Summary
Enhancers are distal regulatory elements that can activate tissue-specific gene expression and are abundant throughout mammalian genomes. While substantial progress has been made towards genome-wide annotation of mammalian enhancers, their temporal activity patterns and global contributions in the context of developmental in vivo processes remain poorly explored. Here we used epigenomic profiling for H3K27ac, a mark of active enhancers, coupled to transgenic mouse assays to examine the genome-wide utilization of enhancers in three different mouse tissues across seven developmental stages. The majority of the ~90,000 enhancers identified exhibited tightly temporally restricted predicted activity windows and were associated with stage-specific biological functions and regulatory pathways in individual tissues. Comparative genomic analysis revealed that evolutionary conservation of enhancers decreases following mid-gestation across all tissues examined. The dynamic enhancer activities uncovered in this study illuminate rapid and pervasive temporal in vivo changes in enhancer usage underlying processes central to development and disease.
Introduction
Distant-acting transcriptional enhancers represent the most abundant class of cis-regulatory sequences in mammalian genomes (Shen et al., 2012), and are predicted to be exceptionally tissue-specific in function (ENCODE Project Consortium et al., 2012; Ernst et al., 2011; Shen et al., 2012; Visel et al., 2009). They are often associated with developmentally expressed genes (Levine, 2010) and can drive spatially highly restricted in vivo activity patterns (Pennacchio et al., 2006; Visel et al., 2009; 2013). Sequence level changes at enhancers underlie evolutionary differences between species (Jones et al., 2012) and significantly contribute to the genetic etiology of human disease (Dickel et al., 2013; ENCODE Project Consortium et al., 2012; Ernst et al., 2011). As such, genome-wide maps of enhancers and their activity patterns provide insight into mechanisms of evolution, development, and disease, and significant progress has been made towards mapping these elements in mammalian genomes (ENCODE Project Consortium et al., 2012; Ernst et al., 2011; Shen et al., 2012). In parallel, in vivo transcriptome profiling of developing tissues has revealed highly dynamic gene expression during tissue ontogenesis (Bruneau, 2008; Kang et al., 2011; Si-Tayeb et al., 2010), and dysregulation of transient developmental gene expression patterns has been linked to congenital defects and pathogenic traits (Garg et al., 2005; Hoerder-Suabedissen et al., 2013). Differences in the chromatin landscape between individual adult and embryonic tissues and in cultured cells (Gifford et al., 2013; Heintzman et al., 2009; Shen et al., 2012; Stergachis et al., 2013; Zhu et al., 2013; Ziller et al., 2013) raise the possibility that, within a given tissue, the genome-wide regulatory architecture might change substantially across developmental stages. While these initial lines of evidence suggest that enhancers may play a significant role in the extensive changes in gene expression observed throughout mammalian development, the in vivo dynamics of enhancer utilization as individual tissues develop pre- and postnatally have been minimally explored. Profiling enhancer activity in developing tissues across a controlled time course has the potential to reveal the temporal dynamics of mammalian enhancer usage in vivo and capture regulatory landscapes orchestrating transient biological processes that are central to human health and disease.
Results
Mapping enhancer activity landscapes via H3K27ac profiling of mouse tissues
To examine genome-wide enhancer activity at a consistent and defined temporal resolution, we performed epigenomic mapping of active enhancers across a developmental time series in three organs with different anatomical and physiological trajectories: forebrain, heart, and liver. The forebrain is the center of many higher brain functions, arising from the ectoderm and undergoing waves of neurogenesis and migration during mid-embryogenesis, with substantial late maturation (Austin and Cepko, 1990; Clinton et al., 2000; Kang et al., 2011). The heart arises from the mesoderm, is one of the earliest organs to form with basic patterning complete by late gestation, and performs the singular function of circulation throughout life (Brand, 2003; Harvey, 2002; Olson, 2006). The liver arises from the endoderm and goes through a major functional transition, switching from fetal hematopoiesis to its mature functions of detoxification, metabolism, and plasma protein and hormone synthesis late in gestation (Zhao and Duncan, 2005; Zorn, 2008). These three tissues are of significant relevance to biomedical research, and pathogenic traits associated with all three systems are closely linked to transient developmental processes.
We generated genome-wide maps of enhancers active in each of these organs via ChIP-seq performed directly on mouse tissue collected at different stages of development (Fig. 1A). The developmental stages (embryonic days [E] 11.5, 14.5, and 17.5; postnatal days [P] 0, 7, 21, and 56) and tissues were selected to capture significant developmental processes in these major organ systems. In total, we profiled 21 unique tissue types collected from pre- and postnatal mice (Supplementary Tab. 1). We assessed the tissue- and stage-specific presence of H3K27ac, a histone modification found at active enhancers (Creyghton et al., 2010; Rada-Iglesias et al., 2011) and at transcription start sites (TSSs). Supplementary Fig. 1 shows a schematic overview of the analysis. ChIP-seq reads were mapped to mm9 and peaks were called for each dataset (see Extended Experimental Procedures for details for all analyses). We separated the H3K27ac-enriched regions into putative distal enhancers, defined as regions positioned at least 1kb from a known TSS, and proximal regions that were within 1kb of or overlapped a TSS. In total, across the three tissues and seven timepoints examined we identified 105,394 H3K27ac-enriched regions, including 16,225 regions that were proximal to known TSSs and 89,169 distal regions representing putative developmental enhancers. Comparison of expression levels of the nearest TSS for both forebrain and heart enhancers showed significantly increased expression in the linked tissue at E11.5 (t-test p-values: forebrain=0.007; heart=0.03). H3K27ac enrichment profiles for biological replicates for a subset of samples showed significant reproducibility across datasets (Supplementary Fig. 2A). The association of enhancers with gene expression, the biological reproducibility of ChIP-seq experiments, and patterns of H3K27ac co-enrichment across tissues and timepoints support the validity of these datasets for genome-wide enhancer analysis (Supplementary Fig. 2).
