX served as the control for translation inhibition. Values shown below the gel represent the mean and the SD of three in vitro translation experiments.
(B and C) The synthetic 18-mer RNA (added at 1–13 μM f.c.) inhibited in vitro translation in yeast extracts in a dose-dependent manner. Product quantification in the absence (no RNA) and in the presence of increasing amounts of synthetic 18-mer RNA is shown. The mean and SDs of three independent experiments are shown. See also Figure S5.
(D) The 18-mer RNA does not inhibit protein synthesis during the elongation phase of translation (condition II) but does so under conditions that allow translation initiation (condition I). The time points at which the radioactive label (35S Met) and the 18-mer RNA were added as well as the incubation temperatures are indicated for each of the three assay conditions (the most important features of the three different conditions are indicated in cursive). Product quantification (cpm of TCA-precipitated proteins) of three independent experiments in the absence (no RNA) or presence of 13 μM (f.c.) 18-mer are shown (mean and SD). Cycloheximide (CHX), a known elongation phase inhibitor, impedes product formation under both conditions I and II. When reactions in the absence of any inhibitor were kept at 0°C throughout the entire experiment (condition III), essentially no product was formed, highlighting that at low temperatures translation initiation is almost completely blocked (III; first bar in the left graph). Background values (in average 3,700 cpm), obtained with samples that were stopped immediately after the complete reactions were assembled on ice, were subtracted from every experimental point. For (C) and (D), the statistical significance of the 18-mer inhibition was determined by a two-tailed unpaired Student’s t test (∗∗p < 0.05).
Ribosome-Bound 18-mer ncRNA Rapidly Attenuates Metabolic Activity Allowing Stress Adaptation
The lack of the 18-mer RNA fragment results in a severely retarded growth under high-salt conditions (Figures 3A and S1). Likely, the 18-mer ncRNA is needed to slow down the metabolic activity in yeast when the environmental conditions become unfavorable thus allowing the adjustment of gene expression. If this assumption is correct, one expects the trm10Δ strain to possess elevated translational activity, reflected by a greater polysome fraction compared to the wild-type (wt) strain in high-salt medium. To test this model, polysome profiling as well as the translational activity of untreated cells were investigated and compared to cells after salt-stress induction. Indeed, the strain lacking the TRM10 gene, and hence the 18-mer ncRNA, shows on average a 2.1-fold higher polysome:80S ratio at elevated salt concentrations (Figure 6A). Although initially both strains had identical polysome profiles and metabolic activities under normal growth conditions, 20 min of high salt stress resulted in an accumulation of 80S ribosomes and a reduced polysomal fraction in the wt strain as compared to the trm10Δ strain, arguing for translation initiation to be affected by the 18-mer RNA. The pool of free ribosomal subunits, however, remained unaltered. These differences in the polysome profiles are also mirrored in the metabolic activities. The trm10Δ strain had a markedly elevated metabolic activity compared to the wt strain within the first 5–20 min (Figure 6B). Similarly, the strain expressing the M2 mutant variant of the 18-mer, an RNA molecule that is unable to efficiently associate with ribosomes (Figure 1C), also possessed an enhanced metabolic rate. These clear differences, however, disappeared rapidly after 45 min (Figure 6B).
Figure 6.
In Vivo Consequences of the Absence of the 18-mer RNA on Polysome Profiles and on Metabolic Activities
(A) Polysome profiles of wt (black trace) and trm10Δ (Δ; pink trace) yeast cells were compared before (unstressed) and after 20 min of high NaCl stress. The location of the 40S, 80S, and polysomal fractions are indicated. Polysome profiles were repeated three times.
(B) Comparison of the translational activities of wt, trm10Δ (Δ), and Δ + p_M2 strains after high-salt-stress induction was monitored by 35S-methionine incorporation. The 0 min time point represents the metabolic activities of unstressed cultures. The translational activity of the wt S. cerevisiae strain after 20 min NaCl stress was taken as 1.00. The mean and SDs of four independent experiments are shown.