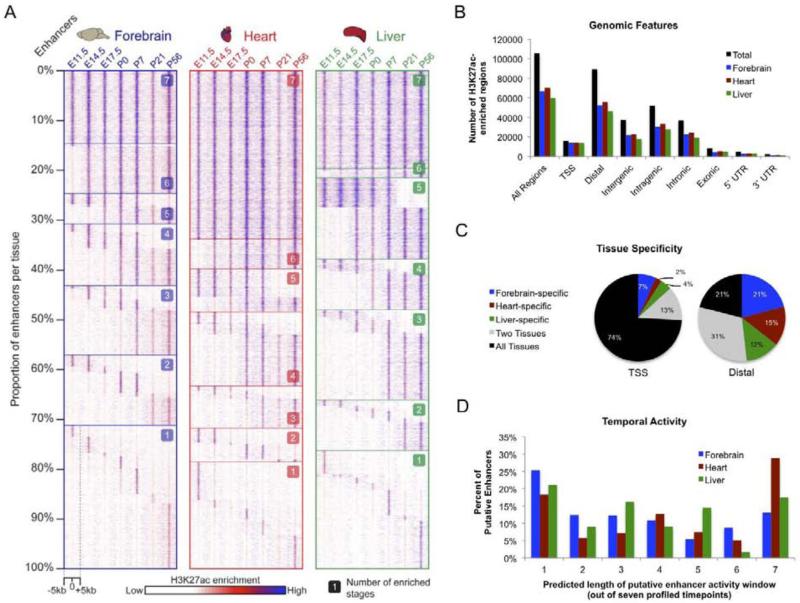
Figure 1. Mapping in vivo enhancers via ChIP-seq performed on mouse forebrain, heart, and liver tissue.
A Schematic of developmental stages and tissues. B Representative examples of putative enhancers exhibiting dynamic H3K27ac signal across tissues and timepoints. Text includes description of loci. (See also Supplementary Figs. 1 & 2; Supplementary Tab. 1).
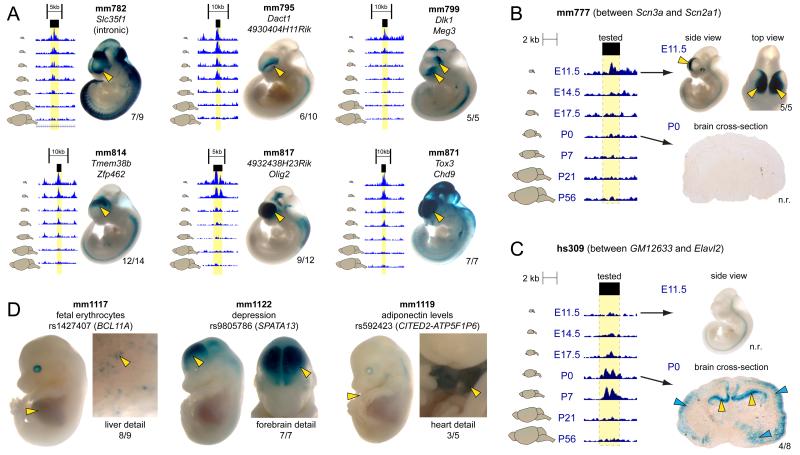
Recent studies of chromatin indicate that H3K27ac is present at enhancers when they are active (Bonn et al., 2012; Cotney et al., 2012; Creyghton et al., 2010; Rada-Iglesias et al., 2011), suggesting a model where dynamic H3K27ac enrichment is associated with transient enhancer activity. This notion is illustrated by examples of differential H3K27ac enrichment across timepoints and tissues at representative putative distal enhancers located near selected developmentally active target genes (Fig. 1B). For instance, an enhancer with early forebrain H3K27ac enrichment was identified near Sox11, a gene critical for prenatal forebrain patterning (Bergsland et al., 2011; Shim et al., 2012; Uwanogho et al., 1995). In contrast, a region with postnatal H3K27ac enrichment in the forebrain was identified near Gnaz, a gene associated with dopamine signaling in the postnatal/adult forebrain (Hendry et al., 2000; Hinton et al., 1990; Leck et al., 2006; Sidhu et al., 1998). In the heart, two enhancers with early and late enrichment peaks were identified near Igf1r and Adcy5, consistent with known roles of these genes in early heart development and later cardiomyocyte survival, respectively (Donath et al., 1994; Holzenberger et al., 2000; Hu et al., 2009; Iwatsubo et al., 2004; Laustsen et al., 2007). Examples in the liver include a prenatal enhancer near Hbb-b1, which encodes a hemoglobin protein expressed in the embryonic liver during fetal hematopoiesis (Whitney, 1977), and Lipc, which encodes a hepatic lipase active in the mature liver that is implicated in cardiovascular disease in humans (Zambon et al., 2003). These examples suggest that dynamic chromatin modification is detectable at enhancers examined across developmental stages.
To investigate dynamic chromatin modification patterns genome-wide, the complete set of peaks called across all datasets was merged by combining peaks where the highest point of enrichment within individual peaks were within 1kb. Each merged peak was then scored for enhancer activity across the 21 datasets based on H3K27ac signal strength. The results from this analysis show that timepoints next to each other and from the same tissue have the most similar H3K27ac enrichment profiles, as expected based on spatial and temporal relationships of the profiled tissues (Supplementary Fig. 2B). Initial clustering analysis indicated that developmental enhancers identified in this study largely exhibited restricted H3K27ac enrichment across tissues and temporally across developmental timepoints (Supplementary Fig. 2C). Using an enrichment classification method robust to false negatives in the ChIP-seq data, we predicted the activity windows of all enhancers identified in the three tissues. In comparison to shuffled data, predicted activity showed significant temporal and spatial correlation structure across timepoints and tissues, indicating that the patterns we observe represent real biological patterns.
While differences in the genome-wide enhancer landscape between developing and mature tissues are known to exist in principle (May et al., 2011; Shen et al., 2012), this time course profiling of an enhancer-specific chromatin mark enables longitudinal examination of predicted in vivo enhancer activities at high temporal resolution. In most cases (forebrain 85%; heart 66%; liver 80%) the predicted tissue-specific temporal activity window of putative enhancers spanned only a subset of the developmental stages examined (Fig. 2A). The three tissues exhibit different patterns with regard to predicted enhancer activity across stages that are in line with their respective developmental trajectories (Fig. 2A-2D, Supplementary Fig. 2). For example, a larger proportion of putative heart enhancers exhibit constitutive predicted activity, consistent with the embryonic heart already attaining many aspects of its mature function at E11.5, the earliest timepoint profiled. Putative distal enhancers map to both intergenic (42%) and intragenic (58%) chromosomal regions, where they overlap with intronic sequence (41%), coding exons (9%), and untranslated regions (8%) (Fig. 2B). In addition to temporal activity restrictions, candidate enhancers were predicted to be predominantly tissue-specific (Fig. 2C), with 42,976 (48%) expected to be active only in one of the three tissues examined. Illustrating the rapidly changing enhancer landscape, 40,696 (45%) of putative distal enhancers identified here are predicted to have highly restricted temporal activity, with enrichment spanning at most two consecutive timepoints in a given tissue (Fig. 2D).
Figure 2. Developmental enhancers exhibit dynamic H3K27ac enrichment associated with in vivo activity.
A. Heatmap displaying H3K27ac enrichment by tissue and timepoint for putative distal enhancers (forebrain n=52,175; heart n=55,869; liver n=46,062). For each tissue, each row of the heatmap shows relative H3K27ac enrichment at one enhancer, with signal across the surrounding 10kb region plotted. Enhancers are organized by the number of timepoints at which the enhancer is active, starting with constitutively active enhancers at the top and proceeding down to single-stage enhancers at the bottom. B. Breakdown on H3K27ac enrichment across genomic features. C. Tissue specificity for TSS and distal H3K27ac enrichment. D. Predicted length of putative distal enhancer activity based on H3K27ac enrichment across seven profiled timepoints. (See also Supplementary Fig. 2).
While many enhancers that exhibit short activity windows are tissue-specific, we also identified clusters of enhancers with predicted activity across multiple tissues (Supplementary Fig. 2) that may control general or shared developmental and/or functional processes. Considering only the ~3% of putative enhancers that showed constitutive H3K27ac enrichment across all tissues and timepoints, we observe strong enrichment near genes associated with hematological traits suggesting that such enhancers are active in blood lineages present in all tissue samples. In contrast to the dynamic epigenomic landscape of enhancers, H3K27ac enrichment at TSSs does not exhibit such tissue- or stage-specific patterns, with 74% of TSSs exhibiting enrichment across all three tissues and the majority of TSS-proximal sites exhibiting constitutive enrichment within a tissue across all timepoints (forebrain 75%; heart 79%; liver 73%) (Fig. 2B and Supplementary Fig. 2). Overall, these results suggest that the genome-wide enhancer landscape active in each of the three organs undergoes extensive and fast-paced turnover during development.
In vivo validation of enhancer activity predictions
The genome-wide changes in H3K27ac enrichment across stages support the prevalence of dynamic enhancer activity based on a known epigenomic signature of active enhancers. To obtain direct evidence of developmentally dynamic enhancer activities, we used an established transgenic mouse enhancer reporter assay (Kothary et al., 1989; Pennacchio et al., 2006) to experimentally validate enhancer activity predictions (Fig. 3). Whole-mount staining of the transgenic mice generated in this assay is possible at E11.5 and E14.5, enabling interrogation of higher numbers of candidate enhancers at these early developmental stages compared to later timepoints, when sectioning is required. As such, we used three strategies to validate H3K27ac-based activity predictions using these assays. First, to establish baseline success rates for in vivo activity predictions made from H3K27ac datasets, we examined sequences predicted to be active forebrain enhancers at E11.5, where 12/18 (67%) drove reproducible expression patterns in vivo (Fig. 3A, Supplementary Fig. 3). This rate of validated in vivo activity is similar to other epigenomic marks of active enhancers, such as p300 (Visel et al., 2009), and is significantly higher than the rate of in vivo forebrain enhancer activity among a control set of tested highly-conserved non-coding presumed functional elements (Pennacchio et al., 2006) (Fisher’s exact test, P = 6×10−6). Next, we examined in vivo activity of a smaller set of eight enhancers at multiple timepoints to validate that change in H3K27ac enrichment corresponded to change in in vivo activity. We examined sequences that were known to be inactive in vivo at E11.5 (Pennacchio et al., 2006), but exhibited an H3K27ac enrichment profile that suggested activity at later timepoints. 