Discussion
The coordinated regulation of protein biosynthesis in response to intra- and extracellular signals is pivotal for the establishment of productive gene expression networks. Translation control typically involves regulatory proteins or small ncRNA molecules of the RNA silencing machinery. With the notable exceptions of the bacterial tmRNA (Felden and Gillet, 2011) and the signal recognition particle RNA (present in all domains) (Akopian et al., 2013), all functionally characterized ncRNAs capable of regulating protein biosynthesis (e.g., miRNAs and siRNAs in eukarya, small antisense RNAs in prokarya) target the mRNA rather than the ribosome directly.
Here, we present evidence that an 18-mer RNA fragment from the TRM10 mRNA in S. cerevisiae associates with ribosomes and regulates protein synthesis under hyperosmotic stress conditions. In vivo and in vitro data demonstrate that the TRM10 18-mer RNA represents a functional ncRNA in S. cerevisiae where it attenuates protein biosynthesis under high-salt conditions. This small ncRNA is remarkable in two ways, namely, that (1) it is a functionally characterized ncRNA deriving from the coding region of an mRNA, and (2) it belongs to an emerging class of ncRNAs regulating translation by directly associating with the ribosome. Association of the 18-mer RNA with the 60S ribosomal particles and reduction of global protein synthesis is crucial for S. cerevisiae under hyperosmotic conditions. Strains lacking the 18-mer RNA or expressing a ribosome-binding-deficient mutant version thereof (M2; Figure 1C) are significantly retarded in downregulating their metabolic activities (Figure 6B). Especially the time window between 5 and 45 min after salt-stress induction appears to be crucial for the fate of S. cerevisiae for adapting to the new environmental conditions. This fits to previous data (Gasch et al., 2000) demonstrating that the transcriptome of yeast cells quickly responses to various stress stimuli within the first 10–45 min, whereas, at later time points, it resembled the unstressed RNome. Although the TRM10 mRNA follows this expression trend (O’Rourke and Herskowitz, 2004), the TRM10-derived 18-mer RNA fragment remains at constant levels independent of stress induction (Figure 1B). Also, the fraction of ribosome-bound 18-mer ncRNA remains constant and does not change during hyperosmotic conditions (Figure 1C). Notably however, upon high salt stress the 18-mer relocates almost quantitatively from nontranslating 80S ribosomes and 60S ribosomal subunits to translating polysomes (Figure 2A). Because only the distribution but not the cellular abundance of the 18-mer RNA changes upon stress induction allows a rapid response to environmental signals without the need for synthesizing new regulatory molecules. It is thus possible that the TRM10 18-mer RNA is involved in regulating this first wave of stress adaptation under hyperosmotic conditions by lowering the efficiency of protein biosynthesis and thus slowing down overall metabolic activity.
From a mechanistic point of view, the 18-mer RNA functions differently than known small ncRNA translation regulators (such as miRNAs, siRNAs, or bacterial antisense RNAs), because it directly binds to 60S ribosomal subunits and does not target mRNAs (Figure 2). The polysome profiles of cells exposed to hyperosmotic stress showed a decrease in the polysome/monosome ratio (Figure 2A), which is indicative of inhibiting translation initiation (Uesono and Toh-E, 2002). In support of this, addition of the 18-mer RNA to an in vitro translation reaction under conditions that allow elongation but prevent reinitiation does not affect protein production (Figure 5D). Even though the 18-mer RNA is less abundant (∼27,000 molecules/cell; Figure S4) than the ribosome (∼200,000/cell), a dynamic relocation between initiating and elongating ribosomes is sufficient to yield global effects on translation (Figure S6).
Although it might appear counterintuitive at the first glance that slowing down protein synthesis (Figures 4 and 5) can in the end stimulate cell growth during stress (Figure 3), very recent evidence suggests that the ribosome serves as a regulatory hub in proteostasis and stress response (reviewed in Pechmann et al., 2013; Sherman and Qian, 2013). The picture that emerges from these recent findings is that slowing down translation, and thereby increasing overall accuracy of protein synthesis and protein folding, actually improves protein homeostasis and stress adaptation. It has been noted that repression of global translation seems faster than changes in regulatory signaling pathways (Sherman and Qian, 2013), such as the TOR pathway that does not appear to be involved in translation inhibition at the onset of hyperosmotic stress (Uesono and Toh-E, 2002). The molecular entity that actually transmits the stress signal to the translating ribosomes, however, remained enigmatic. Thus, the 18-mer ncRNA characterized in this study fulfills the criteria for such a signaling molecule because it is already associated with the translation machinery and can rapidly shift between initiating and elongating ribosomes upon hyperosmotic stress induction to swiftly attenuate the rate of translation. This allows stress-specific adaptation programs to be established, which, in turn, enable cells to survive under challenging environmental conditions.
The origin of the 18-mer RNA (deriving by TRM10 mRNA processing or from an independent transcript unit) has yet to be determined. However, exon-derived RNA fragments have been observed in numerous eukaryal deep-sequencing studies and are likely the result of an evolutionarily conserved mRNA cleavage mechanism (Fejes-Toth, et al., 2009; Mercer et al., 2010). The small RNA transcriptome of S. cerevisiae has been reported to possess an abundant pool of 17- to 19-nucleotide-long RNA molecules, which have been regarded as degradation products of mRNA, tRNA, and rRNA (Drinnenberg et al., 2009). In the light of the accumulating evidence that some of these tRNA-derived (reviewed in Gebetsberger and Polacek, 2013) and mRNA-derived (this study) fragments actually possess cellular functions, it appears more appropriate to refer to these RNA pieces as processing rather than degradation products. The 18-mer RNA described here, together with thousands of other candidates that were picked up in analogous genomic screens in various model organisms spanning all three domains of life (Gebetsberger et al., 2012; Jiao and Meyerowitz, 2010; Zywicki et al., 2012; our unpublished data), reveal the ribosome as a target for small regulatory ncRNAs and suggest the existence of a so far largely unexplored mechanism of translation control. Future work on the small ncRNA interactomes of ribosomes in a variety of model systems will allow deeper insight into the conservation and functional repertoire of this emerging class of regulatory ncRNA molecules.
Experimental Procedures
Yeast Strains, Growth Conditions, and Constructs
S. cerevisiae strain BY4742 and all mutant cells derived from this strain were grown in Sc-Leu medium at 30°C or at various stress conditions (see Supplemental Information for more details). To monitor growth under more stringent hyperosmotic conditions (the “redilution assay”), single colonies were picked and grown in Sc-Leu medium supplemented with NaCl (final concentration [f.c.] 0.7 M) or sorbitol (f.c. 1.025 M) for 72 hr. Subsequently the cultures were rediluted to an OD600 of 0.1 into fresh stress medium, and cell growth was monitored over 20 hr.
Primer Extension Analysis
To monitor the methylation status of m1G9 of tRNAGly, primer extension analysis using the 5′-32P-end-labeled primer 5′-CAACGTTGGATTTTACC-3′ was performed as previously described (Jackman et al., 2003; Polacek and Barta, 1998) (see Supplemental Information for details).
Metabolic Labeling
For metabolic labeling yeast spheroplasts (Russell et al., 1991) were mixed with 10 pmol synthetic 18-mer RNA (or 3′-extended variants thereof) and introduced into the cell via electroporation. After electroporation 1 ml YPD/1 M sorbitol and 1 μl 35S-methionine (1,000 Ci/mmol, 10 mCi/ml) were added, and the reaction was incubated for 1 hr at 30°C. The extent of labeled proteins was monitored by TCA precipitation and subsequent liquid scintillation counting or by SDS PAGE (see Supplemental Information). To monitor the translational activity of S. cerevisiae cells (wt, trm10Δ, or the M2 mutant strain) after NaCl stress induction, 3 ml cultures were inoculated overnight at 30°C in Sc-Leu medium. Stationary phase cultures were diluted into fresh Sc-Leu medium to a final OD600 of 0.3 and incubated at 30°C in the presence of high-salt-stress conditions (f.c.: 0.7 M NaCl). At indicated time points aliquots were taken, and translational activity was monitored for 10 min at 30°C by the addition of 1 μl 35S-methionine (1,000 Ci/mmol, 10 mCi/ml). After 10 min incubation, cells were pelleted and labeled proteins were precipitated by adding 500 μl 20% TCA followed by liquid scintillation counting.