5/8 (63%) of the elements tested showed reproducible in vivo forebrain activity at a later timepoint consistent with the developmental H3K27ac signature (Supplementary Fig. 4). We additionally reexamined two elements where activity was predicted to subside later in development that were active in E11.5. In one of the two we observed no reproducible Lacz staining at E17.5 or P0. Representative staining patterns for two dynamic enhancers are shown in Fig. 3B and 3C. First, an enhancer near Scn2a1, a sodium channel gene expressed in the forebrain between E11.5 and E14.5 (Albrieux et al., 2004) that is required for normal brain development (Planells-Cases et al., 2000) and mutated in autism (Sanders et al., 2012), showed highly reproducible cortical expression at E11.5 but not at P0 (Fig 3B). Second, an enhancer near Elavl2, a gene important to neuronal differentiation (Akamatsu et al., 1999) had no activity at E11.5 but drove reproducible expression in the hippocampus at P0 (Fig. 3C). Finally, we tested six enhancers where the human orthologous region overlapped a lead genome wide association study (GWAS) single nucleotide polymorphism (SNP) associated with a forebrain, heart, or liver phenotype. The lead SNPs overlapped by the tested enhancers were not in linkage disequilibrium with a coding SNP and all the lead SNPs overlapped with putative transcription factor binding motifs predicted using HaploReg (Ward and Kellis, 2012). All six candidate enhancers drove expression in the predicted tissue at E14.5 (Supplementary Fig. 5). Three representative enhancers that overlap GWAS lead SNPs are shown in Figure 3D, including an enhancer active in the fetal mouse liver that contains a SNP associated with levels of blood cells with fetal hemoglobin (F-cells) in adults (Bhatnagar et al., 2011 and Menzel et al., 2007), an enhancer that is active in the developing mouse forebrain that contains a lead SNP for depression and alcohol dependence (Edwards et al., 2012), and a mouse heart and liver enhancer that contains a SNP associated with adiponectin levels (Dastani et al., 2012). These experimentally validated enhancer activity patterns provide in vivo evidence suggesting plausible pathogenic mechanisms of non-coding variation via spatiotemporally restricted impact on target gene expression caused by changes in enhancer sequence. In total, 23/32 (72%) putative enhancers tested in transgenic mice drove H3K27ac-predicted expression patterns in vivo, and many of these enhancers were associated with critical developmental genes or potentially pathogenic variation. Full transgenic results from all experiments performed for this study available on the VISTA website (http://enhancer.lbl.gov/). Together, the genome-wide ChIP-seq data and the complementary transgenic validation of a subset of dynamic activity predictions support the existence of very large numbers of enhancers with restricted activity intervals across development.
Figure 3. In vivo validation of H3K27ac-predicted enhancer activity.
Candidate enhancers were cloned into a vector containing a minimal promoter and the LacZ reporter gene and injected into fertilized mouse oocytes. Multiple transgenic mice with independent enhancer integration events were examined to assess the reproducibility of any given reporter activity pattern. Yellow arrows and numbers next to embryos/sections indicate reproducibility of staining across transgenic individuals. Additional embryo images for each element can be viewed in the VISTA Enhancer Database (http://enhancer.lbl.gov). n.r: not reproducible. A-C. In vivo validation of predicted forebrain enhancers. Forebrain H3K27ac signal across timepoints shown to the left, with yellow highlighting indicating the tested region. A. Six representative enhancers that exhibit diverse forebrain activity patterns at E11.5. B. Enhancer located near Scn2a1 that shows transient H3K27ac enrichment and drives in vivo expression at E11.5, but not P0. C Enhancer upstream of Elavl2 that shows transient enrichment and in vivo activity at P0, but not E11.5.;Blue arrows indicate non-reproducible staining. D. Three representative enhancers active at E14.5 that overlap with lead GWAS SNPs. GWAS phenotype, lead SNP ID, and potential gene of interest are listed. (See also Supplementary Figs. 3-5).
Enhancers control dynamic developmental processes and are enriched for TF binding motifs and disease-associated variation
To assess correlation between the predicted temporal activity of enhancers and biological function beyond anecdotal examples, we examined on a genome-wide scale whether putative enhancers can be linked to biological processes, mouse phenotypes, and regulatory pathways associated with the developmental stages profiled (Heinz et al., 2010; McLean et al., 2010). Putative enhancers are globally enriched near genes that have pertinent tissue- and timepoint-related functional annotations and are enriched for relevant transcription factor binding sites. For example, enhancers predicted to be active early in forebrain development are enriched for annotation terms such as neural precursor cell proliferation and axonogenesis and binding motifs of transcription factors that control neuronal differentiation, such as Lhx3. In contrast, enhancers predicted to be active later in forebrain development are enriched for biological processes such as synaptic transmission and cognition and phenotypes including abnormal learning/memory/conditioning and neurodegeneration. Fig. 4A shows ten representative functions, phenotypes, and binding motifs that exhibit strong differential enrichment patterns across tissues and developmental stages, with such differential patterns recapitulated across the entire set of enriched annotation terms and binding motifs, as shown in Fig 4B. Finally, intersection of putative enhancers identified here with results from genome-wide association studies (Hindorff et al., 2009) showed that disease-associated SNPs are more likely to be located nearby candidate enhancers compared to randomly sampled SNPs (see Extended Experimental Procedures). These results demonstrate that transiently active developmental enhancers are centrally involved in the control of biological processes required for tissue ontogenesis and function, regulating genes essential to developmental and disease phenotypes.
Figure 4. Association of developmental enhancers with functional pathways, mouse phenotypes, and transcription factor binding motifs.
Each heatmap displays results from enrichment analysis performed on forebrain, heart, and liver enhancers active at specified timepoints. A. Ten representative differentially enriched GO biological functions, MGI mouse phenotypes, and known transcription factor binding motifs selected from the complete enrichment datasets. B. Full enrichment dataset heatmaps for GO biological functions (n=827), MGI mouse phenotypes (n=922), and known transcription factor binding motifs (n=215). Annotation terms and TF motifs were hierarchically clustered by enrichment patterns. Differential enrichment across tissues and timepoints occurs widely across the full datasets
Evolutionary pressure on enhancers changes across development
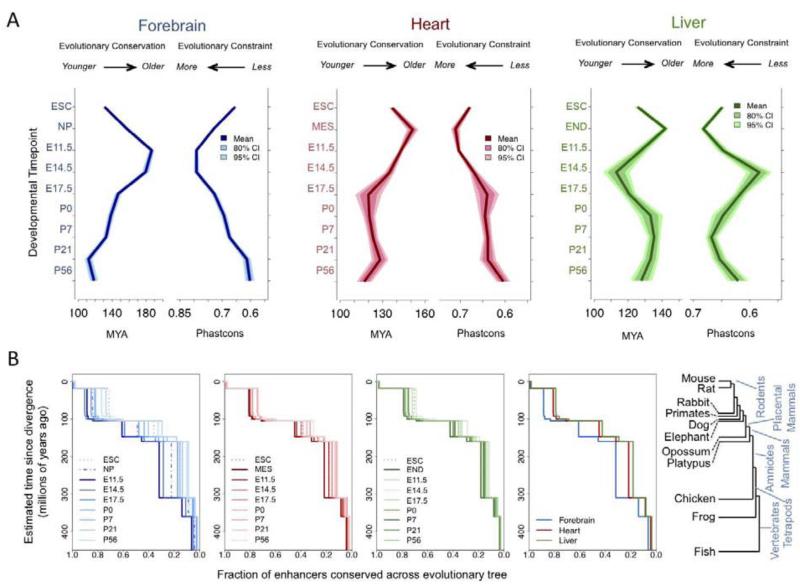
Despite the general utility of evolutionary conservation as a mark of regulatory sequences (Pennacchio and Rubin, 2001), studies in mammalian cell lines and tissues have produced contradictory findings regarding the global conservation level of enhancers (Blow et al., 2010; ENCODE Project Consortium et al., 2012; Pennacchio et al., 2006; Shen et al., 2012). The maps of predicted enhancer activity in the present study, obtained with consistent methodology across tissues and developmental stages, provide an opportunity to examine the evolutionary conservation of enhancers using rigorous comparative genomic measurements. To test whether evolutionary pressure on enhancers varies across tissues or developmental stages, the most constrained core regions of non-coding putative distal enhancers active at different developmental stages were compared using two related measures of sequence evolution, conservation (evolutionary age based on divergence between mouse and most distant vertebrate lineage exhibiting sequence homology to mouse) and constraint (estimate of local sequence conservation across vertebrates). In addition to tissue-derived data, we incorporated in this analysis H3K27ac ChIP-seq data from mouse embryonic stem cells and three cell lineages, neural progenitors, mesoderm, and mesoendoderm, as experimentally accessible proxies for major lineages of the forebrain, heart, and liver at stages prior to E11.5.
Strikingly, we observed substantial differences in evolutionary conservation and sequence constraint of putative enhancers compared both within a given tissue across timepoints and across tissues at the same timepoint (Fig. 5A). Predicted forebrain enhancers exhibit higher overall constraint and are more conserved across the vertebrate tree than enhancers predicted to be active in heart or liver, verifying previous findings comparing enhancers active at E11.5 identified by p300 binding (Blow et al., 2010). However, for all tissues the maximum levels of evolutionary conservation/constraint of putative enhancers were observed in early embryogenesis, with a second phase of temporarily increased conservation/constraint in the liver in early postnatal development. These differences result in distinct tissue-specific evolutionary signatures of in vivo enhancers across development that were robustly reproduced using alternative constraint metrics, phylogenetic comparisons, and expected enhancer core sizes (Supplementary Fig. 6).
Figure 5. Developmental signatures of enhancer evolution.
The tissue-based enhancer set was expanded to include cell lines used as proxies for early development: embryonic stem cells (ESC), neural progenitors (NP), mesoderm (MES), and mesoendoderm (END).A. Mean and 95% and 80% confidence intervals of evolutionary age (left panel) and constraint (right panel) by tissue and timepoint. B. Cumulative proportion of enhancers conserved across the vertebrate tree (shown on right) as defined by enhancer sequence homology. Plots shown for all timepoints in each individual tissue in the first three panels, with higher mean conservation indicated by darker shades. Far right panel shows differences across tissues at the most constrained stage for each tissue. (See also Supplementary Figs. 6 & 7).
To examine the evolutionary history of enhancers, the cumulative percent of enhancers conserved across each transition was determined using sequence homology across the 100 most constrained bases, plotted in Fig. 4B. The color of plotted lines correlates with the summary measures for the dataset (shown in Fig. 4A), with darker color tones indicating stronger overall constraint/conservation. We reasoned that if specific evolutionary transitions disproportionally contributed to the overall differences in conservation observed across tissues and time points, the effect would be reflected by large differences between stages/tissues at specific transitions across the vertebrate tree. For example, the transition to a fully septated heart, present through chicken but not in frogs or more distant vertebrates (Olson, 2006), might be associated with a significantly lower proportion of enhancers conserved across the transition from tetrapods to amniotes relative to the transition from amniotes to placental mammals. Across accessible evolutionary divergence events, the relative proportions of conserved enhancers recapitulated the general patterns observed for mean conservation and constraint (Fig. 5B). At least at the level of whole organ development, these results suggest that the observed tissue- and stage-specific differences represent cumulative effects of increased selective pressure on enhancers active early in embryonic development throughout more than 400 million years of vertebrate evolution.