In Vitro Translation
For in vitro translation, an S30 extract from S. cerevisiae was prepared (see Supplemental Experimental Procedures for details) (Hofbauer et al., 1982). For in vitro translation, 2.5 μl creatine phosphokinase (10 mg/ml; Roche), 7.5 μl CaCl2 (20 mM), 25 μl 10 × translation cocktail (100 mM HEPES/KOH [pH 7.5], 10 mM Mg[OAc]2, 760 mM KCl, 4 mM GTP, 10 mM ATP, 19 amino acid mix [500 μM each; methionine excluded]), 5 μl creatine phosphate (0.6 M; Roche), and 2.5 μl Mg(OAc)2 (100 mM) were mixed with 150 μl S30 and 16 μl 35S-methionine (1,000 Ci/mmol, 10 mCi/ml). The resulting translation mix was aliquoted into 12 μl portions and filled up to 15 μl with sterile H2O, synthetic 18-mer RNA (10–200 pmol) or cycloheximide (7.5 μg/ μl), incubated at 23°C for 30 min, and the products were separated by SDS-PAGE. To test whether translation initiation or elongation are inhibited by the 18-mer RNA, complete reactions, but lacking 35S-methionine and the 18-mer RNA, were assembled and preincubated for 10 min at 23°C. Subsequently the initiated samples were cooled down to 0°C on ice for 5 min followed by the addition of 35S-methionine and 13 μM of the 18-mer RNA. Translation elongation was then carried out for 30 min at 0°C, stopped by TCA precipitation, and the products were quantified by liquid scintillation counting (see Supplemental Experimental Procedures for details).
Polysome Profiling
For polysome profiling, 150 A260 units of S. cerevisiae cell extracts (see Supplemental Information) were layered on top of a 10%–40% sucrose gradient and centrifuged for 5 hr at 25,000 rpm in an SW28 swing-out rotor. Polysomal, monosomal, and subunit fractions were isolated, and the ribosomal particles in the collected fractions were precipitated by adding 2.5 vol EtOH. The ribosome-associated RNA were purified by phenol/chlorophorm/isoamyl-alcohol (PCI) extraction and finally used for northern blot analyses.
Northern Blot Analyses
For northern blotting, 15 μg total RNA extracted from wt or mutant cells under different stress conditions was separated by denaturating polyacrylamide gels (7 M urea) and subsequently electroblotted onto nylon membranes (Amersham Hybond N+, GE Healthcare) as described (Gebetsberger et al., 2012). All membranes were hybridized to a 32P-5′-end-labeled LNA probe (Exiqon) complementary to the TRM10 18-mer RNA sequence.