We observed two further relationships with enhancer constraint: positive correlation between constraint and distance from the nearest TSS and a TSS distance-independent effect where intergenic enhancers exhibited increased constraint versus intronic enhancers (Supplementary Fig. 7). These patterns are associated with a larger proportion of enhancers predicted to be active in mature tissues to be located nearby the TSS and within gene bodies relative to the same tissues at earlier stages, partially consistent with recent findings comparing adult tissues to differentiating cell lineages (Zhu et al., 2013). The strength of these patterns varied across tissues, with forebrain exhibiting increase in the difference in distance to the nearest TSS and proportion of intronic enhancers between timepoints relative to the other two tissues. These patterns indicate that constraint and position in the genome are interconnected with regard to stage of enhancer activity, and suggest that these patterns may be driven by general aspects of genome evolution and structure.
Discussion
We report the developmental activity annotation of nearly 90,000 candidate distal enhancers across three major mammalian organ systems. These genome-wide enhancer activity profiles obtained directly from ex vivo tissues across multiple stages of the mammalian lifespan provide insight into the temporal utilization of enhancers as occurring in vivo in the developing organism. Mapping dynamic H3K27ac enrichment alone is a basic model for enhancer identification and activity prediction, as H3K27ac is unlikely to be present at all active enhancers, may not correlate universally with enhancer activity, and may be present at other non-coding genomic features. In the future, concurrent analysis of additional informative chromatin marks and genome-wide binding or transcription datasets is likely to refine enhancer activity maps further, enabling increased sensitivity and specificity with regard to enhancer activity predictions and a corresponding increase in the success rate of in vivo validation assays. These limitations notwithstanding, the strong global signatures of dynamic predicted enhancer activity in our results coupled with transgenic validation of in vivo activity predictions demonstrate the power of interrogating relevant tissues across developmental transitions. Most of the candidate enhancers identified here are predicted to have tightly restricted temporal activity windows, indicating that dynamic processes in mammalian development and tissue ontogenesis are regulated by the transient activities of large numbers of temporally and spatially restricted developmental enhancers. The rapid temporal changes in enhancer landscape identified via time course profiling mirror patterns of dynamic gene expression across development, suggesting that regulatory control of spatiotemporal gene expression patterns is accomplished through the combinatorial activity of regulatory elements that far outnumber coding genes. These findings have major ramifications in the context of predicting regulatory elements controlling clinically relevant tissue- and stage-specific processes, and regarding efforts to systematically map enhancers in the human genome. While validating the importance and impact of large-scale efforts to annotate functional genomic elements, our observations also highlight a substantial challenge in producing a complete functional annotation of all distant-acting enhancers in the human genome. The large numbers of putative enhancers identified here with predicted short activity windows in specific tissues suggest that tightly spaced developmental time series from diverse panels of tissues may be required to capture a truly comprehensive picture of the genome-wide enhancer landscape.
The observation of very high constraint of enhancers predicted to be active at mid-gestation corroborates and partially explains reports that as many as half of all extremely conserved non-coding sequences may act as enhancers in vivo at E11.5, with particular enrichment for neuronal tissue activities (Pennacchio et al., 2006; Visel et al., 2008). These findings are in line with recent whole-embryo transcriptome studies of zebrafish and Drosophila and with evolutionary signatures observed at regulatory sequences across human cell lineages that support the evolutionary hourglass model of development (Domazet-Lošo and Tautz, 2010; Kalinka et al., 2010; Stergachis et al., 2013). This model posits that increased evolutionary constraint at critical stages of embryogenesis produces high levels of similarity across evolutionary lineages during early development, with relaxed constraint and increased evolutionary divergence before and after these critical stages (Raff, 1996). The results from this study show that distinct patterns of sequence evolution apply to enhancers with transient in vivo activities in mammalian development and identify tissue-specific variation in the timing and level of maximum constraint that suggest differences in the evolutionary history of different organ systems. The tissue-specific differences in the timing of maximal enhancer constraint coincide with transitional phases during the ontogenesis of these three organs. Enhancer constraint in the developing forebrain peaks at E11.5 and continues to be high at E14.5, spanning critical stages of forebrain patterning and neuronal migration (Austin and Cepko, 1990). In contrast, maximal constraint in the heart is observed at E11.5, consistent with early maturation of the heart during embryogenesis (Harvey, 2002), and average enhancer constraint is relatively stable from E17.5 through P56. In liver a similar early maximum in enhancer constraint is present, with a secondary peak around postnatal day seven, which tracks the transition from fetal hematopoiesis to the change to assume the predominantly metabolic functions of the mature liver (Zorn, 2008). While these explanations are speculative, it is clear that evolutionary pressure on enhancers changes in a tissue-specific manner across development. These dynamic evolutionary signatures of active enhancers reconcile previous contradicting findings regarding constraint of mammalian enhancers, illuminate evolutionary forces shaping development, and reinforce the long-held notion that regulatory DNA is a primary substrate upon which evolution acts (King and Wilson, 1975).
Experimental Procedures
Extended Experimental Procedures contains detailed methods and references for all analyses described in the text and below. All custom analysis scripts are available from the authors at request.
ChIP-Seq
Tissues from pre- and postnatal CD-1 mice were collected on ice, crosslinked using formaldehyde, lysed with SDS-based reagents, and chromatin was sonicated on a Diagenode Bioruptor instrument using ChIP-seq protocols optimized for mouse tissues (Visel et al., 2009). Chromatin immunoprecipitation was performed using antibodies for H3K27ac (Abcam Ab4729). Prepared libraries from ChIP and input DNA were sequenced on an Illumina HiSeq instrument. For all experiments, reads were mapped to mm9 using BWA (Li and Durbin, 2009) and peaks were called using MACS (Zhang et al., 2008). H3K27ac ChIP-seq data from this study is available in GEO (http://www.ncbi.nlm.nih.gov/geo/, accession GSE52386).
Enhancer Activity Prediction and In Vivo Validation
Using the merged set of H3K27ac peaks, enrichment for each region in each dataset was scored based on comparison of coverage within the candidate enhancer versus experiment background after input correction. Enrichment scores across experiments were analyzed using both unsupervised and supervised approaches to determine the tissue- and stage-specific activity profiles for each putative enhancer. Activity predictions were validated using an established mouse transgenic system (Kothary et al., 1989; Pennacchio et al., 2006), where a vector containing a candidate enhancer, a minimal promoter, and the LacZ gene are stably integrated into the mouse genome via standard pronuclear injection. An enhancer was considered validated if the LacZ staining pattern driven by the enhancer in F0 transgenic mice was consistent with the H3K27ac predicted activity across independent transgenic mice representing independent insertion events in the mouse genome.
Functional and evolutionary analysis
Functional annotation of putative enhancers was performed using the GREAT tool (McLean et al., 2010), which tests for global enhancer enrichment near annotated gene classes. Motif analysis performed using the HOMER tool (Heinz et al., 2010). Overlap with GWAS SNPs (Hindorff et al., 2009) was compared to overlap with non-GWAS SNPs present on standard genotyping arrays, and individual candidate enhancers harboring GWAS SNPs were selected for validation of enhancer activity via the transgenic assay. Evolutionary analysis included six additional cell-derived H3K27ac ChIP-seq datasets that were processed using the same methods. Base-wise sequence homology and evolutionary constraint were compared for the core enhancer region (defined as the 100bp within the enhancer regions exhibiting maximal constraint) across accessible vertebrate genomes. Additional details reported in Extended Experimental Procedures.
Access to full data sets
Complete data files are available online at http://enhancer.lbl.gov/mouse_timecourse, including enhancer predictions mapped to the mouse reference genome (mm9) in BED and TXT format, predicted enhancer coordinates lifted over to the human reference genome (hg19), overlap between predicted enhancers and lead GWAS SNPs, and results from functional annotation and motif enrichment analyses as text files and labeled heatmaps.
Supplementary Material
Highlights.
Enhancer-associated chromatin landscapes change rapidly in developing tissues
Transgenic mouse assays validate predicted dynamic activities of enhancers
Dynamic enhancer activity controls processes central to development and disease
Evolutionary pressure on enhancers changes across tissues and developmental stages
Acknowledgements
The authors thank Chia-lin Wei and Cindy Choi for help with chromatin immunoprecipitation from embryonic mouse tissue; Diane Dickel and Han Wu for guidance and advice regarding enhancer activity analysis. A.S.N. was supported by NIH/NIGMS NRSA F32 fellowship GM105202. C.A. was supported by a SNSF advanced researchers fellowship. A.V. and L.A.P. were supported by NIH grants R01NS062859A, R01HG003988 and U01DE020060. J.L.R.R. was supported by NIMH grant R37MH049428. Research was conducted at the E.O. Lawrence Berkeley National Laboratory and performed under Department of Energy Contract DE-AC02-05CH11231, University of California.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc. Natl. Acad. Sci. U.S.a. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrieux M, Platel J-C, Dupuis A, Villaz M, Moody WJ. Early expression of sodium channel transcripts and sodium current by cajal-retzius cells in the preplate of the embryonic mouse neocortex. J. Neurosci. 2004;24:1719–1725. doi: 10.1523/JNEUROSCI.3548-02.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CP, Cepko CL. Cellular migration patterns in the developing mouse cerebral cortex. Development. 1990;110:713–732. doi: 10.1242/dev.110.3.713. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Ramsköld D, Zaouter C, Klum S, Sandberg R, Muhr J. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25:2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar P, Purvis S, Barron-Casella E, DeBaun MR, Casella JF, Arking DE, Keefer JR. Genome-wide association study identifies genetic variants influencing F-cell levels in sickle-cell patients. J. Hum. Genet. 2011;56:316–323. doi: 10.1038/jhg.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczyński B, Riddell A, Furlong EEM. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev. Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- Clinton M, Manson J, McBride D, Miele G. Gene expression changes during murine postnatal brain development. Genome Biol. 2000;1:RESEARCH0005. doi: 10.1186/gb-2000-1-3-research0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Oh S, DeMare LE, Reilly SK, Gerstein MB, Noonan JP. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Research. 2012;22:1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.a. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastani Z, Hivert M-F, Timpson N, Perry JRB, Yuan X, Scott RA, Henneman P, Heid IM, Kizer JR, Lyytikäinen L-P, DIAGRAM+ Consortium. MAGIC Consortium. GLGC Investigators. MuTHER Consortium. DIAGRAM Consortium. GIANT Consortium. Global B Pgen Consortium. Procardis Consortium. MAGIC investigators. GLGC Consortium et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8:e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickel DE, Visel A, Pennacchio LA. Functional anatomy of distant-acting mammalian enhancers. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2013;368:20120359. doi: 10.1098/rstb.2012.0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Lošo T, Tautz D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468:815–818. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- Donath MY, Zapf J, Eppenberger-Eberhardt M, Froesch ER, Eppenberger HM. Insulin-like growth factor I stimulates myofibril development and decreases smooth muscle alpha-actin of adult cardiomyocytes. Proc. Natl. Acad. Sci. U.S.a. 1994;91:1686–1690. doi: 10.1073/pnas.91.