Acknowledgments
We thank D. Teis for yeast materials and for insightful discussions and Jennifer Gebetsberger, Simon Spiegl, Selma Tuzlak, and Simon Rohrbach for experimental help. Sean Connell, Miriam Koch, Roland Micura, Oliver Mühlemann, André Schneider, and Nora Vasquez-Laslop are acknowledged for comments on the manuscript. Grant support derives from the Swiss National Science Foundation (31003A_143388/1) and the Austrian Science Fund FWF (project number: Y315) to N.P.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
References
- Aalto A.P., Pasquinelli A.E. Small non-coding RNAs mount a silent revolution in gene expression. Curr. Opin. Cell Biol. 2012;24:333–340. doi: 10.1016/j.ceb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian D., Shen K., Zhang X., Shan S.O. Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Fageeh M.B., Smales C.M. Control and regulation of the cellular responses to cold shock: the responses in yeast and mammalian systems. Biochem. J. 2006;397:247–259. doi: 10.1042/BJ20060166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral P.P., Dinger M.E., Mercer T.R., Mattick J.S. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- Crick F.H. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- del Prete M.J., Vernal R., Dolznig H., Müllner E.W., Garcia-Sanz J.A. Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. RNA. 2007;13:414–421. doi: 10.1261/rna.79407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg I.A., Weinberg D.E., Xie K.T., Mower J.P., Wolfe K.H., Fink G.R., Bartel D.P. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes-Toth K., Sotirova V., Sachidanandam R., Assaf G., Hannon G.J., Kapranov P., Foissac S., Willingham A.T., Duttagupta R., Dumais E., Affymetrix ENCODE Transcriptome Project. Cold Spring Harbor Laboratory ENCODE Transcriptome Project Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felden B., Gillet R. SmpB as the handyman of tmRNA during trans-translation. RNA Biol. 2011;8:440–449. doi: 10.4161/rna.8.3.15387. [DOI] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F., Hentze M.W. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J., Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J., Zywicki M., Künzi A., Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson M., Ronne H. Evidence that tRNA modifying enzymes are important in vivo targets for 5-fluorouracil in yeast. RNA. 2008;14:666–674. doi: 10.1261/rna.966208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer R., Fessl F., Hamilton B., Ruis H. Preparation of a mRNA-dependent cell-free translation system from whole cells of Saccharomyces cerevisiae. Eur. J. Biochem. 1982;122:199–203. doi: 10.1111/j.1432-1033.1982.tb05867.x. [DOI] [PubMed] [Google Scholar]
- Hofmann S., Cherkasova V., Bankhead P., Bukau B., Stoecklin G. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol. Biol. Cell. 2012;23:3786–3800. doi: 10.1091/mbc.E12-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J., Tollervey D. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochim. Biophys. Acta. 2008;1779:239–246. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Huntzinger E., Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A., Schattner P., Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Jackman J.E., Montange R.K., Malik H.S., Phizicky E.M. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9:574–585. doi: 10.1261/rna.5070303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Meyerowitz E.M. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 2010;6:419. doi: 10.1038/msb.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Mattick J.S. RNA regulation: a new genetics? Nat. Rev. Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Conte D., Jr. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- Mercer T.R., Dinger M.E., Bracken C.P., Kolle G., Szubert J.M., Korbie D.J., Askarian-Amiri M.E., Gardiner B.B., Goodall G.J., Grimmond S.M., Mattick J.S. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 2010;20:1639–1650. doi: 10.1101/gr.112128.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S.M., Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell. 2004;15:532–542. doi: 10.1091/mbc.E03-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S., Willmund F., Frydman J. The ribosome as a hub for protein quality control. Mol. Cell. 2013;49:411–421. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petelenz-Kurdziel E., Eriksson E., Smedh M., Beck C., Hohmann S., Goksör M. Quantification of cell volume changes upon hyperosmotic stress in Saccharomyces cerevisiae. Integr Biol (Camb) 2011;3:1120–1126. doi: 10.1039/c1ib00027f. [DOI] [PubMed] [Google Scholar]
- Polacek N., Barta A. Metal ion probing of rRNAs: evidence for evolutionarily conserved divalent cation binding pockets. RNA. 1998;4:1282–1294. doi: 10.1017/s1355838298980347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P.J., Hambidge S.J., Kirkegaard K. Direct introduction and transient expression of capped and non-capped RNA in Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:4949–4953. doi: 10.1093/nar/19.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M.Y., Qian S.B. Less is more: improving proteostasis by translation slow down. Trends Biochem. Sci. 2013;38:585–591. doi: 10.1016/j.tibs.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Steitz T.A., Moore P.B. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem. Sci. 2003;28:411–418. doi: 10.1016/S0968-0004(03)00169-5. [DOI] [PubMed] [Google Scholar]
- Tuck A.C., Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Uesono Y., Toh-E A. Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 2002;277:13848–13855. doi: 10.1074/jbc.M108848200. [DOI] [PubMed] [Google Scholar]
- Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Zywicki M., Bakowska-Zywicka K., Polacek N. Revealing stable processing products from ribosome-associated small RNAs by deep-sequencing data analysis. Nucleic Acids Res. 2012;40:4013–4024. doi: 10.1093/nar/gks020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.