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Aliev F, Bierut LJ, Bucholz KK, Edenberg H, Hesselbrock V, Kramer J, Kuperman S, Nurnberger JI, Jr., Schuckit MA, et al. Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr. Genet. 2012;22:31–41. doi: 10.1097/YPG.0b013e32834acd07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RP. Patterning the vertebrate heart. Nat. Rev. Genet. 2002;3:544–556. doi: 10.1038/nrg843. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry IA, Kelleher KL, Bartlett SE, Leck KJ, Reynolds AJ, Heydon K, Mellick A, Megirian D, Matthaei KI. Hypertolerance to morphine in G(z alpha)-deficient mice. Brain Res. 2000;870:10–19. doi: 10.1016/s0006-8993(00)02387-8. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U.S.a. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton DR, Blanks JC, Fong HK, Casey PJ, Hildebrandt E, Simons MI. Novel localization of a G protein, Gz-alpha, in neurons of brain and retina. J. Neurosci. 1990;10:2763–2770. doi: 10.1523/JNEUROSCI.10-08-02763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerder-Suabedissen A, Oeschger FM, Krishnan ML, Belgard TG, Wang WZ, Lee S, Webber C, Petretto E, Edwards AD, Molnár Z. Expression profiling of mouse subplate reveals a dynamic gene network and disease association with autism and schizophrenia. Proc. Natl. Acad. Sci. U.S.a. 2013;110:3555–3560. doi: 10.1073/pnas.1218510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M, Leneuve P, Hamard G, Ducos B, Perin L, Binoux M, Le Bouc Y. A targeted partial invalidation of the insulin-like growth factor I receptor gene in mice causes a postnatal growth deficit. Endocrinology. 2000;141:2557–2566. doi: 10.1210/endo.141.7.7550. [DOI] [PubMed] [Google Scholar]
- Hu C-L, Chandra R, Ge H, Pain J, Yan L, Babu G, Depre C, Iwatsubo K, Ishikawa Y, Sadoshima J, et al. Adenylyl cyclase type 5 protein expression during cardiac development and stress. Am. J. Physiol. Heart Circ. Physiol. 2009;297:H1776–H1782. doi: 10.1152/ajpheart.00050.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsubo K, Minamisawa S, Tsunematsu T, Nakagome M, Toya Y, Tomlinson JE, Umemura S, Scarborough RM, Levy DE, Ishikawa Y. Direct inhibition of type 5 adenylyl cyclase prevents myocardial apoptosis without functional deterioration. J. Biol. Chem. 2004;279:40938–40945. doi: 10.1074/jbc.M314238200. [DOI] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, Ohler U, Bergman CM, Tomancak P. Gene expression divergence recapitulates the developmental hourglass model. Nature. 2010;468:811–814. doi: 10.1038/nature09634. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Laustsen PG, Russell SJ, Cui L, Entingh-Pearsall A, Holzenberger M, Liao R, Kahn CR. Essential role of insulin and insulin-like growth factor 1 receptor signaling in cardiac development and function. Mol. Cell. Biol. 2007;27:1649–1664. doi: 10.1128/MCB.01110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leck KJ, Blaha CD, Matthaei KI, Forster GL, Holgate J, Hendry IA. Gz proteins are functionally coupled to dopamine D2-like receptors in vivo. Neuropharmacology. 2006;51:597–605. doi: 10.1016/j.neuropharm.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Curr. Biol. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, et al. Large-scale discovery of enhancers from human heart tissue. Nat. Genet. 2011;44:89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel S, Garner C, Gut I, Matsuda F, Yamaguchi M, Heath S, Foglio M, Zelenika D, Boland A, Rooks H, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat. Genet. 2007;39:1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Below JE, Krithika S, Valladares A, Barta JL, Cox NJ, Hanis CL, Wacher N, Garcia-Mena J, Hu P, et al. Genome-wide association study of type 2 diabetes in a sample from Mexico City and a meta-analysis of a Mexican-American sample from Starr County, Texas. Diabetologia. 2011;54:2038–2046. doi: 10.1007/s00125-011-2172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Planells-Cases R, Caprini M, Zhang J, Rockenstein EM, Rivera RR, Murre C, Masliah E, Montal M. Neuronal death and perinatal lethality in voltage-gated sodium channel alpha(II)-deficient mice. Biophys. J. 2000;78:2878–2891. doi: 10.1016/S0006-3495(00)76829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff RA. The Shape of Life: Genes, Development, and the Evolution of Animal Form. University of Chicago Press; 1996. [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, Dilullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, Sestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev. Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Sidhu A, Kimura K, Uh M, White BH, Patel S. Multiple coupling of human D5 dopamine receptors to guanine nucleotide binding proteins Gs and Gz. J. Neurochem. 1998;70:2459–2467. doi: 10.1046/j.1471-4159.1998.70062459.x. [DOI] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. Developmental Fate and Cellular Maturity Encoded in Human Regulatory DNA Landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech. Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat. Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JB. Differential control of the synthesis of two hemoglobin beta chains in normal mice. Cell. 1977;12:863–871. doi: 10.1016/0092-8674(77)90150-7. [DOI] [PubMed] [Google Scholar]
- Zambon A, Deeb SS, Pauletto P, Crepaldi G, Brunzell JD. Hepatic lipase: a marker for cardiovascular disease risk and response to therapy. Curr. Opin. Lipidol. 2003;14:179–189. doi: 10.1097/00041433-200304000-00010. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Duncan SA. Embryonic development of the liver. Hepatology. 2005;41:956–967. doi: 10.1002/hep.20691. [DOI] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT-Y, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM. Liver development. Harvard Stem Cell Institute; Cambridge (MA): 2008. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.